S-72, a Novel Orally Available Tubulin Inhibitor, Overcomes Paclitaxel Resistance via Inactivation of the STING Pathway in Breast Cancer
Abstract
1. Introduction
2. Results
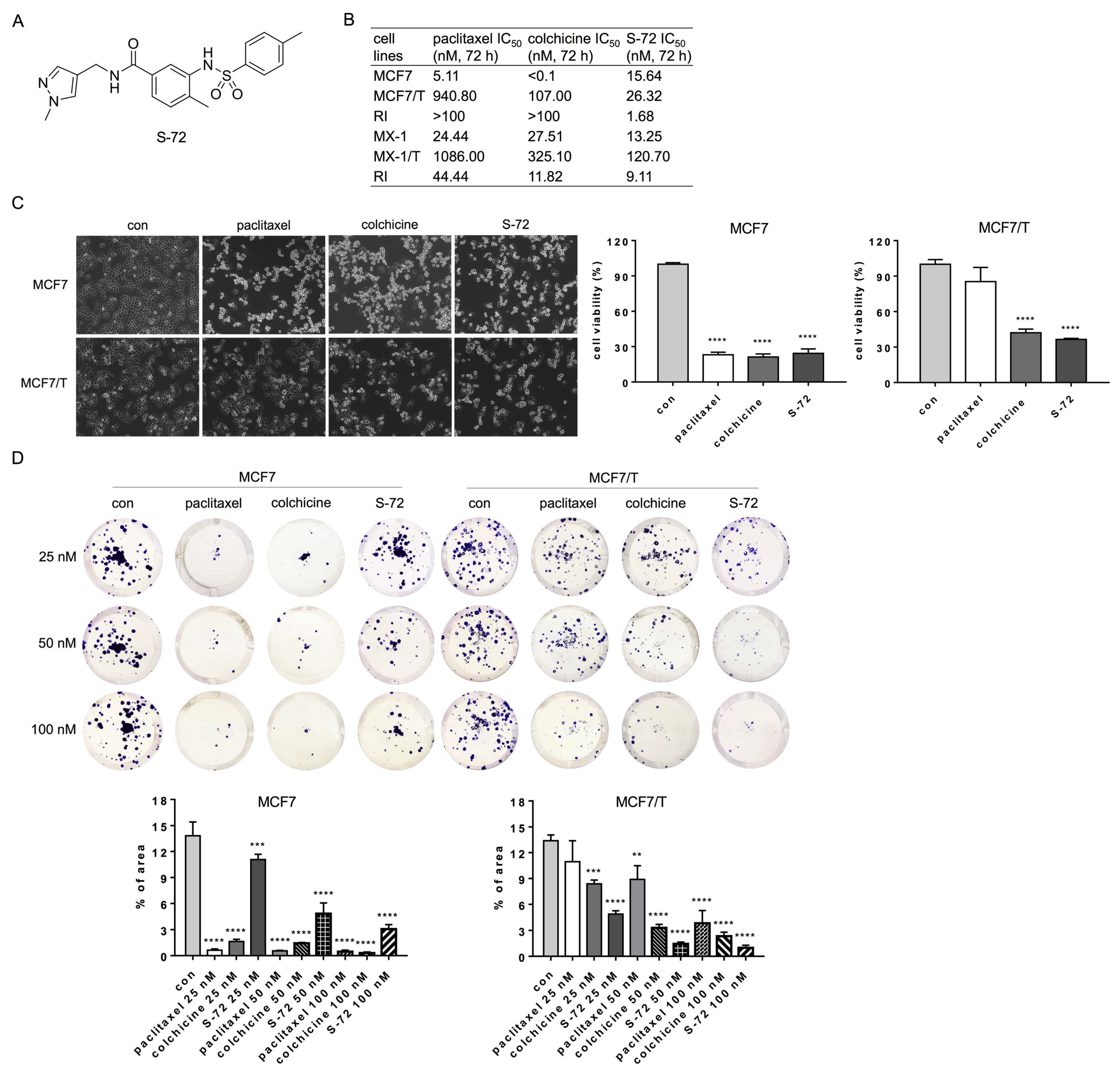
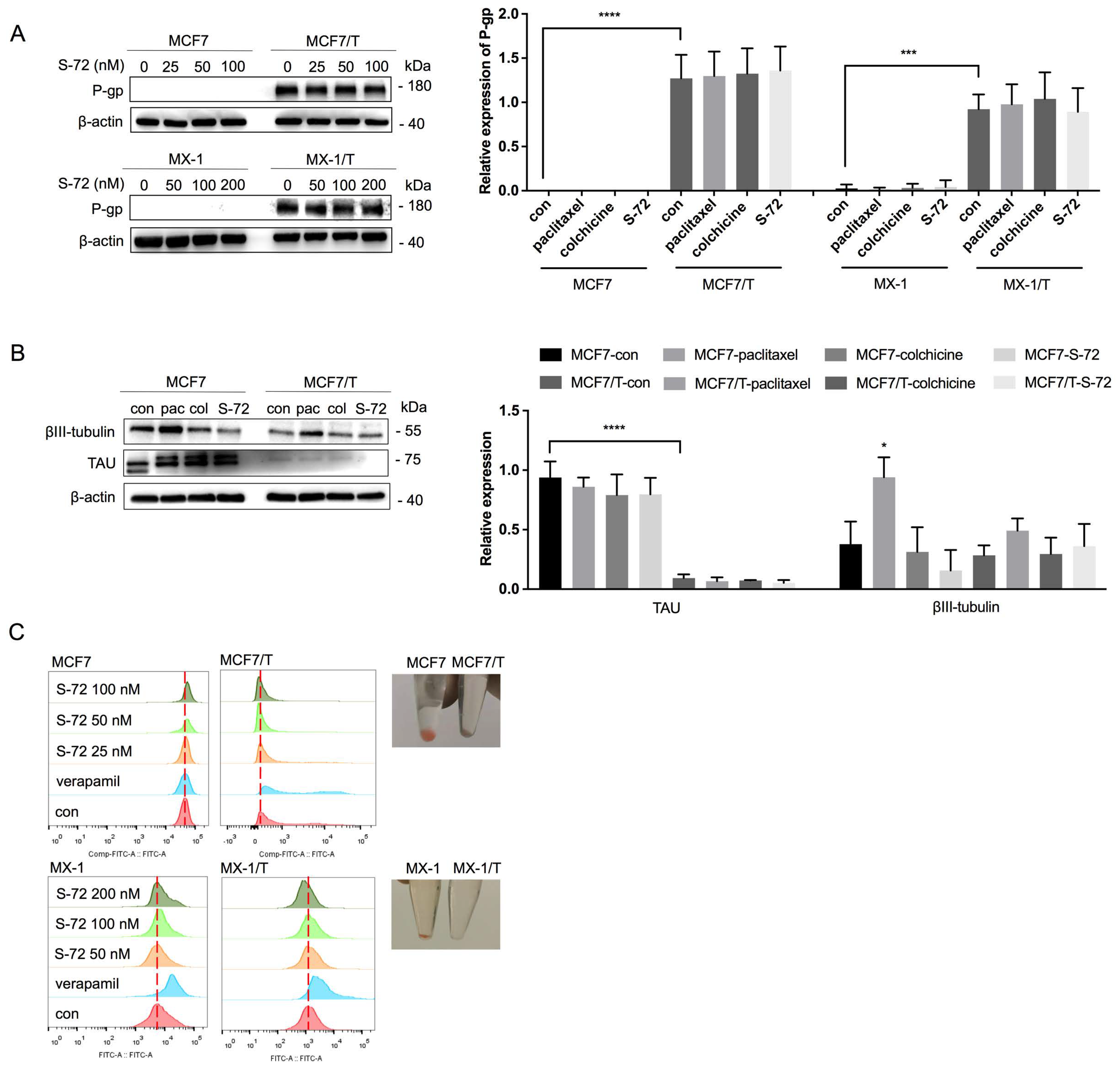
2.1. S-72 Exhibited Potent Cytotoxicity in Paclitaxel-Resistant Breast Cancer Cells
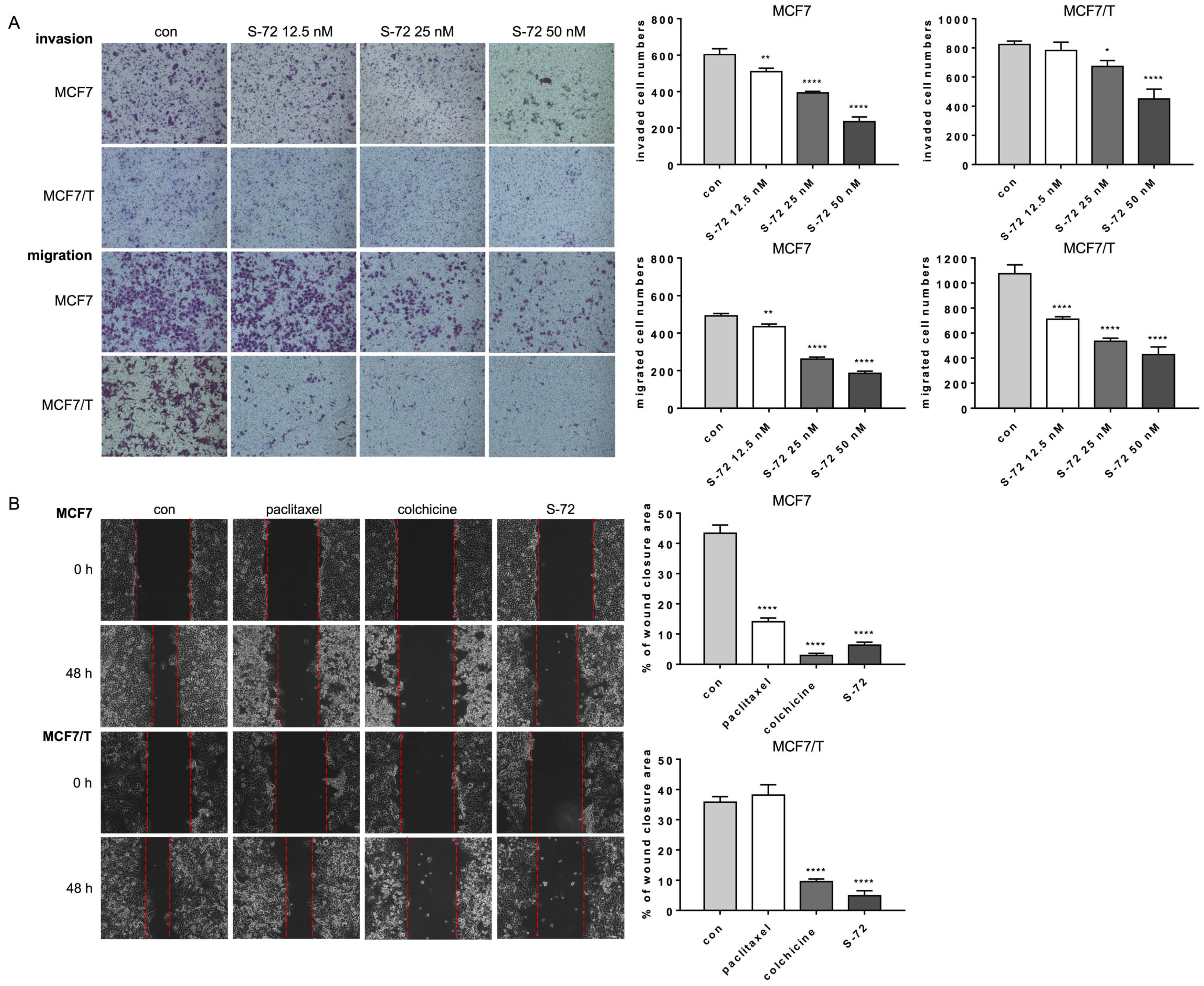
2.2. S-72 Suppressed Both the Invasion and Migration of MCF7 and MCF7/T Cells
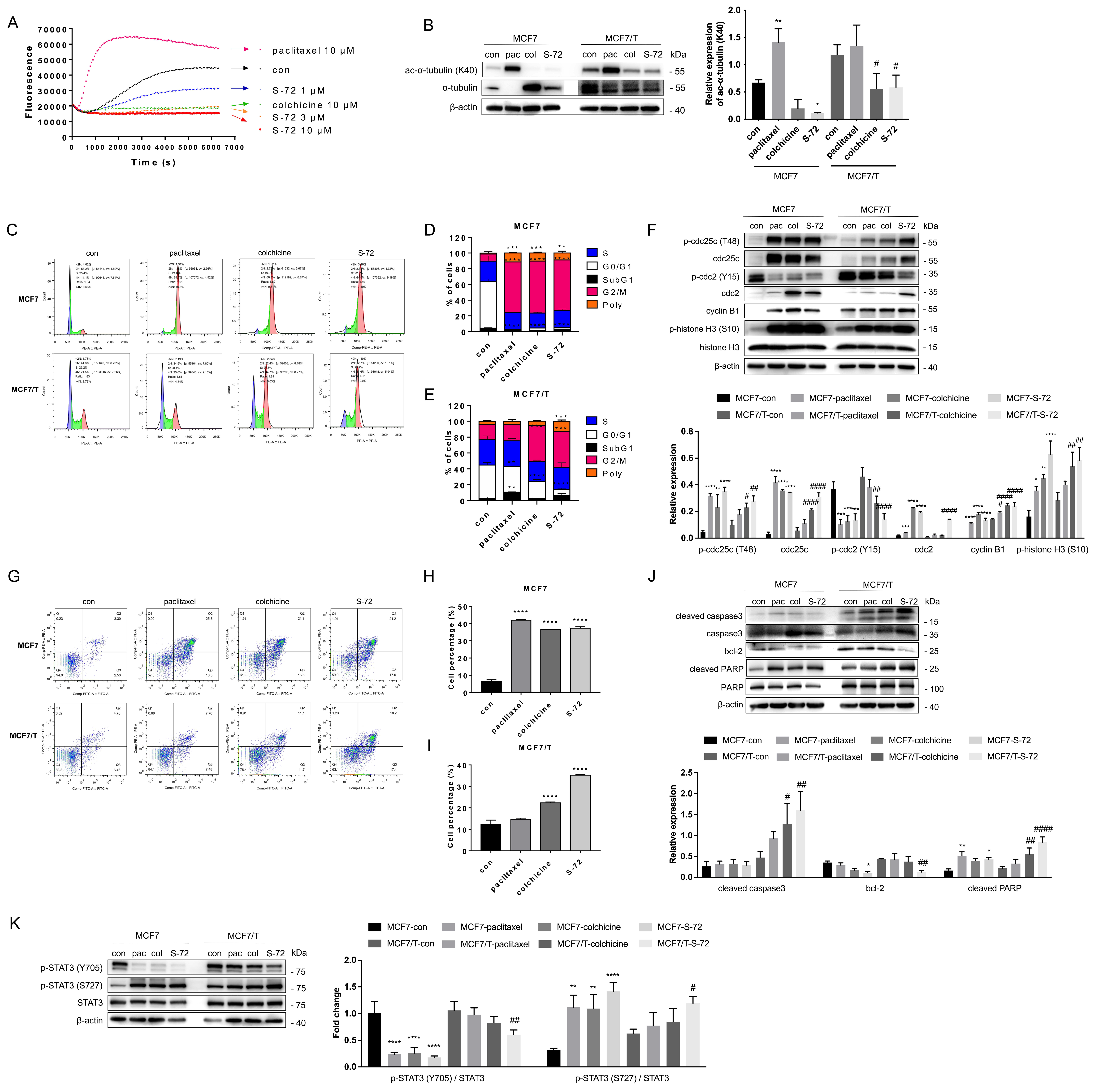
2.3. S-72 Inhibited Tubulin Polymerization, Triggered Mitosis-Phase Cell Cycle Arrest and Cell Apoptosis and Disrupted STAT3 Signaling Activation
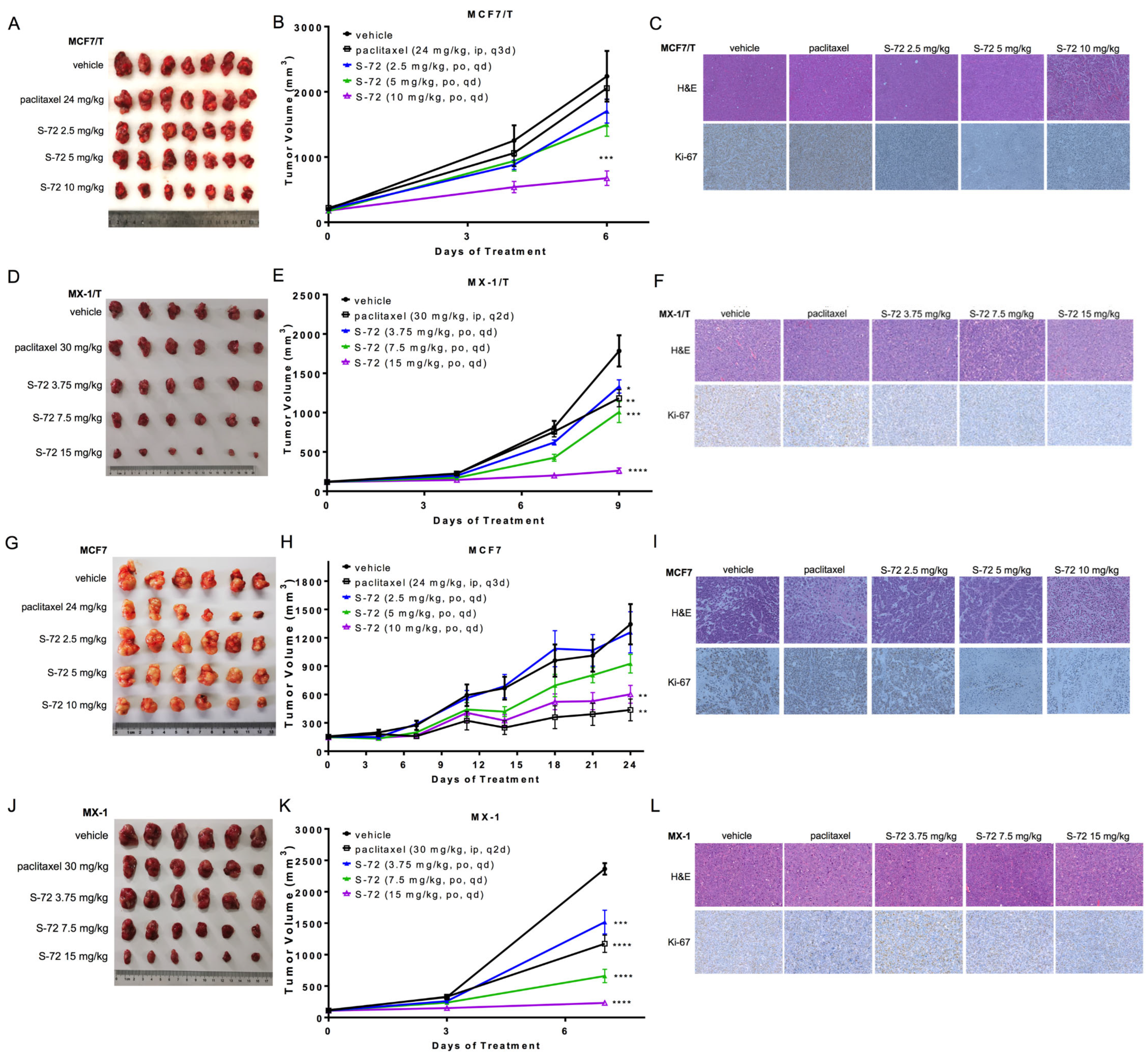
2.4. S-72 Repressed the Growth of Both Paclitaxel-Sensitive and Paclitaxel-Resistant Human Tumor Xenografts In Vivo
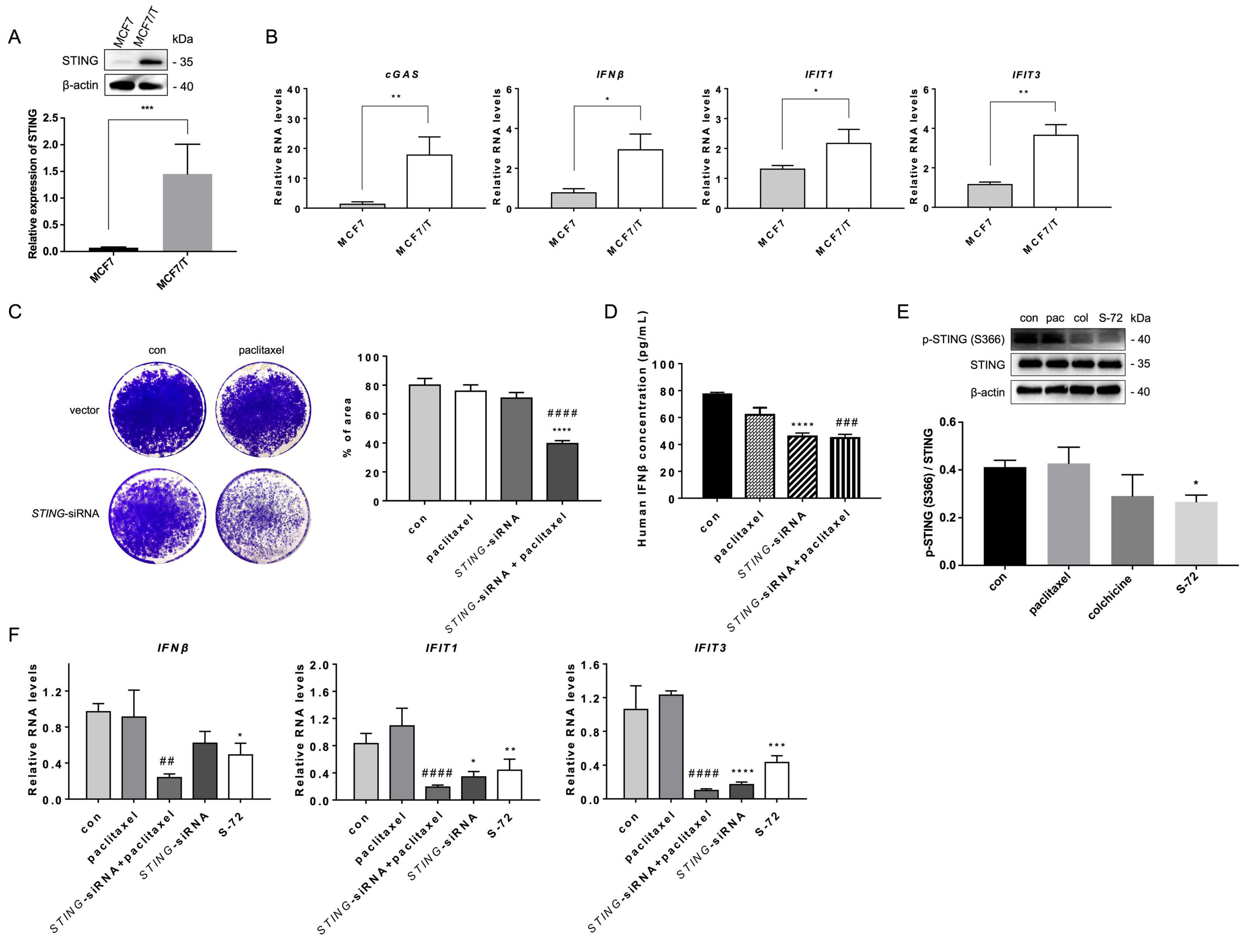
2.5. S-72 Inhibited STING Activation in Paclitaxel-Resistant Cancer Cells
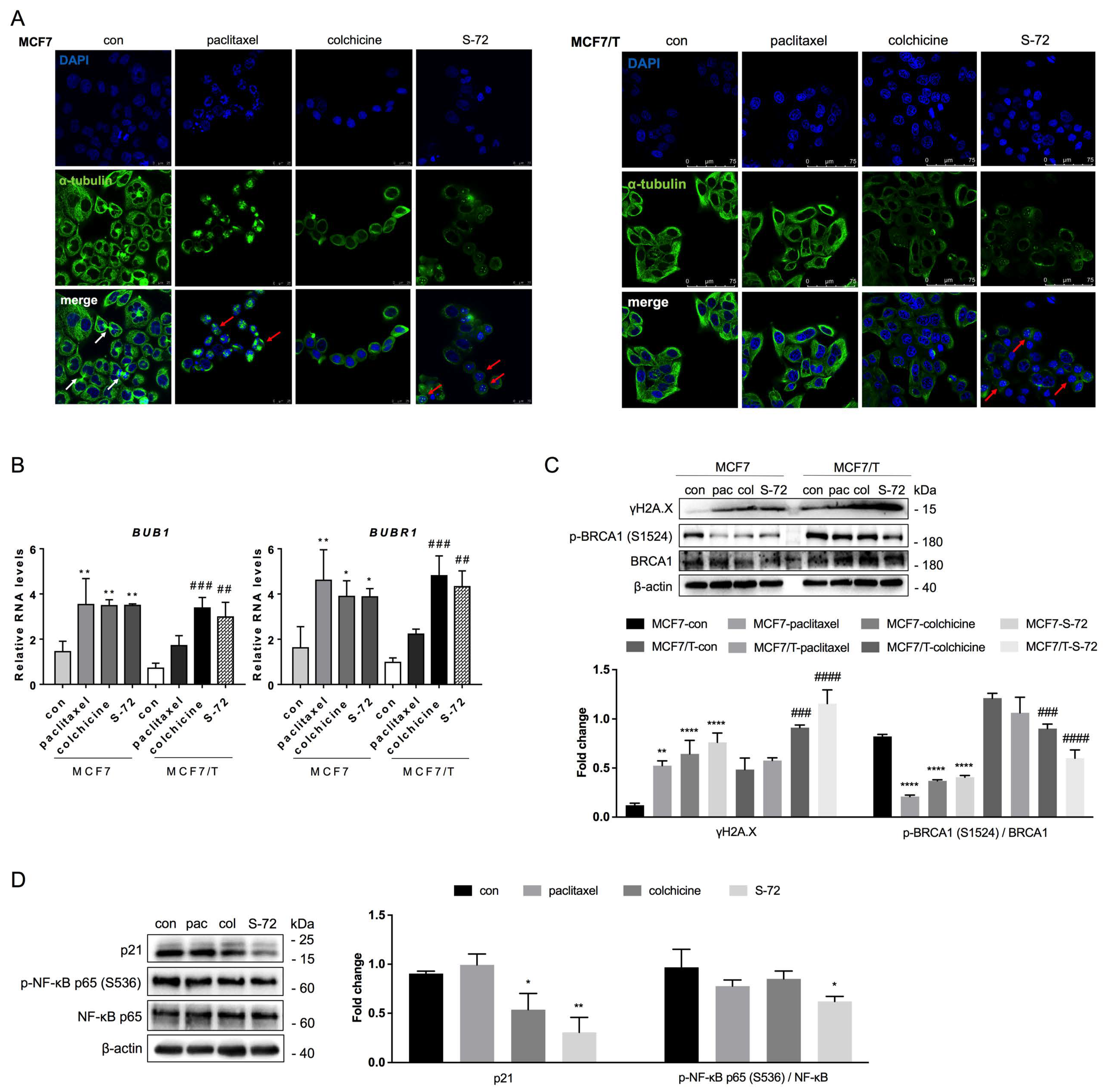
2.6. S-72-Induced STING Inactivation Led to Chromosomal Instability
3. Discussion
4. Materials and Methods
4.1. Chemicals and Cell Lines
4.2. Cell Viability Assay
4.3. Colony Formation Assay
4.4. Cell Invasion and Migration Assay
4.5. In Vivo Xenograft Model
4.6. In Vitro Tubulin Polymerization Assay
4.7. Flow Cytometry
4.8. Western Blot
4.9. Quantitative Real-Time PCR
4.10. Immunofluorescence
4.11. Plasmid Construction and Transfection
4.12. Sandwich Enzyme Linked Immuno-Sorbent Assay (ELISA)
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Crown, J.; O’Leary, M.; Ooi, W.S. Docetaxel and paclitaxel in the treatment of breast cancer: A review of clinical experience. Oncologist 2004, 9 (Suppl. 2), 24–32. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.; Briasoulis, E.; Linardou, H.; Bafaloukos, D.; Papadimitriou, C. Taxane resistance in breast cancer: Mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 2012, 38, 890–903. [Google Scholar] [CrossRef]
- Orr, G.A.; Verdier-Pinard, P.; McDaid, H.; Horwitz, S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene 2003, 22, 7280–7295. [Google Scholar] [CrossRef]
