Influence of Bifidobacterium breve on the Glycaemic Control, Lipid Profile and Microbiome of Type 2 Diabetic Subjects: A Preliminary Randomized Clinical Trial
Abstract
1. Introduction
2. Results
2.1. The Efficacy of B. breve Supplementation on Blood Biochemical Parameters
2.2. Microbiome Analysis
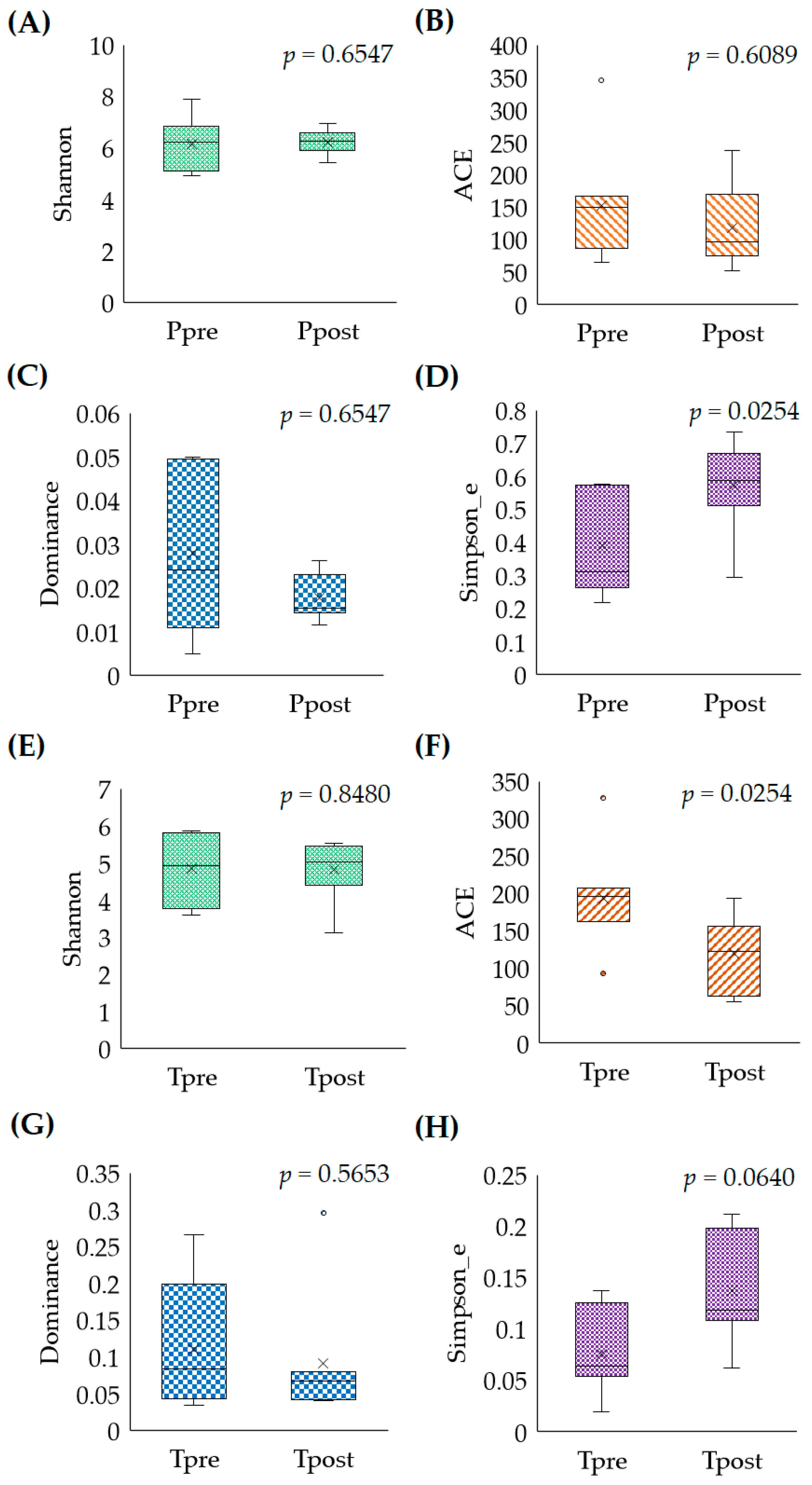
2.2.1. Microbial Abundances (Alpha-Diversity)
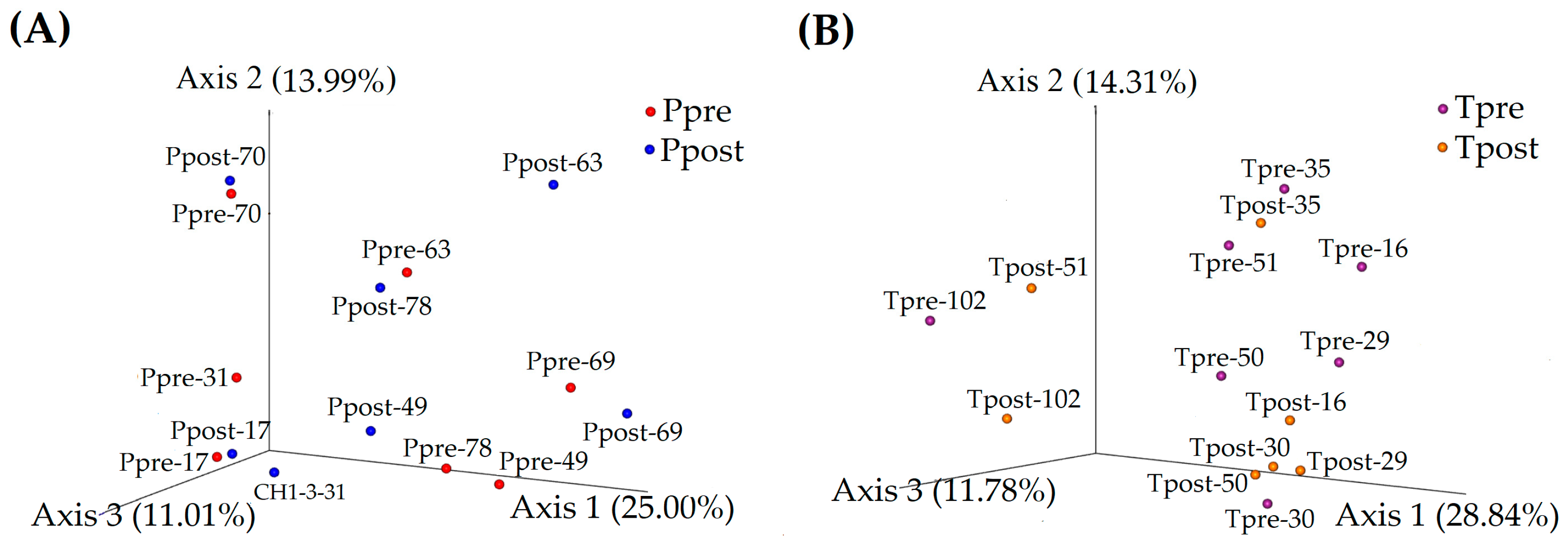
2.2.2. Microbiome Similarity and Variance (Beta-Diversity)
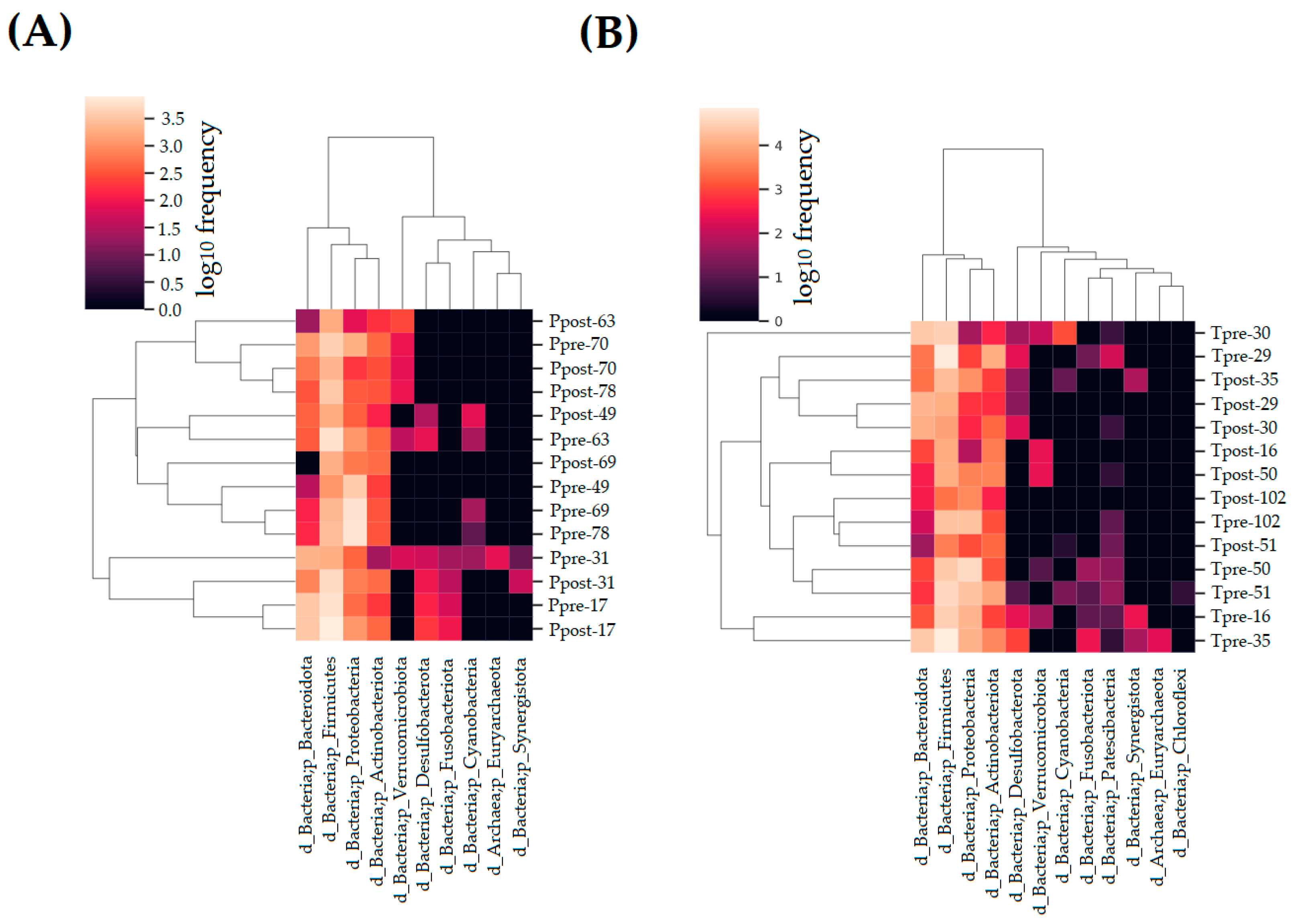
2.3. Taxonomy Assignment
2.3.1. Estimation of the Phylum
2.3.2. Estimation of the Genera
2.3.3. Estimation of the Species
3. Discussion
4. Materials and Methods
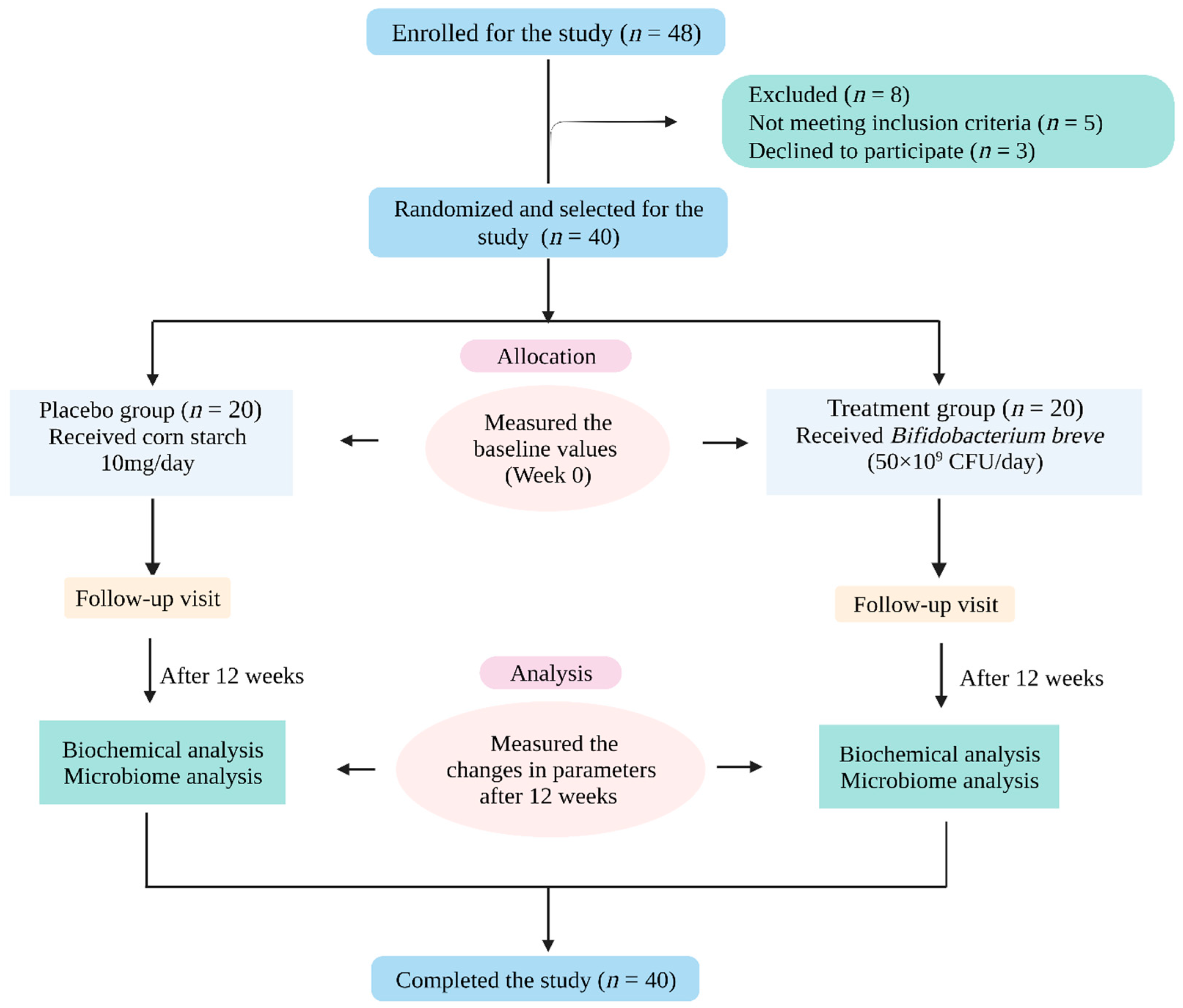
4.1. Study Subjects
4.2. Study Protocol
4.3. Demographic Assessments
4.4. Blood Biochemical Analysis
4.5. Next-Generation Sequencing
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Larsen, N.; Vogensen, F.K.; Van den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; AI-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from nondiabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Hilliard, M.E.; Isaacs, D.; Johnson, E.L.; et al. 2. Classification and Diagnosis of Diabetes: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46, S19–S40. [Google Scholar] [PubMed]
- Nie, Q.X.; Chen, H.H.; Hu, J.L.; Fan, S.T.; Nie, S.P. Dietary compounds and traditional Chinese medicine ameliorate type 2 diabetes by modulating gut microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 848–863. [Google Scholar] [CrossRef]
- Mooradian, A. Dyslipidemia in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2009, 5, 150–159. [Google Scholar] [CrossRef]
- Zhao, L.P.; Zhang, F.; Ding, X.Y.; Wu, G.J.; Lam, Y.Y.; Wang, X.J.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef]
- Aw, W.; Fukuda, S. Understanding the role of the gut ecosystem in diabetes mellitus. J. Diabetes Investig. 2018, 9, 5–12. [Google Scholar] [CrossRef]
- Grigorescu, I.; Dumitrascu, D.L. Implication of gut microbiota in diabetes mellitus and obesity. Acta Endocrinol. 2016, 12, 206–214. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. The Influence of Supplementation of Anthocyanins on Obesity-Associated Comorbidities: A Concise Review. Foods 2020, 9, 687. [Google Scholar] [CrossRef]
