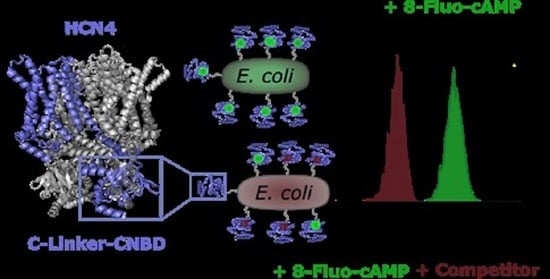
A Novel Flow Cytometry-Based Assay for the Identification of HCN4 CNBD Ligands
Abstract
1. Introduction
2. Results
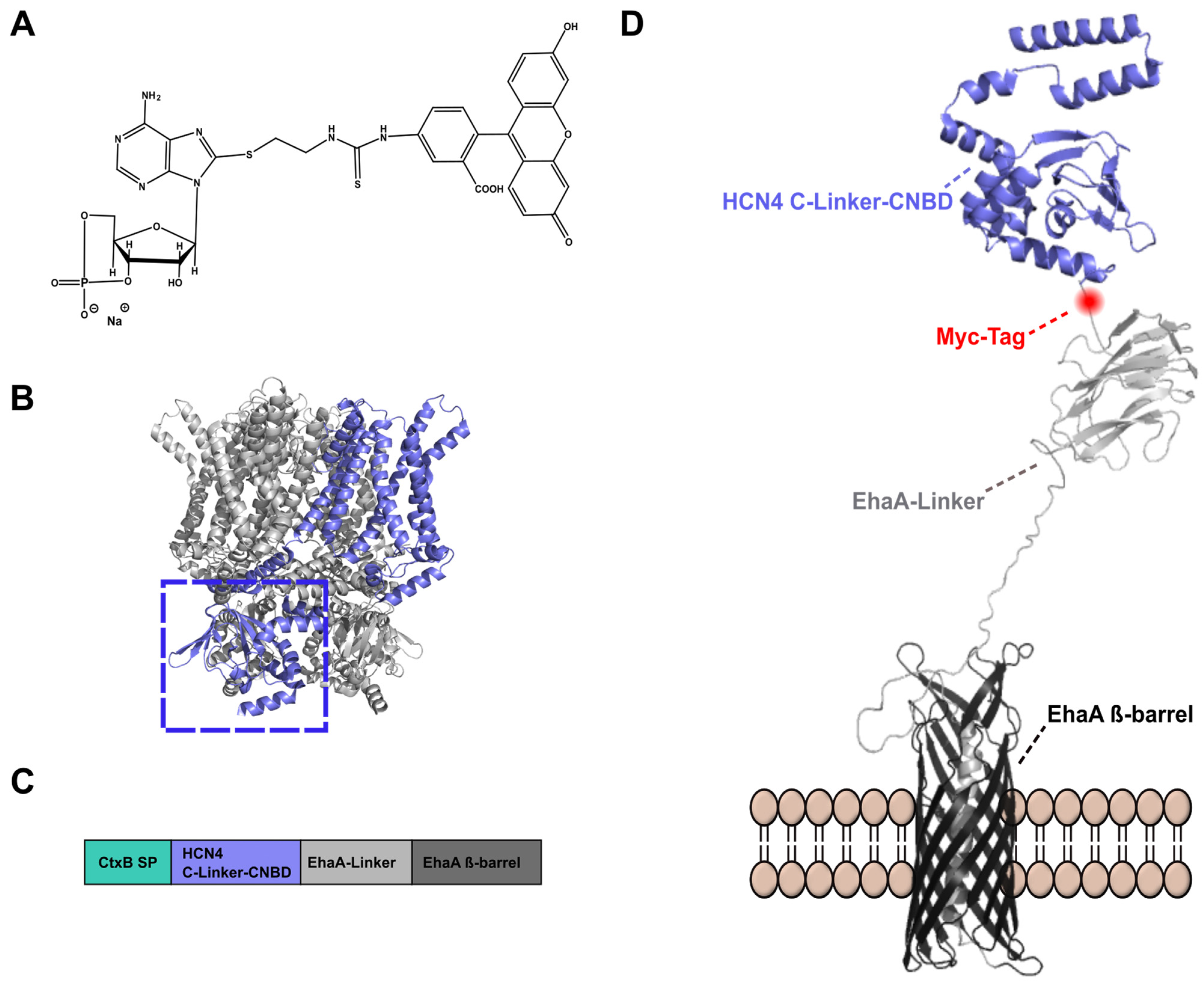
2.1. Autodisplay of the HCN4 C-Linker-CNBD
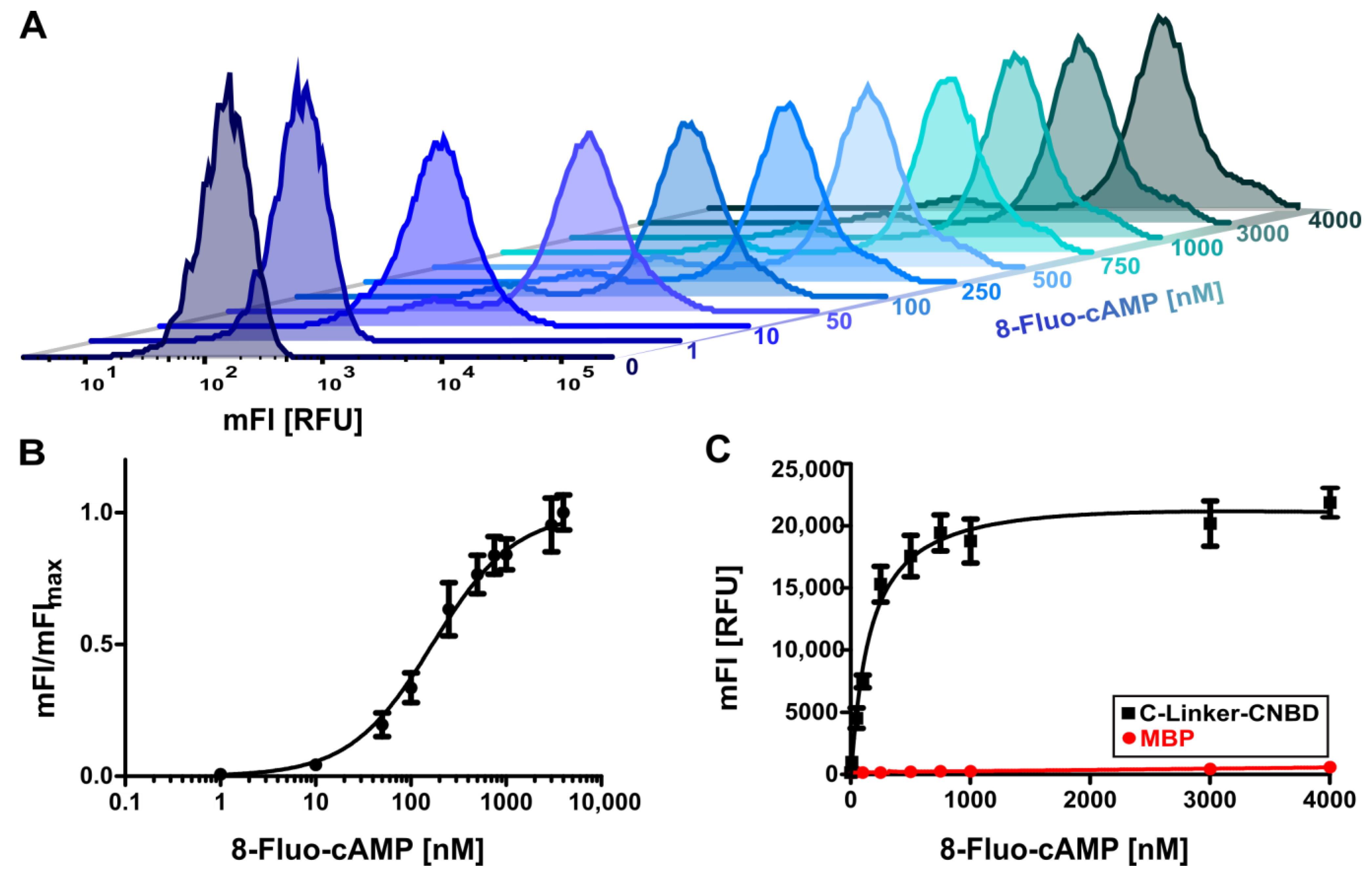
2.2. Binding of 8-Fluo-cAMP to Surface-Displayed HCN4 C-Linker-CNBD
2.3. Evaluation of Assay Conditions
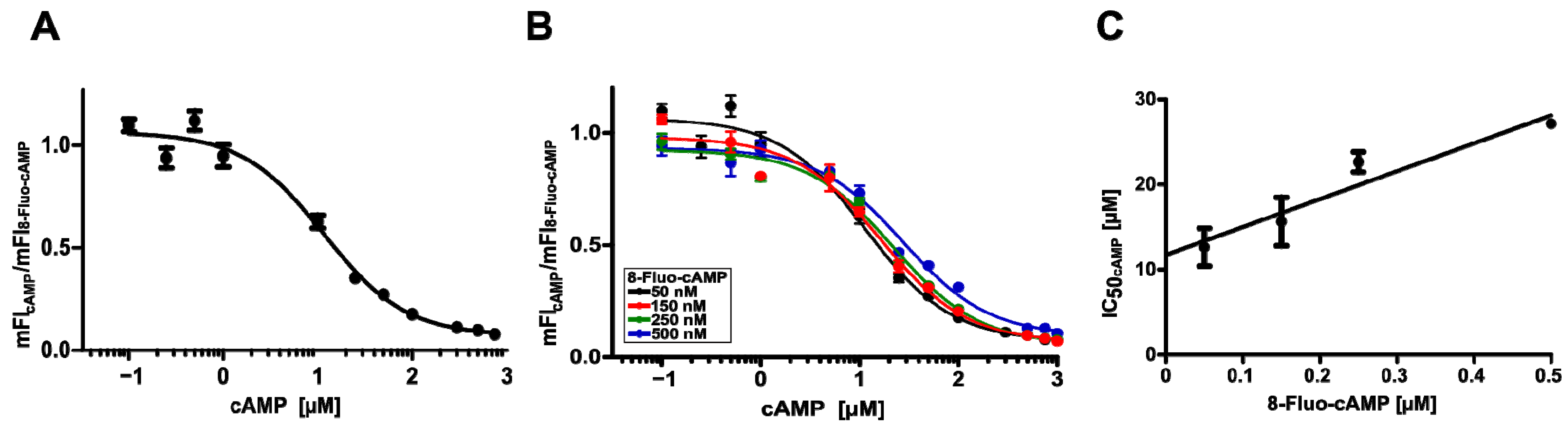
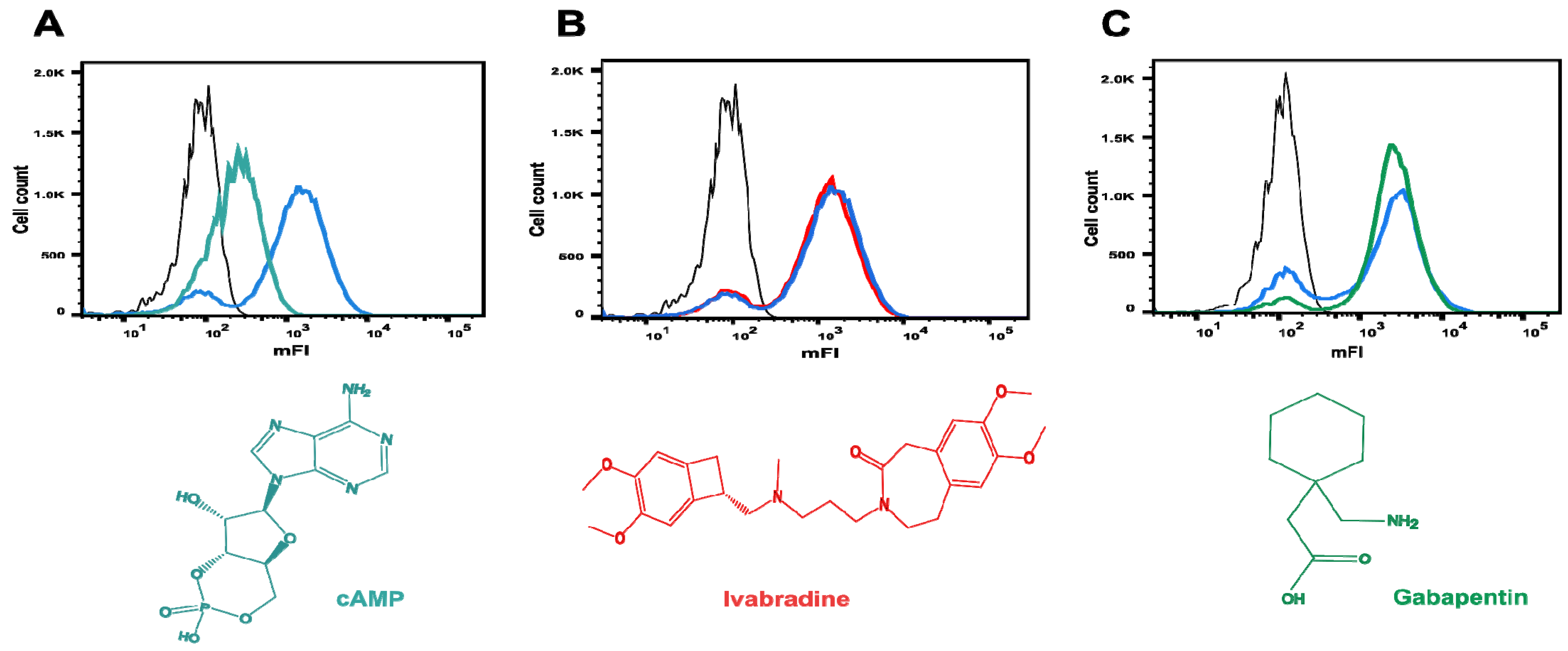
2.4. Binding of cAMP to Surface-Displayed HCN4 C-Linker-CNBD
2.5. Further Analysis of the Competitive Binding between 8-Fluo-cAMP and cAMP
2.6. Investigating Ivabradine and Gabapentin in the C-Linker-CNBD Binding Assay
2.7. Screening for Inhibitors
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Bacterial Strains and Plasmid Construction
4.3. Culture Conditions and Sample Preparation
4.4. Proteinase K Digestion
4.5. Immunolabeling
4.6. Ligand-Binding Assays
4.7. Flow Cytometry Analysis
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, F.; Yarov-Yarovoy, V.; Gutman, G.A.; Catterall, W.A. Overview of Molecular Relationships in the Voltage-Gated Ion Channel Superfamily. Pharmacol. Rev. 2005, 57, 387–395. [Google Scholar] [CrossRef]
- Wahl-Schott, C.; Biel, M. HCN channels: Structure, cellular regulation and physiological function. Cell. Mol. Life Sci. 2009, 66, 470–494. [Google Scholar] [CrossRef]
- Santoro, B.; Liu, D.T.; Yao, H.; Bartsch, D.; Kandel, E.R.; Siegelbaum, S.; Tibbs, G.R. Identification of a Gene Encoding a Hyperpolarization-Activated Pacemaker Channel of Brain. Cell 1998, 93, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.; Zong, X.; Jeglitsch, M.; Hofmann, F.; Biel, M. A family of hyperpolarization-activated mammalian cation channels. Nature 1998, 393, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J.; Siegelbaum, S.A. Properties of Hyperpolarization-Activated Pacemaker Current Defined by Coassembly of Hcn1 and Hcn2 Subunits and Basal Modulation by Cyclic Nucleotide. J. Gen. Physiol. 2001, 117, 491–504. [Google Scholar] [CrossRef]
- Lee, C.-H.; MacKinnon, R. Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell 2017, 168, 111–120.E11. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; MacKinnon, R. Voltage Sensor Movements during Hyperpolarization in the HCN Channel. Cell 2019, 179, 1582–1589.E7. [Google Scholar] [CrossRef]