- Barbolina, M.V. Dichotomous role of microtubule associated protein tau as a biomarker of response to and a target for increasing efficacy of taxane treatment in cancers of epithelial origin. Pharmacol. Res. 2021, 168, 105585. [Google Scholar] [CrossRef]
- Xiang, S.; Dauchy, R.T.; Hoffman, A.E.; Pointer, D.; Frasch, T.; Blask, D.E.; Hill, S.M. Epigenetic inhibition of the tumor suppressor ARHI by light at night-induced circadian melatonin disruption mediates STAT3-driven paclitaxel resistance in breast cancer. J. Pineal Res. 2019, 67, e12586. [Google Scholar] [CrossRef] [PubMed]
- Scribano, C.M.; Wan, J.; Esbona, K.; Tucker, J.B.; Lasek, A.; Zhou, A.S.; Zasadil, L.M.; Molini, R.; Fitzgerald, J.; Lager, A.M.; et al. Chromosomal instability sensitizes patient breast tumors to multipolar divisions induced by paclitaxel. Sci. Transl. Med. 2021, 13, eabd4811. [Google Scholar] [CrossRef] [PubMed]
- Lukow, D.A.; Sheltzer, J.M. Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer 2022, 8, 43–53. [Google Scholar] [CrossRef]
- Bronder, D.; Bakhoum, S.F. A CINful way to overcome addiction: How chromosomal instability enables cancer to overcome its oncogene addiction. EMBO Mol. Med. 2020, 12, e12017. [Google Scholar] [CrossRef]
- Ranoa, D.R.E.; Widau, R.C.; Mallon, S.; Parekh, A.D.; Nicolae, C.M.; Huang, X.; Bolt, M.J.; Arina, A.; Parry, R.; Kron, S.J.; et al. STING promotes homeostasis via regulation of cell proliferation and chromosomal stability. Cancer Res. 2019, 79, 1465–1479. [Google Scholar] [CrossRef]
- Hong, C.; Schubert, M.; Tijhuis, A.E.; Requesens, M.; Roorda, M.; Brink, A.V.D.; Ruiz, L.A.; Bakker, P.L.; van der Sluis, T.; Pieters, W.; et al. cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 2022, 607, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, E.; Wu, J.; Wang, T.L.; Hsieh, H.P.; Liu, X. An antimitotic and antivascular agent BPR0L075 overcomes multidrug resistance and induces mitotic catastrophe in paclitaxel-resistant ovarian cancer cells. PLoS ONE 2013, 8, e65686. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Gao, M.; Xu, X.; Zhang, H.; Xu, J.; Guan, Q.; Wang, Q.; Du, J.; Li, Z.; Zuo, D.; et al. Overcoming resistance to mitochondrial apoptosis by BZML-induced mitotic catastrophe is enhanced by inhibition of autophagy in A549/Taxol cells. Cell Prolif. 2018, 51, e12450. [Google Scholar] [CrossRef]