- Gavin, P.G.; Hamilton-Williams, E.E. The Gut Microbiota in Type 1 Diabetes: Friend or Foe? Curr. Opin. Endocrinol. Diabetes Obes. 2019, 26, 207–212. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, L.; Zhang, S.; Zhao, X.; Gang, X.; Wang, G. Evaluating the Causal Role of Gut Microbiota in Type 1 Diabetes and Its Possible Pathogenic Mechanisms. Front. Endocrinol. 2020, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Sircana, A.; Framarin, L.; Leone, N.; Berrutti, M.; Castellino, F.; Parente, R.; De Michieli, F.; Paschetta, E.; Musso, G. Altered gut microbiota in type 2 diabetes: Just a coincidence? Curr. Diabetes Rep. 2018, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Canfora, E.E.; Meex, R.; Venema, K.; Blaak, E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol. 2019, 15, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Sivamaruthi, B.S. A comprehensive review on clinical outcome of probiotic and synbiotic therapy for inflammatory bowel diseases. Asian Pac. J. Trop. Biomed. 2018, 8, 179–186. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. A review on anti-aging properties of probiotics. Int. J. Appl. Pharm. 2018, 10, 23–27. [Google Scholar] [CrossRef]
- Tenorio-Jiménez, C.; Martínez-Ramírez, M.J.; Gil, Á.; Gómez-Llorente, C. Effects of Probiotics on Metabolic Syndrome: A Systematic Review of Randomized Clinical Trials. Nutrients 2020, 12, 124. [Google Scholar] [CrossRef]
- Akhgarjand, C.; Vahabi, Z.; Shab-Bidar, S.; Etesam, F.; Djafarian, K. Effects of probiotic supplements on cognition, anxiety, and physical activity in subjects with mild and moderate Alzheimer’s disease: A randomized, double-blind, and placebo-controlled study. Front. Aging Neurosci. 2022, 14, 1032494. [Google Scholar] [CrossRef]
- Thangaleela, S.; Sivamaruthi, B.S.; Kesika, P.; Bharathi, M.; Chaiyasut, C. Role of the Gut-Brain Axis, Gut Microbial Composition, Diet, and Probiotic Intervention in Parkinson’s Disease. Microorganisms 2022, 10, 1544. [Google Scholar] [CrossRef]
- Kesika, P.; Sivamaruthi, B.S.; Chaiyasut, C. Do Probiotics Improve the Health Status of Individuals with Diabetes Mellitus? A Review on Outcomes of Clinical Trials. BioMed Res. Int. 2019, 2019, 1531567. [Google Scholar] [CrossRef]
- Toejing, P.; Khampithum, N.; Sirilun, S.; Chaiyasut, C.; Lailerd, N. Influence of Lactobacillus paracasei HII01 Supplementation on Glycemia and Inflammatory Biomarkers in Type 2 Diabetes: A Randomized Clinical Trial. Foods 2021, 10, 1455. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S.; Zeng, Z.; Qin, Y.; Shen, Q.; Li, P. Anti-diabetic effects of Bifidobacterium animalis 01 through improving hepatic insulin sensitivity in type 2 diabetic rat model. J. Funct. Foods. 2020, 67, 103843. [Google Scholar] [CrossRef]