- Saponaro, A.; Bauer, D.; Giese, M.H.; Swuec, P.; Porro, A.; Gasparri, F.; Sharifzadeh, A.S.; Chaves-Sanjuan, A.; Alberio, L.; Parisi, G.; et al. Gating movements and ion permeation in HCN4 pacemaker channels. Mol. Cell 2021, 81, 2929–2943.E6. [Google Scholar] [CrossRef]
- Xu, X.; Vysotskaya, Z.V.; Liu, Q.; Zhou, L. Structural Basis for the cAMP-dependent Gating in the Human HCN4 Channel. J. Biol. Chem. 2010, 285, 37082–37091. [Google Scholar] [CrossRef]
- Wainger, B.J.; DeGennaro, M.; Santoro, B.; Siegelbaum, S.A.; Tibbs, G.R. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 2001, 411, 805–810. [Google Scholar] [CrossRef]
- Almanza, A.; Luis, E.; Mercado, F.; Vega, R.; Soto, E. Molecular identity, ontogeny, and cAMP modulation of the hyperpolarization-activated current in vestibular ganglion neurons. J. Neurophysiol. 2012, 108, 2264–2275. [Google Scholar] [CrossRef] [PubMed]
- Moosmang, S.; Stieber, J.; Zong, X.; Biel, M.; Hofmann, F.; Ludwig, A. Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur. J. Biochem. 2001, 268, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Scholten, A.; Ivanova, E.; Haverkamp, S.; Kremmer, E.; Kaupp, U.B. HCN channels are expressed differentially in retinal bipolar cells and concentrated at synaptic terminals. Eur. J. Neurosci. 2003, 17, 2084–2096. [Google Scholar] [CrossRef]
- Tellez, J.O.; Dobrzynski, H.; Greener, I.D.; Graham, G.M.; Laing, E.; Honjo, H.; Hubbard, S.J.; Boyett, M.R.; Billeter, R. Differential Expression of Ion Channel Transcripts in Atrial Muscle and Sinoatrial Node in Rabbit. Circ. Res. 2006, 99, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Wahl-Schott, C.; Michalakis, S.; Zong, X. Hyperpolarization-Activated Cation Channels: From Genes to Function. Physiol. Rev. 2009, 89, 847–885. [Google Scholar] [CrossRef]
- Milanesi, R.; Baruscotti, M.; Gnecchi-Ruscone, T.; DiFrancesco, D. Familial Sinus Bradycardia Associated with a Mutation in the Cardiac Pacemaker Channel. N. Engl. J. Med. 2006, 354, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Nakamura, K.; Hayashi, T.; Inagaki, N.; Takahashi, M.; Arimura, T.; Morita, H.; Higashiuesato, Y.; Hirano, Y.; Yasunami, M.; et al. Functional Characterization of a Trafficking-Defective HCN4 Mutation, D553N, Associated with Cardiac Arrhythmia. J. Biol. Chem. 2004, 279, 27194–27198. [Google Scholar] [CrossRef]