- Mahmud, F.; Deng, S.; Chen, H.; Miller, D.D.; Li, W. Orally available tubulin inhibitor VERU-111 enhances antitumor efficacy in paclitaxel-resistant lung cancer. Cancer Lett. 2020, 495, 76–88. [Google Scholar] [CrossRef]
- Tian, C.; Wang, M.; Shi, X.; Chen, X.; Wang, X.; Zhang, Z.; Liu, J. Discovery of (2-(pyrrolidin-1-yl)thieno[3,2-d]pyrimidin-4-yl)(3,4,5-trimethoxyphenyl)methanone as a novel potent tubulin depolymerizing and vascular disrupting agent. Eur. J. Med. Chem. 2022, 238, 114466. [Google Scholar] [CrossRef]
- Deng, S.; Krutilina, R.I.; Hartman, K.L.; Chen, H.; Parke, D.N.; Wang, R.; Mahmud, F.; Ma, D.; Lukka, P.B.; Meibohm, B.; et al. Colchicine-binding site agent CH-2-77 as a potent tubulin inhibitor suppressing triple-negative breast cancer. Mol. Cancer Ther. 2022, 21, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Egharevba, G.O.; Kamal, A.; Dosumu, O.O.; Routhu, S.; Fadare, O.A.; Oguntoye, S.O.; Njinga, S.N.; Oluyori, A.P. Synthesis and characterization of novel combretastatin analogues of 1,1-diaryl vinyl sulfones, with antiproliferative potential via in-silico and in-vitro studies. Sci. Rep. 2022, 12, 1901. [Google Scholar] [CrossRef] [PubMed]
- Cogle, C.R.; Collins, B.; Turner, D.; Pettiford, L.C.; Bossé, R.; Hawkins, K.E.; Beachamp, Z.; Wise, E.; Cline, C.; May, W.S.; et al. Safety, feasibility and preliminary efficacy of single agent combretastatin A1 diphosphate (OXi4503) in patients with relapsed or refractory acute myeloid leukemia or myelodysplastic syndromes. Br. J. Haematol. 2020, 189, e211–e213. [Google Scholar] [CrossRef]
- Markowski, M.C.; Eisenberger, M.A.; Tutrone, R.F.; Pieczonka, C.M.; Getzenberg, R.H.; Rodriguez, D.; Barnette, K.G.; Steiner, M.S.; Saltzstein, D.R.; Antonarakis, E.S. Clinical study of VERU-111, an oral cytoskeletal disruptor, in metastatic castrationresistant prostate cancer (mCRPC) who failed an androgen receptor targeting agent. J. Clin. Oncol. 2021, 39, 131. [Google Scholar] [CrossRef]