- Park, Y.; Jin, H.S.; Aki, D.; Lee, J.; Liu, Y.C. The ubiquitin system in immune regulation. Adv. Immunol. 2014, 124, 17–66. [Google Scholar]
- Qian, X.; Si, Q.; Lin, G.; Zhu, M.; Lu, J.; Zhang, H.; Wang, G.; Chen, W. Bifidobacterium adolescentis Is Effective in Relieving Type 2 Diabetes and May Be Related to Its Dominant Core Genome and Gut Microbiota Modulation Capacity. Nutrients 2022, 14, 2479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chu, J.; Hao, W.; Zhang, J.; Li, H.; Yang, C.; Yang, J.; Chen, X.; Wang, H. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediat. Inflamm. 2021, 2021, 5110276. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining Dysbiosis and Its Influence on Host Immunity and Disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Harsch, I.; Konturek, P. The Role of Gut Microbiota in Obesity and Type 2 and Type 1 Diabetes Mellitus: New Insights into “Old” Diseases. Med. Sci. 2018, 6, 32. [Google Scholar] [CrossRef]
- Liaqat, I.; Ali, N.M.; Arshad, N.; Sajjad, S.; Rashid, F.; Hanif, U.; Ara, C.; Ulfat, M.; Andleeb, S.; Awan, U.F.; et al. Gut dysbiosis, inflammation and type 2 diabetes in mice using synthetic gut microbiota from diabetic humans. Braz. J. Biol. 2021, 83, e242818. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Wang, J.S.; Liu, J.C. Intestinal microbiota in the treatment of metabolically associated fatty liver disease. World J. Clin. Cases 2022, 10, 11240–11251. [Google Scholar] [CrossRef]
- Venkataraman, R.; Jose, P.; Jose, J. Impact of probiotics on health-related quality of life in type II diabetes mellitus: A randomized, single-blind, placebo-controlled study. J. Nat. Sci. Biol. Med. 2019, 10, 2–7. [Google Scholar] [CrossRef]
- Kassaian, N.; Feizi, A.; Aminorroaya, A.; Jafari, P.; Ebrahimi, M.T.; Amini, M. The effects of probiotics and synbiotic supplementation on glucose and insulin metabolism in adults with prediabetes: A double-blind randomized clinical trial. Acta Diabetol. 2018, 55, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Ketema, E.B.; Kibret, K.T. Correlation of fasting and postprandial plasma glucose with HbA1c in assessing glycemic control; systematic review and meta-analysis. Arch. Public Health 2015, 73, 43. [Google Scholar] [CrossRef] [PubMed]
- Weykamp, C.; Garry John, W.; Mosca, A. A review of the challenge in measuring hemoglobin A1c. J. Diabetes Sci. Technol. 2009, 3, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Pasupathi, P.; Manivannan, P.M.; Uma, M.; Deepa, M. Glycated haemoglobin (HbA1c) as a stable indicator of type 2 diabetes. Int. J. Pharm. Biomed. Res. 2010, 1, 53–56. [Google Scholar]