- Marini, C.; Porro, A.; Rastetter, A.; Dalle, C.; Rivolta, I.; Bauer, D.; Oegema, R.; Nava, C.; Parrini, E.; Mei, D.; et al. HCN1 mutation spectrum: From neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain 2018, 141, 3160–3178. [Google Scholar] [CrossRef]
- Nava, C.; Dalle, C.; Rastetter, A.; Striano, P.; de Kovel, C.G.; Nabbout, R.; Cances, C.; Ville, D.; Brilstra, E.H.; Gobbi, G.; et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat. Genet. 2014, 46, 640–645. [Google Scholar] [CrossRef]
- Luo, L.; Chang, L.; Brown, S.; Ao, H.; Lee, D.; Higuera, E.; Dubin, A.; Chaplan, S. Role of peripheral hyperpolarization-activated cyclic nucleotide-modulated channel pacemaker channels in acute and chronic pain models in the rat. Neuroscience 2007, 144, 1477–1485. [Google Scholar] [CrossRef]
- Emery, E.C.; Young, G.T.; Berrocoso, E.M.; Chen, L.; McNaughton, P.A. HCN2 Ion Channels Play a Central Role in Inflammatory and Neuropathic Pain. Science 2011, 333, 1462–1466. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.F.; Difrancesco, D.; Noble, S.J. How does adrenaline accelerate the heart? Nature 1979, 280, 235–236. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.F.; Giles, W.; Noble, S.J. Membrane currents underlying activity in frog sinus venosus. J. Physiol. 1977, 271, 783–816. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, D. A new interpretation of the pace-maker current in calf Purkinje fibres. J. Physiol. 1981, 314, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Kessi, M.; Peng, J.; Duan, H.; He, H.; Chen, B.; Xiong, J.; Wang, Y.; Yang, L.; Wang, G.; Kiprotich, K.; et al. The Contribution of HCN Channelopathies in Different Epileptic Syndromes, Mechanisms, Modulators, and Potential Treatment Targets: A Systematic Review. Front. Mol. Neurosci. 2022, 15, 807202. [Google Scholar] [CrossRef]
- Rivolta, I.; Binda, A.; Masi, A.; DiFrancesco, J.C. Cardiac and neuronal HCN channelopathies. Pflug. Arch. 2020, 472, 931–951. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, C.S.; Owens, R.E.; Bolorunduro, O.B.; Jha, S.K. Ivabradine: A Review of Labeled and Off-Label Uses. Am. J. Cardiovasc. Drugs 2016, 16, 337–347. [Google Scholar] [CrossRef]
- Balducci, V.; Credi, C.; Sacconi, L.; Romanelli, M.N.; Sartiani, L.; Cerbai, E. The HCN channel as a pharmacological target: Why, where, and how to block it. Prog. Biophys. Mol. Biol. 2021, 166, 173–181. [Google Scholar] [CrossRef]