- Pooler, D.B.; Ness, D.B.; Danilov, A.V.; Labrie, B.M.; Tosteson, T.D.; Eastman, A.; Lewis, L.D.; Lansigan, F. A phase I trial of BNC105P and Ibrutinib in patients with relapsed/refractory chronic lymphocytic leukemia. EJHaem 2022, 3, 1445–1448. [Google Scholar] [CrossRef]
- Karahalil, B.; Yardim-Akaydin, S.; Nacak Baytas, S. An overview of microtubule targeting agents for cancer therapy. Arh. Hig. Rada Toksikol. 2019, 70, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Sherbet, G.V. Combretastatin analogues in cancer biology: A prospective view. J. Cell Biochem. 2020, 121, 2127–2138. [Google Scholar] [CrossRef]
- Tian, C.; Chen, X.; Zhang, Z.; Wang, X.; Liu, J. Design and synthesis of (2-(phenylamino)thieno[3,2-d]pyrimidin-4-yl)(3,4,5-trimethoxyphenyl)methanone analogues as potent anti-tubulin polymerization agents. Eur. J. Med. Chem. 2019, 183, 111679. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Gao, M.; Zhang, H.; Guan, Q.; Xu, J.; Li, Y.; Qi, H.; Li, Z.; Zuo, D.; Zhang, W.; et al. BZML, a novel colchicine binding site inhibitor, overcomes multidrug resistance in A549/Taxol cells by inhibiting P-gp function and inducing mitotic catastrophe. Cancer Lett. 2017, 402, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Banerjee, S.; Chen, H.; Pochampally, S.; Wang, Y.; Yun, M.-K.; White, S.W.; Parmar, K.; Meibohm, B.; Hartman, K.L.; et al. SB226, an inhibitor of tubulin polymerization, inhibits paclitaxel-resistant melanoma growth and spontaneous metastasis. Cancer Lett. 2023, 555, 216046. [Google Scholar] [CrossRef]
- Mei, M.; Xie, D.; Zhang, Y.; Jin, J.; You, F.; Li, Y.; Dai, J.; Chen, X. A New 2a,5a,10b,14b-tetraacetoxy-4(20),11-taxadiene (SIA) derivative overcomes paclitaxel resistance by inhibiting MAPK signaling and increasing paclitaxel accumulation in breast cancer cells. PLoS ONE 2014, 9, e104317. [Google Scholar] [CrossRef]
- Basit, A.; Cho, M.G.; Kim, E.Y.; Kwon, D.; Kang, S.J.; Lee, J.H. The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp. Mol. Med. 2020, 52, 643–657. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Cantley, L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef]