- Wang, C.H.; Yen, H.R.; Lu, W.L.; Ho, H.H.; Lin, W.Y.; Kuo, Y.W.; Huang, Y.Y.; Tsai, S.Y.; Lin, H.C. Adjuvant Probiotics of Lactobacillus salivarius subsp. salicinius AP-32, L. johnsonii MH-68, and Bifidobacterium animalis subsp. lactis CP-9 Attenuate Glycemic levels and inflammatory cytokines in patients with Type 1 Diabetes Mellitus. Front. Endocrinol. 2022, 13, 754401. [Google Scholar] [CrossRef] [PubMed]
- Asemi, Z.; Zare, Z.; Shakeri, H.; Sabihi, S.S.; Esmaillzadeh, A. Effect of Multispecies Probiotic Supplements on Metabolic Profiles, Hs-CRP, and Oxidative Stress in Patients with Type 2 Diabetes. Ann. Nutr. Metab. 2013, 63, 1–9. [Google Scholar] [CrossRef]
- Ejtahed, H.S.; Mohtadi-Nia, J.; Homayouni-Rad, A.; Niafar, M.; Asghari-Jafarabadi, M.; Mofid, V.; Akbarian-Moghari, A. Effect of probiotic yogurt containing Lactobacillus acidophilus and Bifidobacterium lactis on lipid profile in individuals with type 2 diabetes mellitus. J. Dairy Sci. 2011, 94, 3288–3294. [Google Scholar] [CrossRef]
- Ahmadian, F.; Razmpoosh, E.; Ejtahed, H.S.; Javadi, M.; Mirmiran, P.; Azizi, F. Effects of probiotic supplementation on major cardiovascular-related parameters in patients with type-2 diabetes mellitus: A secondary-data analysis of a randomized double-blind controlled trial. Diabetol. Metab. Syndr. 2022, 14, 52. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, Y.; Xu, D.; Wang, Q. Probiotics ameliorates glycemic control of patients with diabetic nephropathy: A randomized clinical study. J. Clin. Lab. Anal. 2021, 35, e23650. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Luo, L.; Le, Y.; Li, Y.; Yuan, F.; Wu, Y.; Xu, P. The Beneficial Effects of a Multispecies Probiotic Supplement on Glycaemic Control and Metabolic Profile in Adults with Type 1 Diabetes: A Randomised, Double-Blinded, Placebo-Controlled Pilot-Study. Diabetes Metab. Syndr. Obes. 2023, 16, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Solito, A.; Bozzi Cionci, N.; Calgaro, M.; Caputo, M.; Vannini, L.; Hasballa, I.; Archero, F.; Giglione, E.; Ricotti, R.; Walker, G.E.; et al. Supplementation with Bifidobacterium breve BR03 and B632 strains improved insulin sensitivity in children and adolescents with obesity in a cross-over, randomized double-blind placebo-controlled trial. Clin. Nutr. 2021, 40, 4585–4594. [Google Scholar] [CrossRef] [PubMed]