- Romanelli, M.N.; Del Lungo, M.; Guandalini, L.; Zobeiri, M.; Gyökeres, A.; Árpádffy-Lovas, T.; Koncz, I.; Sartiani, L.; Bartolucci, G.; Dei, S.; et al. EC18 as a Tool To Understand the Role of HCN4 Channels in Mediating Hyperpolarization-Activated Current in Tissues. ACS Med. Chem. Lett. 2019, 10, 584–589. [Google Scholar] [CrossRef]
- Del Lungo, M.; Melchiorre, M.; Guandalini, L.; Sartiani, L.; Mugelli, A.; Koncz, I.; Szel, T.; Varro, A.; Romanelli, M.N.; Cerbai, E. Novel blockers of hyperpolarization-activated current with isoform selectivity in recombinant cells and native tissue. Br. J. Pharmacol. 2012, 166, 602–616. [Google Scholar] [CrossRef]
- Nakashima, K.; Nakao, K.; Matsui, H. Discovery of Novel HCN4 Blockers with Unique Blocking Kinetics and Binding Properties. SLAS Discov. 2021, 26, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.N.; Sartiani, L.; Masi, A.; Mannaioni, G.; Manetti, D.; Mugelli, A.; Cerbai, E. HCN Channels Modulators: The Need for Selectivity. Curr. Top. Med. Chem. 2016, 16, 1764–1791. [Google Scholar] [CrossRef] [PubMed]
- Tanguay, J.; Callahan, K.M.; D’avanzo, N. Characterization of drug binding within the HCN1 channel pore. Sci. Rep. 2019, 9, 465. [Google Scholar] [CrossRef] [PubMed]
- Postea, O.; Biel, M. Exploring HCN channels as novel drug targets. Nat. Rev. Drug Discov. 2011, 10, 903–914. [Google Scholar] [CrossRef]
- Möller, S.; Alfieri, A.; Bertinetti, D.; Aquila, M.; Schwede, F.; Lolicato, M.; Rehmann, H.; Moroni, A.; Herberg, F.W. Cyclic Nucleotide Mapping of Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) Channels. ACS Chem. Biol. 2014, 9, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Hayoz, S.; Tiwari, P.B.; Piszczek, G.; Uren, A.; Brelidze, T.I. Investigating cyclic nucleotide and cyclic dinucleotide binding to HCN channels by surface plasmon resonance. PLoS ONE 2017, 12, e0185359. [Google Scholar] [CrossRef]
- Goldenberg, D.P. Genetic Studies of Protein Stability and Mechanisms of Folding. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 481–507. [Google Scholar] [CrossRef]
- Butt, T.R.; Edavettal, S.C.; Hall, J.P.; Mattern, M.R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 2005, 43, 1–9. [Google Scholar] [CrossRef]
- Routzahn, K.M.; Waugh, D.S. Differential effects of supplementary affinity tags on the solubility of MBP fusion proteins. J. Struct. Funct. Genom. 2002, 2, 83–92. [Google Scholar] [CrossRef]