- Müsch, A. Microtubule organization and function in epithelial cells. Traffic 2004, 5, 1–9. [Google Scholar] [CrossRef]
- Mosca, L.; Ilari, A.; Fazi, F.; Assaraf, Y.G.; Colotti, G. Taxanes in cancer treatment: Activity, chemoresistance and its overcoming. Drug Resist. Updat. 2021, 54, 100742. [Google Scholar] [CrossRef]
- Raab, M.; Sanhaji, M.; Zhou, S.; Rödel, F.; El-Balat, A.; Becker, S.; Strebhardt, K. Blocking mitotic exit of ovarian cancer cells by pharmaceutical inhibition of the anaphase-promoting complex reduces chromosomal instability. Neoplasia 2019, 21, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Raab, M.; Kobayashi, N.F.; Becker, S.; Kurunci-Csacsko, E.; Krämer, A.; Strebhardt, K.; Sanhaji, M. Boosting the apoptotic response of high-grade serous ovarian cancers with CCNE1 amplification to paclitaxel in vitro by targeting APC/C and the pro-survival protein MCL-1. Int. J. Cancer 2020, 146, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Ferreira, S.; Moindjie, H.; Haykal, M.M.; Nahmias, C. Predicting and overcoming taxane chemoresistance. Trends Mol. Med. 2021, 27, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 2012, 29, 2943–2971. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Lin, S.; Ji, M.; Xue, N.; Liu, Y.; Zhang, Z.; Zhang, K.; Zhang, J.; Zhang, Y.; Wang, Q.; et al. A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity. Cancer Lett. 2020, 495, 22–32. [Google Scholar] [CrossRef]
- Zhang, C.; Min, L.; Zhang, L.; Ma, Y.; Yang, Y.; Shou, C. Combined analysis identifies six genes correlated with augmented malignancy from non-small cell to small cell lung cancer. Tumour Biol. 2016, 37, 2193–2207. [Google Scholar] [CrossRef]
- Chang, L.-C.; Yu, Y.-L.; Liu, C.-Y.; Cheng, Y.-Y.; Chou, R.-H.; Hsieh, M.-T.; Lin, H.-Y.; Hung, H.-Y.; Huang, L.-J.; Wu, Y.-C.; et al. The newly synthesized 2-arylnaphthyridin-4-one, CSC-3436, induces apoptosis of non-small cell lung cancer cells by inhibiting tubulin dynamics and activating CDK1. Cancer Chemother. Pharmacol. 2015, 75, 1303–1315. [Google Scholar] [CrossRef]