- Madempudi, R.S.; Ahire, J.J.; Neelamraju, J.; Tripathi, A.; Nanal, S. Efficacy of UB0316, a multi-strain probiotic formulation in patients with type 2 diabetes mellitus: A double blind, randomized, placebo-controlled study. PLoS ONE 2019, 14, e0225168. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.C.; Tsai, W.H.; Jheng, Y.P.; Su, S.L.; Wang, S.Y.; Lin, C.C.; Chen, Y.H.; Chang, W.W. The beneficial effects of Lactobacillus reuteri ADR-1 or ADR-3 consumption on type 2 diabetes mellitus: A randomized, double-blinded, placebo-controlled trial. Sci. Rep. 2018, 8, 16791. [Google Scholar] [CrossRef]
- Tlaskalová-Hogenová, H.; Stěpánková, R.; Kozáková, H.; Hudcovic, T.; Vannucci, L.; Tučková, L.; Rossmann, P.; Hrnčíř, T.; Kverka, M.; Zákostelská, Z.; et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 2011, 8, 110–120. [Google Scholar]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar, R.D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Landman, C.; Quévrain, E. Gut microbiota: Description, role and pathophysiologic implications. Rev. Med. Interne 2016, 37, 418–423. [Google Scholar] [CrossRef]
- Li, W.; Ma, Z.S. FBA ecological guild: Trio of firmicutes-bacteroidetes alliance against actinobacteria in human oral microbiome. Sci. Rep. 2020, 10, 287. [Google Scholar] [CrossRef]
- Wu, X.; Ma, C.; Han, L.; Nawaz, M.; Gao, F.; Zhang, X.; Yu, P.; Zhao, C.; Li, L.; Zhou, A.; et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr. Microbiol. 2010, 61, 69–78. [Google Scholar] [CrossRef]
- Zhao, L.; Lou, H.; Peng, Y.; Chen, S.; Zhang, Y.; Li, X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.C.; Hung, W.W.; Tsai, H.J.; Chang, C.C.; Chiu, Y.W.; Hwang, S.J.; Kuo, M.C.; Chen, S.C.; Dai, C.Y.; Tsai, Y.C. The Association of Targeted Gut Microbiota with Body Composition in Type 2 Diabetes Mellitus. Int. J. Med. Sci. 2021, 18, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Egshatyan, L.; Kashtanova, D.; Popenko, A.; Tkacheva, O.; Tyakht, A.; Alexeev, D.; Karamnova, N.; Kostryukova, E.; Babenko, V.; Vakhitova, M.; et al. Gut microbiota and diet in patients with different glucose tolerance. Endocr. Connect. 2016, 5, 1–9. [Google Scholar] [CrossRef]
- Pataky, Z.; Genton, L.; Spahr, L.; Lazarevic, V.; Terraz, S.; Gaïa, N.; Rubbia-Brandt, L.; Golay, A.; Schrenzel, J.; Pichard, C. Impact of hypocaloric hyper proteic diet on gut microbiota in overweight or obese patients with nonalcoholic fatty liver disease: A pilot study. Digest Dis. Sci. 2016, 61, 2721–2731. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef]
- Kampoo, K.; Teanpaisan, R.; Ledder, R.G.; McBain, A.J. Oral bacterial communities in individuals with type 2 diabetes who live in southern Thailand. Appl. Environ. Microbiol. 2014, 80, 662–671. [Google Scholar] [CrossRef]