- Jose, J.; Maas, R.M.; Teese, M.G. Autodisplay of enzymes—Molecular basis and perspectives. J. Biotechnol. 2012, 161, 92–103. [Google Scholar] [CrossRef]
- Jose, J.; Meyer, T.F. The Autodisplay Story, from Discovery to Biotechnical and Biomedical Applications. Microbiol. Mol. Biol. Rev. 2007, 71, 600–619. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef] [PubMed]
- Nickelsen, A.; Jose, J. Label-free flow cytometry-based enzyme inhibitor identification. Anal. Chim. Acta 2021, 1179, 338826. [Google Scholar] [CrossRef]
- Gercke, D.; Furtmann, C.; Tozakidis, I.E.P.; Jose, J. Highly Crystalline Post-Consumer PET Waste Hydrolysis by Surface Displayed PETase Using a Bacterial Whole-Cell Biocatalyst. Chemcatchem 2021, 13, 3479–3489. [Google Scholar] [CrossRef]
- Sichwart, S.; Tozakidis, I.E.P.; Teese, M.G.; Jose, J. Maximized Autotransporter Mediated Expression (MATE) for Surface Display and Secretion of Recombinant Proteins in Escherichia coli. Food Technol. Biotechnol. 2015, 53, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, S.D.; Hannemann, F.; Teese, M.G.; Bernhardt, R.; Jose, J. Autodisplay of functional CYP106A2 in Escherichia coli. J. Biotechnol. 2012, 161, 104–112. [Google Scholar] [CrossRef]
- Lolicato, M.; Nardini, M.; Gazzarrini, S.; Möller, S.; Bertinetti, D.; Herberg, F.W.; Bolognesi, M.; Martin, H.; Fasolini, M.; Bertrand, J.A.; et al. Tetramerization Dynamics of C-Terminal Domain Underlies Isoform-Specific cAMP Gating in Hyperpolarization-Activated Cyclic Nucleotide-Gated Channels. J. Biol. Chem. 2011, 286, 44811–44820. [Google Scholar] [CrossRef]
- Hunter, S.; Cochran, J. Cell-Binding Assays for Determining the Affinity of Protein–Protein Interactions: Technologies and Considerations. Methods Enzymol. 2016, 580, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Hulme, E.C.; Trevethick, M.A. Ligand binding assays at equilibrium: Validation and interpretation. Br. J. Pharmacol. 2010, 161, 1219–1237. [Google Scholar] [CrossRef]
- Tian, H.; Furtmann, C.; Lenz, F.; Srinivasamurthy, V.; Bornscheuer, U.T.; Jose, J. Enzyme cascade converting cyclohexanol into epsilon-caprolactone coupled with NADPH recycling using surface displayed alcohol dehydrogenase and cyclohexanone monooxygenase on E. coli. Microb. Biotechnol. 2022, 15, 2235–2249. [Google Scholar] [CrossRef]