- Sur, S.; Agrawal, D.K. Phosphatases and kinases regulating CDC25 activity in the cell cycle: Clinical implications of CDC25 overexpression and potential treatment strategies. Mol. Cell Biochem. 2016, 416, 33–46. [Google Scholar] [CrossRef]
- Bulavin, D.V.; Higashimoto, Y.; Demidenko, Z.N.; Meek, S.; Graves, P.; Phillips, C.; Zhao, H.; Moody, S.A.; Appella, E.; Piwnica-Worms, H.; et al. Dual phosphorylation controls Cdc25 phosphatases and mitotic entry. Nat. Cell Biol. 2003, 5, 545–551. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, F.-L.; Lu, Z.-N.; Wang, H.-Y.; Cheng, Y.-N.; Liu, Z.-P.; Yu, L.-G.; Zhang, H.-H.; Guo, X.-L. DHPAC, a novel synthetic microtubule destabilizing agent, possess high anti-tumor activity in vincristine-resistant oral epidermoid carcinoma in vitro and in vivo. Int. J. Biochem. Cell Biol. 2017, 93, 1–11. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.; Cao, J.; Xu, P.; Chen, Z.; Wang, S.; Li, B.; Zhang, L.; Xie, L.; Fang, L.; et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis. 2021, 12, 910. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.R.; Chaudhury, M.; Nelson, E.A.; Frank, D.A. Microtubule-targeted chemotherapeutic agents inhibit signal transducer and activator of transcription 3 (STAT3) signaling. Mol. Pharmacol. 2010, 78, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Xie, S.; Liu, Z.; Luo, Y.; Zhou, J.; Li, D.; Liu, M. STAT3 association with microtubules and its activation are independent of HDAC6 activity. DNA Cell Biol. 2015, 34, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Csomós, I.; Nagy, P.; Filep, C.; Rebenku, I.; Nizsalóczki, E.; Kovács, T.; Vámosi, G.; Mátyus, L.; Bodnár, A. Opposing effects of chelidonine on tyrosine and serine phosphorylation of STAT3 in human uveal melanoma cells. Int. J. Mol. Sci. 2021, 22, 12974. [Google Scholar] [CrossRef]
- Yokogami, K.; Wakisaka, S.; Avruch, J.; Reeves, S.A. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 2000, 10, 47–50. [Google Scholar] [CrossRef]