- Zeng, H.; Ishaq, S.L.; Zhao, F.Q.; Wright, A.D. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016, 35, 30–36. [Google Scholar] [CrossRef]
- Muralidharan, J.; Moreno-Indias, I.; Bulló, M.; Lopez, J.V.; Corella, D.; Castañer, O.; Vidal, J.; Atzeni, A.; Fernandez-García, J.C.; Torres-Collado, L.; et al. Effect on gut microbiota of a 1-y lifestyle intervention with Mediterranean diet compared with energy-reduced Mediterranean diet and physical activity promotion: PREDIMED-Plus Study. Am. J. Clin. Nutr. 2021, 114, 1148–1158. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Chaiyasut, C.; Sirilun, S.; Juntarachot, N.; Tongpong, P.; Ouparee, W.; Sivamaruthi, B.S.; Peerajan, S.; Waditee-Sirisattha, R.; Prombutara, P.; Klankeo, P.; et al. Effect of Dextranase and Dextranase-and-Nisin-Containing Mouthwashes on Oral Microbial Community of Healthy Adults—A Pilot Study. Appl. Sci. 2022, 12, 1650. [Google Scholar] [CrossRef]
- Conteville, L.C.; Oliveira-Ferreira, J.; Vicente, A.C.P. Gut Microbiome Biomarkers and Functional Diversity Within an Amazonian Semi-Nomadic Hunter-Gatherer Group. Front. Microbiol. 2019, 30, 1743. [Google Scholar] [CrossRef] [PubMed]

| No | Parameters | Placebo (n = 20) | Treatment (n = 20) | p-Value |
|---|---|---|---|---|
| 1 | Age (year) | 61.05 ± 1.83 | 63.90 ± 1.44 | 0.228 a |
| 2 | Gender | 1.000 b | ||
| Male | 3 (15.00) | 3 (15.00) | ||
| Female | 17 (85.00) | 17 (85.00) | ||
| 3 | Smoking | |||
| No | 20 (100.00) | 20 (100.00) | ||
| Yes | 0 (00.00) | 0 (00.00) | ||
| 4 | Alcohol consumption | 1.000 b | ||
| No | 19 (95.00) | 19 (95.00) | ||
| Yes | 1 (5.00) | 1 (5.00) | ||
| 5 | Body weight (kg) | 58.99 ± 2.37 | 54.86 ± 2.17 | 0.207 a |
| 6 | BMI (kg/m2) | 25.73 ± 1.27 | 23.80 ± 0.74 | 0.196 a |
| 7 | Body fat (%) | 28.21 ± 1.59 | 26.11 ± 1.27 | 0.308 a |
| 8 | Visceral fat (%) | 12.05 ± 0.93 | 10.00 ± 0.67 | 0.082 a |
| 9 | BUN (mg/dL) | 18.44 ± 2.07 | 14.69 ± 0.86 | 0.102 a |
| 10 | Creatinine (mg/dL) | 1.45 ± 0.13 | 1.28 ± 0.06 | 0.235 a |
| 11 | AST (IU/L) | 21.89 ± 1.91 | 24.65 ± 3.12 | 0.988 c |
| 12 | ALT (IU/L) | 22.11 ± 2.93 | 21.60 ± 3.30 | 0.403 c |
| 13 | ALP (IU/L) | 102.17 ± 4.82 | 98.67 ± 5.84 | 0.647 a |
| 14 | FBS (mg/dL) | 142.40 ± 9.86 | 148.25 ± 10.74 | 0.691 a |
| 15 | HbA1c (%) | 6.85 ± 0.37 | 7.44 ± 0.49 | 0.343 a |
| 16 | TC (mg/dL) | 192.85 ± 9.69 | 196.40 ± 8.20 | 0.781 a |
| 17 | TG (mg/dL) | 158.80 ± 18.38 | 156.25 ± 16.51 | 0.978 c |
| 18 | HDL (mg/dL) | 57.85 ± 2.55 | 63.75 ± 4.40 | 0.253 a |
| 19 | LDL (mg/dL) | 102.94 ± 7.14 | 113.61 ± 10.04 | 0.392 a |