- Jose, J.; Chung, J.-W.; Jeon, B.-J.; Maas, R.M.; Nam, C.-H.; Pyun, J.-C. Escherichia coli with autodisplayed Z-domain of protein A for signal amplification of SPR biosensor. Biosens. Bioelectron. 2009, 24, 1324–1329. [Google Scholar] [CrossRef]
- Newton, P.; Harrison, P.; Clulow, S. A novel method for determination of the affinity of protein: Protein interactions in homogeneous assays. J. Biomol. Screen 2008, 13, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, M.; Zhang, Z.; Boulton, S.; Selvaratnam, R.; VanSchouwen, B.; Gloyd, M.; Accili, E.A.; Lange, O.F.; Melacini, G. A Mechanism for the Auto-Inhibition of Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) Channel Opening and Its Relief by cAMP. J. Biol. Chem. 2014, 289, 22205–22220. [Google Scholar] [CrossRef] [PubMed]
- Lazareno, S. Quantification of receptor interactions using binding methods. J. Recept. Signal Transduct. Res. 2001, 21, 139–165. [Google Scholar] [CrossRef]
- Bucchi, A.; Baruscotti, M.; Nardini, M.; Barbuti, A.; Micheloni, S.; Bolognesi, M.; DiFrancesco, D. Identification of the Molecular Site of Ivabradine Binding to HCN4 Channels. PLoS ONE 2013, 8, e53132. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.W.; Atkinson, L.Z.; Al-Juffali, N.; Awad, A.; Geddes, J.R.; Tunbridge, E.M.; Harrison, P.J.; Cipriani, A. Gabapentin and pregabalin in bipolar disorder, anxiety states, and insomnia: Systematic review, meta-analysis, and rationale. Mol. Psychiatry 2022, 27, 1339–1349. [Google Scholar] [CrossRef]
- Tae, H.-S.; Smith, K.M.; Phillips, A.M.; Boyle, K.A.; Li, M.; Forster, I.C.; Hatch, R.J.; Richardson, R.; Hughes, D.I.; Graham, B.A.; et al. Gabapentin Modulates HCN4 Channel Voltage-Dependence. Front. Pharmacol. 2017, 8, 554. [Google Scholar] [CrossRef] [PubMed]
- Algul, O.; Kaessler, A.; Apcin, Y.; Yilmaz, A.; Jose, J. Comparative Studies on Conventional and Microwave Synthesis of Some Benzimidazole, Benzothiazole and Indole Derivatives and Testing on Inhibition of Hyaluronidase. Molecules 2008, 13, 736–748. [Google Scholar] [CrossRef]
- Lindenblatt, D.; Nickelsen, A.; Applegate, V.M.; Hochscherf, J.; Witulski, B.; Bouaziz, Z.; Marminon, C.; Bretner, M.; Le Borgne, M.; Jose, J.; et al. Diacritic Binding of an Indenoindole Inhibitor by CK2α Paralogs Explored by a Reliable Path to Atomic Resolution CK2α′ Structures. ACS Omega 2019, 4, 5471–5478. [Google Scholar] [CrossRef]
