Discovery of Small-Molecule Antagonists of Orexin 1/2 Receptors from Traditional Chinese Medicinal Plants with a Hypnotic Effect
Abstract
1. Introduction
2. Results
2.1. In-Home Ligand Library
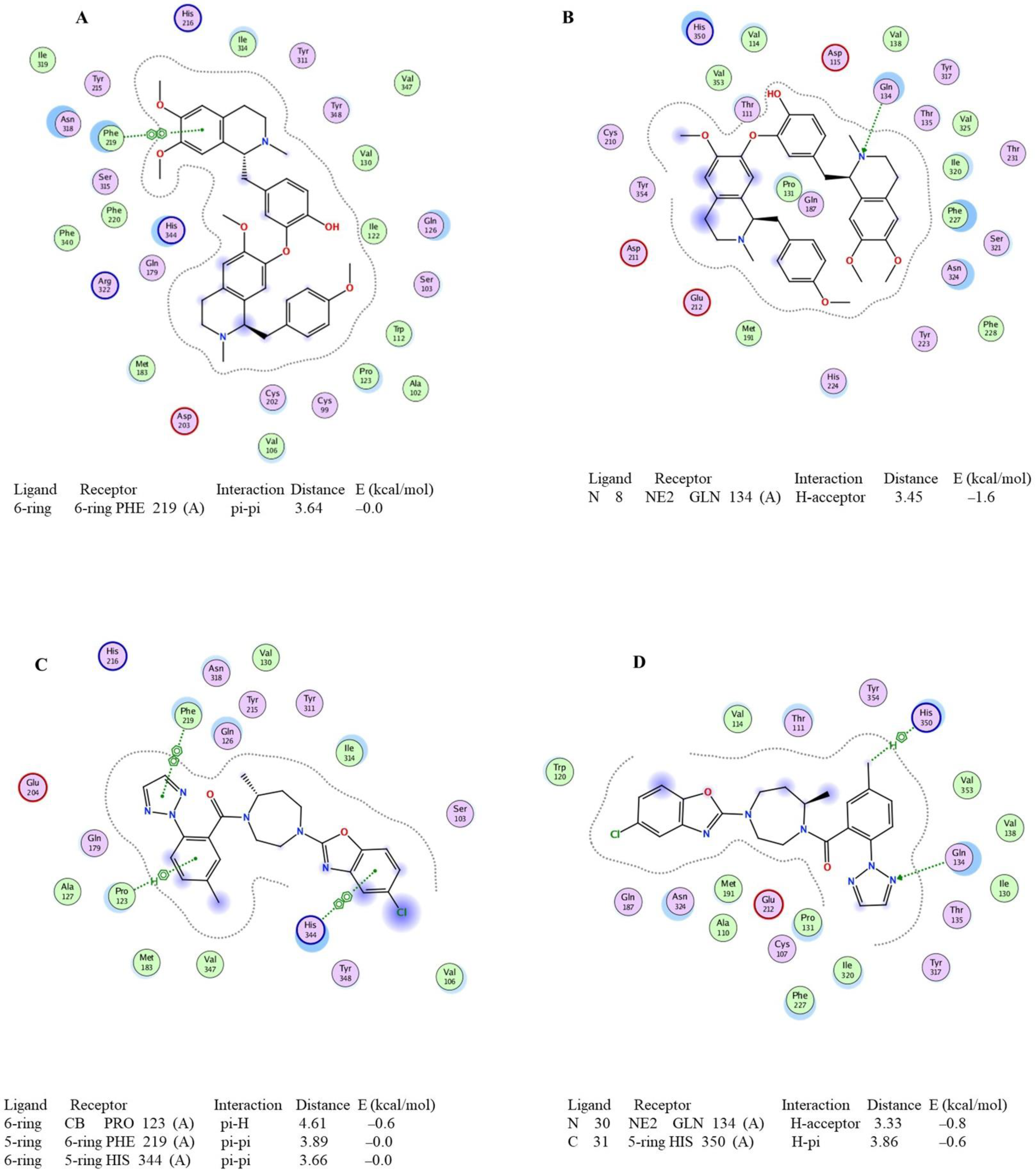
2.2. Molecular-Docking-Based Virtual Screening of Orexin Receptor Antagonists from the In-Home Ligand Library
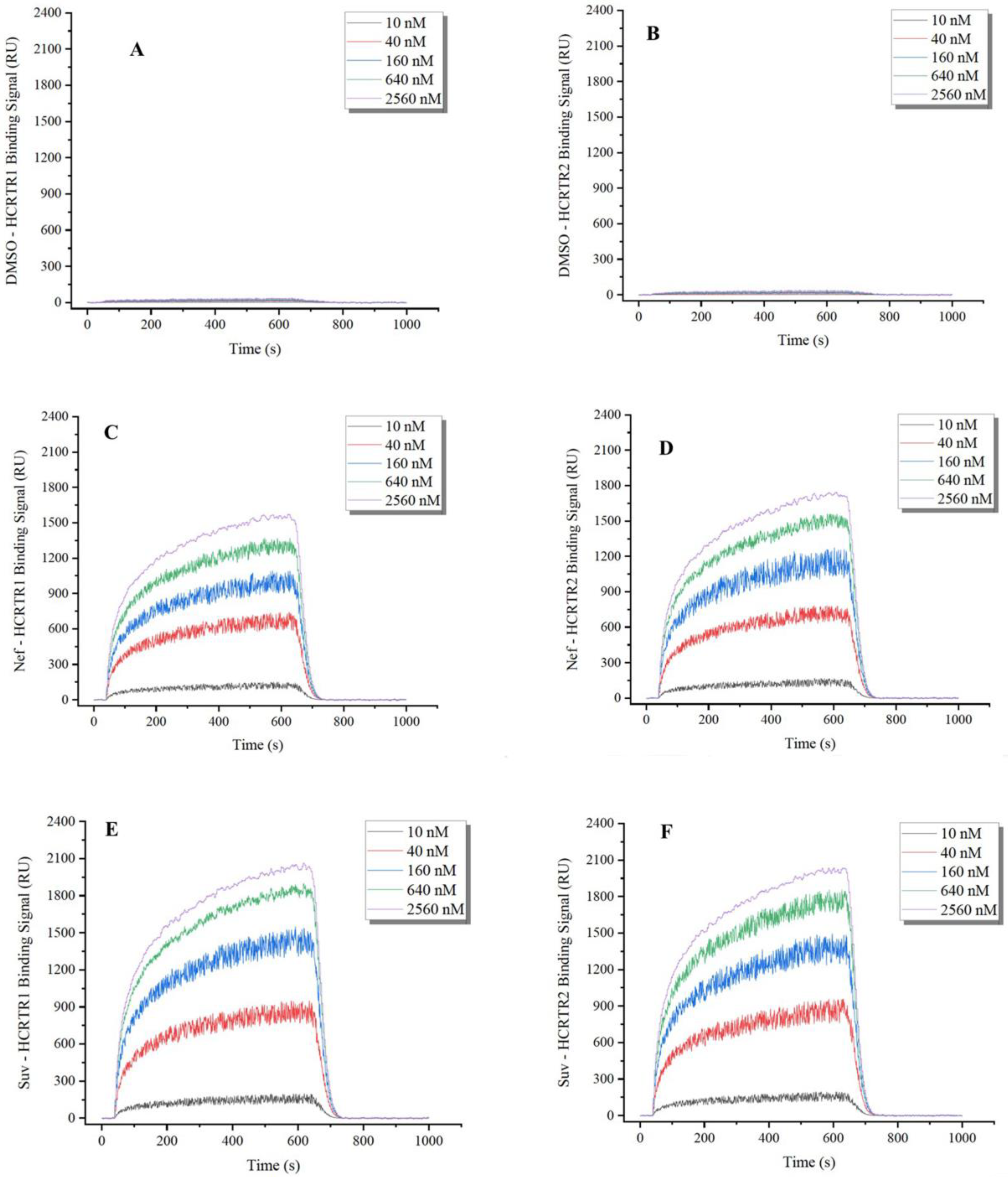
2.3. SPR Analysis of the Binding Affinity between Bisbenzylisoquinoline Alkaloids and OX1R and OX2R
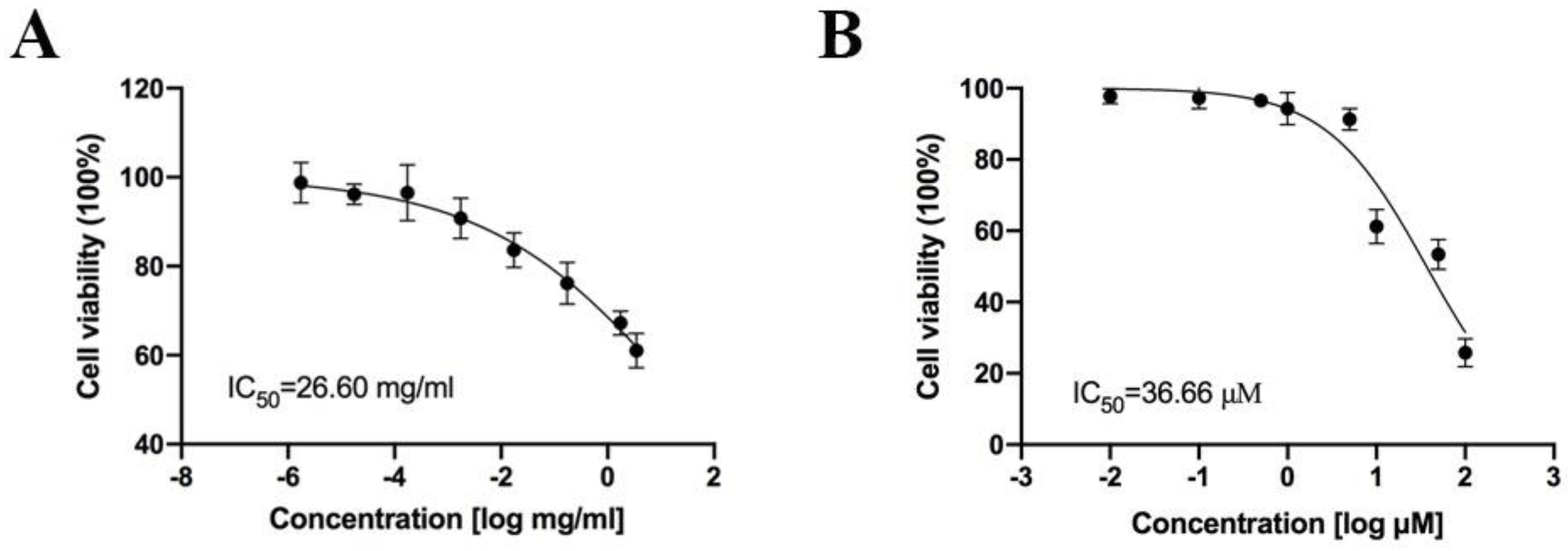
2.4. Inhibitory Activity of Neferine on OX1R and OX2R
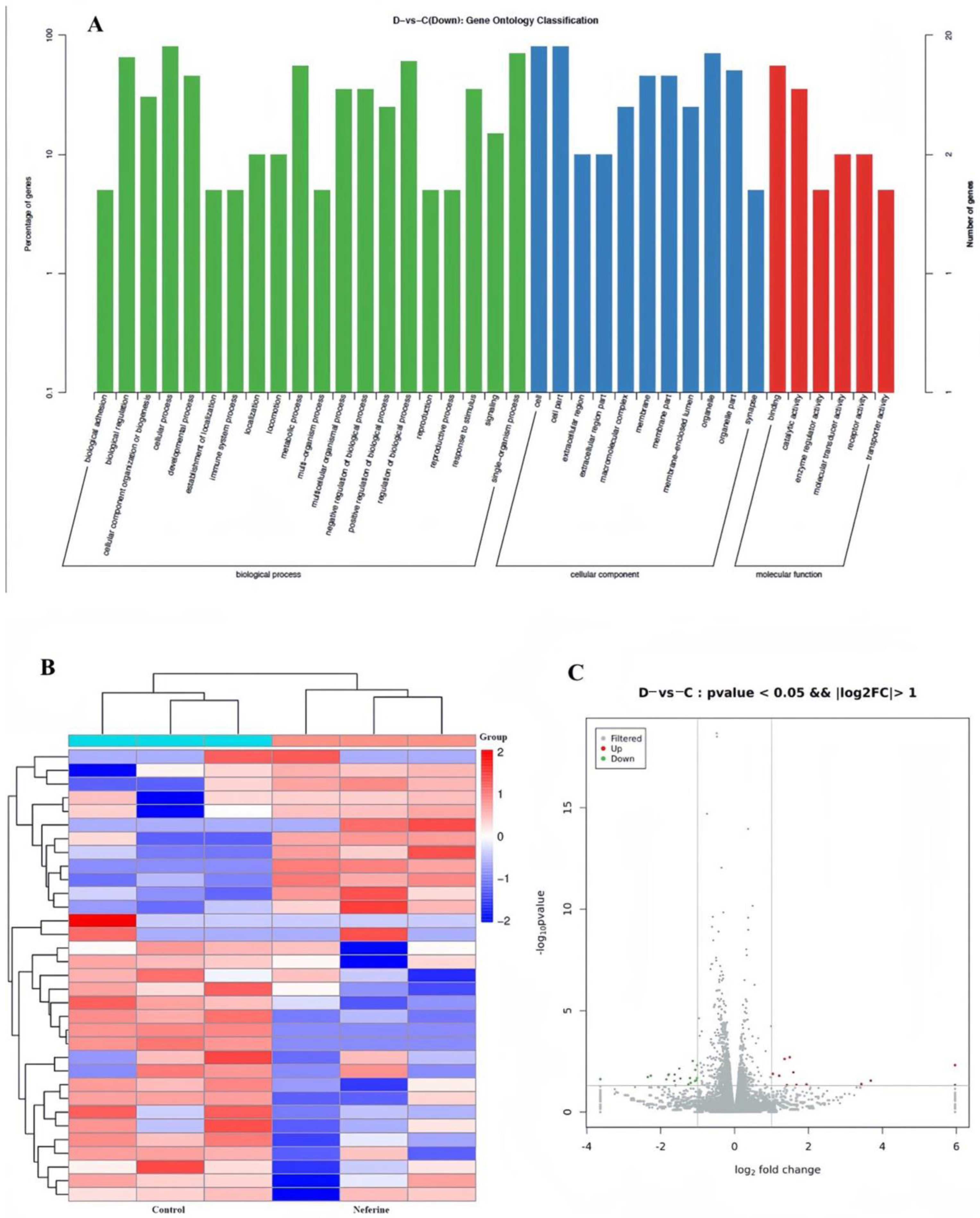
2.4.1. Systemically Exploring the Candidate Gene Mechanism of Neferine through RNA Sequencing
2.4.2. Quantitative Reverse Transcription–Polymerase Chain Reaction and Western Blotting of the Orexin Receptor Antagonist Neferine