- Wakahara, R.; Kunimoto, H.; Tanino, K.; Kojima, H.; Inoue, A.; Shintaku, H.; Nakajima, K. Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells 2012, 17, 132–145. [Google Scholar] [CrossRef]
- Shi, X.Q.; Zhang, H.; Paddon, H.; Lee, G.; Cao, X.M.; Pelech, S. Phosphorylation of STAT3 serine-727 by cyclin-dependent kinase 1 is critical for nocodazole-induced mitotic arrest. Biochemistry 2006, 45, 5857–5867. [Google Scholar] [CrossRef]
- Lukow, D.A.; Sausville, E.L.; Suri, P.; Chunduri, N.K.; Wieland, A.; Leu, J.; Smith, J.C.; Girish, V.; Kumar, A.A.; Kendall, J.; et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell 2021, 56, 2427–2439.e2424. [Google Scholar] [CrossRef]
- Ippolito, M.R.; Martis, V.; Martin, S.; Tijhuis, A.E.; Hong, C.; Wardenaar, R.; Dumont, M.; Zerbib, J.; Spierings, D.C.; Fachinetti, D.; et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev. Cell 2021, 56, 2440–2454.e2446. [Google Scholar] [CrossRef]
- Shoshani, O.; Brunner, S.F.; Yaeger, R.; Ly, P.; Nechemia-Arbely, Y.; Kim, D.H.; Fang, R.; Castillon, G.A.; Yu, M.; Li, J.S.Z.; et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature 2021, 591, 137–141. [Google Scholar] [CrossRef]
- Replogle, J.M.; Zhou, W.; Amaro, A.E.; McFarland, J.M.; Villalobos-Ortiz, M.; Ryan, J.; Letai, A.; Yilmaz, O.; Sheltzer, J.; Lippard, S.J.; et al. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc. Natl. Acad. Sci. USA 2020, 117, 30566–30576. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, A.; Baruah, P.S.; Smith, J.C.; Wang, Z.; Sayles, N.M.; Andrews, P.; Kendall, J.; Leu, J.; Chunduri, N.K.; Levy, D.; et al. Single-chromosomal gains can function as metastasis suppressors and promoters in colon cancer. Dev. Cell 2020, 52, 413–428.e416. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, A.; Schukken, K.M.; Sausville, E.L.; Girish, V.; Adebambo, O.A.; Sheltzer, J.M. Aneuploidy as a promoter and suppressor of malignant growth. Nat. Rev. Cancer 2021, 21, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Elstrodt, F.; Hollestelle, A.; Nagel, J.H.; Gorin, M.; Wasielewski, M.; van den Ouweland, A.; Merajver, S.D.; Ethier, S.P.; Schutte, M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006, 66, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Cheon, H.; Holvey-Bates, E.G.; McGrail, D.J.; Stark, G.R. PD-L1 sustains chronic, cancer cell-intrinsic responses to type I interferon, enhancing resistance to DNA damage. Proc. Natl. Acad. Sci. USA 2021, 118, e2112258118. [Google Scholar] [CrossRef]
- Liang, H.; Deng, L.; Hou, Y.; Meng, X.; Huang, X.; Rao, E.; Zheng, W.; Mauceri, H.; Mack, M.; Xu, M.; et al. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 2017, 8, 1736. [Google Scholar] [CrossRef]
- Lin, S.; Du, T.; Zhang, J.; Wu, D.; Tian, H.; Zhang, K.; Jiang, L.; Lu, D.; Sheng, L.; Li, Y.; et al. Optimization of benzamide derivatives as potent and orally active tubulin inhibitors targeting the colchicine binding site. J. Med. Chem. 2022, 65, 16372–16391. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Z.; Lin, S.; Du, T.; Wang, M.; Wang, W.; You, S.; Xue, N.; Liu, Y.; Ji, M.; Xu, H.; et al. S-72, a Novel Orally Available Tubulin Inhibitor, Overcomes Paclitaxel Resistance via Inactivation of the STING Pathway in Breast Cancer. Pharmaceuticals 2023, 16, 749. https://doi.org/10.3390/ph16050749
Hou Z, Lin S, Du T, Wang M, Wang W, You S, Xue N, Liu Y, Ji M, Xu H, et al. S-72, a Novel Orally Available Tubulin Inhibitor, Overcomes Paclitaxel Resistance via Inactivation of the STING Pathway in Breast Cancer. Pharmaceuticals. 2023; 16(5):749. https://doi.org/10.3390/ph16050749
Chicago/Turabian StyleHou, Zhenyan, Songwen Lin, Tingting Du, Mingjin Wang, Weida Wang, Shen You, Nina Xue, Yichen Liu, Ming Ji, Heng Xu, and et al. 2023. "S-72, a Novel Orally Available Tubulin Inhibitor, Overcomes Paclitaxel Resistance via Inactivation of the STING Pathway in Breast Cancer" Pharmaceuticals 16, no. 5: 749. https://doi.org/10.3390/ph16050749
APA StyleHou, Z., Lin, S., Du, T., Wang, M., Wang, W., You, S., Xue, N., Liu, Y., Ji, M., Xu, H., & Chen, X. (2023). S-72, a Novel Orally Available Tubulin Inhibitor, Overcomes Paclitaxel Resistance via Inactivation of the STING Pathway in Breast Cancer. Pharmaceuticals, 16(5), 749. https://doi.org/10.3390/ph16050749