- Hundsdörfer, C.; Hemmerling, H.-J.; Götz, C.; Totzke, F.; Bednarski, P.; Le Borgne, M.; Jose, J. Indeno[1,2-b]indole derivatives as a novel class of potent human protein kinase CK2 inhibitors. Bioorg. Med. Chem. 2012, 20, 2282–2289. [Google Scholar] [CrossRef]
- Di Pietro, A.; Gozzi, G.J.; Bouaziz, Z.; Winter, E.; Daflon-Yunes, N.; Honorat, M.; Guragossian, N.; Marminon, C.; Valdameri, G.; Bollacke, A.; et al. Phenolic indeno[1,2-b]indoles as ABCG2-selective potent and non-toxic inhibitors stimulating basal ATPase activity. Drug Des. Dev. Ther. 2015, 9, 3481–3495. [Google Scholar] [CrossRef] [PubMed]
- Hundsdörfer, C.; Hemmerling, H.-J.; Hamberger, J.; Le Borgne, M.; Bednarski, P.; Götz, C.; Totzke, F.; Jose, J. Novel indeno[1,2-b]indoloquinones as inhibitors of the human protein kinase CK2 with antiproliferative activity towards a broad panel of cancer cell lines. Biochem. Biophys. Res. Commun. 2012, 424, 71–75. [Google Scholar] [CrossRef]
- Prinz, H.; Chamasmani, B.; Vogel, K.; Böhm, K.J.; Aicher, B.; Gerlach, M.; Günther, E.G.; Amon, P.; Ivanov, I.; Müller, K. N-Benzoylated Phenoxazines and Phenothiazines: Synthesis, Antiproliferative Activity, and Inhibition of Tubulin Polymerization. J. Med. Chem. 2011, 54, 4247–4263. [Google Scholar] [CrossRef]
- Prinz, H.; Ridder, A.-K.; Vogel, K.; Böhm, K.J.; Ivanov, I.; Ghasemi, J.B.; Aghaee, E.; Müller, K. N-Heterocyclic (4-Phenylpiperazin-1-yl)methanones Derived from Phenoxazine and Phenothiazine as Highly Potent Inhibitors of Tubulin Polymerization. J. Med. Chem. 2017, 60, 749–766. [Google Scholar] [CrossRef]
- Waltemate, J.; Ivanov, I.; Ghasemi, J.B.; Aghaee, E.; Daniliuc, C.G.; Müller, K.; Prinz, H. 10-(4-Phenylpiperazine-1-carbonyl)acridin-9(10H)-ones and related compounds: Synthesis, antiproliferative activity and inhibition of tubulin polymerization. Bioorg. Med. Chem. Lett. 2021, 32, 127687. [Google Scholar] [CrossRef] [PubMed]
- Bannwitz, S.; Krane, D.; Vortherms, S.; Kalin, T.; Lindenschmidt, C.; Golpayegani, N.Z.; Tentrop, J.; Prinz, H.; Müller, K. Synthesis and Structure–Activity Relationships of Lapacho Analogues. 2. Modification of the Basic Naphtho[2,3-b]furan-4,9-dione, Redox Activation, and Suppression of Human Keratinocyte Hyperproliferation by 8-Hydroxynaphtho[2,3-b]thiophene-4,9-diones. J. Med. Chem. 2014, 57, 6226–6239. [Google Scholar] [CrossRef]