| Parameters | Placebo (n = 20) | p-Value | Treatment (n = 20) | p-Value | ||
|---|---|---|---|---|---|---|
| Ppre | Ppost | Tpre | Tpost | |||
| Body weight (kg) | 58.99 ± 2.37 | 60.35 ± 2.23 | 0.061 a | 54.86 ± 2.17 | 54.45 ± 2.15 | 0.466 a |
| BMI (kg/m2) | 25.73 ± 1.27 | 26.24 ± 1.15 | 0.095 a | 23.80 ± 0.74 | 23.54 ± 0.69 | 0.269 a |
| Body fat (%) | 28.21 ± 1.59 | 32.06 ± 1.54 | <0.001 *a | 26.11 ± 1.27 | 26.09 ± 1.18 | 0.989 a |
| Visceral fat (%) | 12.05 ± 0.93 | 12.45 ± 0.87 | 0.119 a | 10.00 ± 0.67 | 9.40 ± 0.50 | 0.244 a |
| BUN (mg/dL) | 18.44 ± 2.07 | 17.65 ± 2.34 | 0.412 b | 14.69 ± 0.86 | 13.15 ± 0.56 | 0.083 b |
| Creatinine (mg/dL) | 1.45 ± 0.13 | 1.25 ± 0.12 | 0.226 a | 1.28 ± 0.06 | 1.04 ± 0.05 | <0.001 *a |
| AST (IU/L) | 21.89 ± 1.91 | 24.61 ± 2.85 | 0.230 b | 24.65 ± 3.12 | 23.45 ± 2.40 | 0.561 b |
| ALT (IU/L) | 22.11 ± 2.93 | 29.61 ± 10.08 | 0.391 b | 21.60 ± 3.30 | 20.35 ± 2.76 | 0.464 b |
| ALP (IU/L) | 102.17 ± 4.82 | 90.61 ± 10.87 | 0.003 *b | 98.67 ± 5.84 | 81.56 ± 3.49 | 0.001 *b |
| FBS (mg/dL) | 142.40 ± 9.86 | 153.65 ± 9.69 | 0.114 a | 148.25 ± 10.74 | 141.65 ± 8.84 | 0.381 a |
| HbA1c (%) | 6.85 ± 0.37 | 7.61 ± 0.47 | 0.016 *a | 7.44 ± 0.49 | 7.34 ± 0.55 | 0.732 a |
| TC (mg/dL) | 192.85 ± 9.69 | 195.50 ± 9.21 | 0.770 a | 196.40 ± 8.20 | 192.10 ± 9.97 | 0.467 a |
| TG (mg/dL) | 158.80 ± 18.38 | 183.30 ± 19.56 | 0.065 b | 156.25 ± 16.51 | 157.95 ± 13.82 | 0.896 b |
| HDL (mg/dL) | 57.85 ± 2.55 | 65.85 ± 4.14 | 0.026 *a | 63.75 ± 4.40 | 77.00 ± 5.25 | 0.027 *a |
| LDL (mg/dL) | 102.94 ± 7.14 | 104.97 ± 9.61 | 0.601 b | 113.61 ± 10.04 | 95.64 ± 10.61 | 0.009 *b |
| Variables | Difference | p-Value | |
|---|---|---|---|
| Placebo (n = 20) | Treatment (n = 20) | ||
| Body weight (kg) | 1.36 ± 0.68 | −0.41 ± 0.55 | 0.023 * |
| BMI (kg/m2) | 0.51 ± 0.29 | −0.26 ± 0.23 | 0.060 |
| Body fat (%) | 3.86 ± 0.59 | −0.01 ± 1.05 | 0.004 * |
| Visceral fat (%) | 0.40 ± 0.24 | −0.60 ± 0.50 | 0.080 |
| BUN (mg/dL) | −0.80 ± 1.56 | −1.55 ± 0.81 | 0.735 |
| Creatinine (mg/dL) | −0.20 ± 0.16 | −0.25 ± 0.04 | 0.715 |
| AST (IU/L) | 2.72 ± 1.94 | −1.20 ± 2.75 | 0.628 |
| ALT (IU/L) | 7.50 ± 10.25 | −1.25 ± 2.70 | 0.326 |
| ALP (IU/L) | −11.56 ± 11.34 | −17.11 ± 3.37 | 0.635 |
| FBS (mg/dL) | 11.25 ± 6.79 | −6.60 ± 7.36 | 0.044 * |
| HbA1c (%) | 0.76 ± 0.28 | −0.10 ± 0.29 | 0.023 * |
| TC (mg/dL) | 2.65 ± 8.94 | −4.30 ± 5.79 | 0.351 |
| TG (mg/dL) | 24.50 ± 12.82 | 1.70 ± 21.00 | 0.449 |
| HDL (mg/dL) | 8.00 ± 3.32 | 13.25 ± 5.53 | 0.808 |
| LDL (mg/dL) | 2.03 ± 11.69 | −17.97 ± 5.88 | 0.337 |