- Basoglu, A.; Dirkmann, S.; Golpayegani, N.Z.; Vortherms, S.; Tentrop, J.; Nowottnik, D.; Prinz, H.; Fröhlich, R.; Müller, K. Oxadiazole-substituted naphtho[2,3-b]thiophene-4,9-diones as potent inhibitors of keratinocyte hyperproliferation. Structure−activity relationships of the tricyclic quinone skeleton and the oxadiazole substituent. Eur. J. Med. Chem. 2017, 134, 119–132. [Google Scholar] [CrossRef]
- Reichstein, A.; Vortherms, S.; Bannwitz, S.; Tentrop, J.; Prinz, H.; Müller, K. Synthesis and Structure–Activity Relationships of Lapacho Analogues. 1. Suppression of Human Keratinocyte Hyperproliferation by 2-Substituted Naphtho[2,3-b]furan-4,9-diones, Activation by Enzymatic One- and Two-Electron Reduction, and Intracellular Generation of Superoxide. J. Med. Chem. 2012, 55, 7273–7284. [Google Scholar] [CrossRef]
- Jose, J.; Bernhardt, R.; Hannemann, F. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J. Biotechnol. 2002, 95, 257–268. [Google Scholar] [CrossRef]
- Bopp, B.; Ciglia, E.; Ouald-Chaib, A.; Groth, G.; Gohlke, H.; Jose, J. Design and biological testing of peptidic dimerization inhibitors of human Hsp90 that target the C-terminal domain. Biochim. Biophys. Acta 2016, 1860, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Gratz, A.; Bollacke, A.; Stephan, S.; Nienberg, C.; Le Borgne, M.; Götz, C.; Jose, J. Functional display of heterotetrameric human protein kinase CK2 on Escherichia coli: A novel tool for drug discovery. Microb. Cell Fact. 2015, 14, 74. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Grodberg, J.; Dunn, J.J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 1988, 170, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wojciechowski, M.N.; Schreiber, S.; Jose, J. A Novel Flow Cytometry-Based Assay for the Identification of HCN4 CNBD Ligands. Pharmaceuticals 2023, 16, 710. https://doi.org/10.3390/ph16050710
Wojciechowski MN, Schreiber S, Jose J. A Novel Flow Cytometry-Based Assay for the Identification of HCN4 CNBD Ligands. Pharmaceuticals. 2023; 16(5):710. https://doi.org/10.3390/ph16050710
Chicago/Turabian StyleWojciechowski, Magdalena N., Sebastian Schreiber, and Joachim Jose. 2023. "A Novel Flow Cytometry-Based Assay for the Identification of HCN4 CNBD Ligands" Pharmaceuticals 16, no. 5: 710. https://doi.org/10.3390/ph16050710
APA StyleWojciechowski, M. N., Schreiber, S., & Jose, J. (2023). A Novel Flow Cytometry-Based Assay for the Identification of HCN4 CNBD Ligands. Pharmaceuticals, 16(5), 710. https://doi.org/10.3390/ph16050710