| Parameters | Coefficient | 95% Confidence Interval | p-Value |
|---|---|---|---|
| Body weight (kg) | −2.52 | −6.50 to 1.46 | 0.198 |
| BMI (kg/m2) | −1.08 | −2.71 to 0.55 | 0.178 |
| Body fat (%) | −3.32 | −7.19 to 0.55 | 0.088 |
| Visceral fat (%) | −1.62 | −2.92 to 0.31 | 0.018 * |
| BUN (mg/dL) | −3.08 | −7.60 to 1.45 | 0.171 |
| Creatinine (mg/dL) | −0.12 | −0.33 to 0.08 | 0.218 |
| AST (IU/L) | −1.82 | −8.01 to 4.37 | 0.553 |
| ALT (IU/L) | −6.48 | −27.69 to 14.73 | 0.538 |
| ALP (IU/L) | −4.88 | −29.86 to 20.09 | 0.692 |
| FBS (mg/dL) | −22.95 | −42.25 to −3.66 | 0.021 * |
| HbA1c (%) | −1.35 | −2.43 to −0.28 | 0.016 * |
| TC (mg/dL) | −28.89 | −56.61 to −1.17 | 0.042 * |
| TG (mg/dL) | −46.44 | −101.33 to 8.46 | 0.094 |
| HDL (mg/dL) | 8.61 | −4.71 to 21.92 | 0.198 |
| LDL (mg/dL) | −28.84 | −56.48 to −1.20 | 0.041 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaiyasut, C.; Sivamaruthi, B.S.; Lailerd, N.; Sirilun, S.; Thangaleela, S.; Khongtan, S.; Bharathi, M.; Kesika, P.; Saelee, M.; Choeisoongnern, T.; et al. Influence of Bifidobacterium breve on the Glycaemic Control, Lipid Profile and Microbiome of Type 2 Diabetic Subjects: A Preliminary Randomized Clinical Trial. Pharmaceuticals 2023, 16, 695. https://doi.org/10.3390/ph16050695
Chaiyasut C, Sivamaruthi BS, Lailerd N, Sirilun S, Thangaleela S, Khongtan S, Bharathi M, Kesika P, Saelee M, Choeisoongnern T, et al. Influence of Bifidobacterium breve on the Glycaemic Control, Lipid Profile and Microbiome of Type 2 Diabetic Subjects: A Preliminary Randomized Clinical Trial. Pharmaceuticals. 2023; 16(5):695. https://doi.org/10.3390/ph16050695
Chicago/Turabian StyleChaiyasut, Chaiyavat, Bhagavathi Sundaram Sivamaruthi, Narissara Lailerd, Sasithorn Sirilun, Subramanian Thangaleela, Suchanat Khongtan, Muruganantham Bharathi, Periyanaina Kesika, Manee Saelee, Thiwanya Choeisoongnern, and et al. 2023. "Influence of Bifidobacterium breve on the Glycaemic Control, Lipid Profile and Microbiome of Type 2 Diabetic Subjects: A Preliminary Randomized Clinical Trial" Pharmaceuticals 16, no. 5: 695. https://doi.org/10.3390/ph16050695
APA StyleChaiyasut, C., Sivamaruthi, B. S., Lailerd, N., Sirilun, S., Thangaleela, S., Khongtan, S., Bharathi, M., Kesika, P., Saelee, M., Choeisoongnern, T., Fukngoen, P., Peerajan, S., & Sittiprapaporn, P. (2023). Influence of Bifidobacterium breve on the Glycaemic Control, Lipid Profile and Microbiome of Type 2 Diabetic Subjects: A Preliminary Randomized Clinical Trial. Pharmaceuticals, 16(5), 695. https://doi.org/10.3390/ph16050695