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Chemicals, and Reagents
4.2. Establishment of an In-Home Ligand Library
4.3. Virtual Screening Using Molecular Docking Technology
4.4. Surface Plasmon Resonance Assay
4.5. Cell Lines and Cell Culture
4.6. Cell Counting Kit-8 Assay
4.7. Quantitative Reverse Transcription and Real-Time Polymerase Chain Reaction
4.8. Western Blotting
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perez, M.-N.; Salas, R.-M.-E. Insomnia. Contin. Minneap. Minn. 2020, 26, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Steinberg, J.; Patel, P. Insomnia in the Elderly: A Review. J. Clin. Sleep Med. 2018, 14, 1017–1024. [Google Scholar] [CrossRef]
- Crowley, K. Sleep and Sleep Disorders in Older Adults. Neuropsychol. Rev. 2011, 21, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Brownlow, J.A.; Miller, K.E.; Gehrman, P.R. Insomnia and Cognitive Performance. Sleep Med. Clin. 2020, 15, 71–76. [Google Scholar] [CrossRef]
- Ileri, I.; Borazan, F.Y.; Cavusoglu, C.; Göker, B. The relationship between the severity of insomnia and falls in the elderly. Psychogeriatrics 2022, 22, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Jaussent, I.; Bouyer, J.; Ancelin, M.-L.; Akbaraly, T.; Peres, K.; Ritchie, K.; Besset, A.; Dauvilliers, Y. Insomnia and Daytime Sleepiness Are Risk Factors for Depressive Symptoms in the Elderly. Sleep 2011, 34, 1103–1110. [Google Scholar] [CrossRef]
- Schneider, F.; Brose, U. Beyond diversity: How nested predator effects control ecosystem functions. J. Anim. Ecol. 2013, 82, 64–71. [Google Scholar] [CrossRef]
- Seppi, K.; Ray Chaudhuri, K.; Coelho, M.; Fox, S.-H.; Katzenschlager, R.; Perez Lloret, S.; Weintraub, D.-C.; Sampaio, C.-C. The collaborators of the Parkinson’s Disease Update on Non-Motor Symptoms Study Group on behalf of the Movement Disorders Society Evidence-Based Medicine, Update on treatments for nonmotor symptoms of Parkinson’s disease—An evidence-based medicine review. Mov. Disord. 2019, 34, 180–198. [Google Scholar] [CrossRef]
- Rosenberg, R.; Citrome, L.; Drake, C.L. Advances in the Treatment of Chronic Insomnia: A Narrative Review of New Nonpharmacologic and Pharmacologic Therapies. Neuropsychiatr. Dis. Treat. 2021, 17, 2549–2566. [Google Scholar] [CrossRef]
- Sateia, M.J.; Buysse, D.J.; Krystal, A.D.; Neubauer, D.N.; Heald, J.L. Clinical Practice Guideline for the Pharmacologic Treatment of Chronic Insomnia in Adults: An American Academy of Sleep Medicine Clinical Practice Guideline. J. Clin. Sleep Med. 2017, 13, 307–349. [Google Scholar] [CrossRef]
- Riemann, D.; Nissen, C.; Palagini, L.; Otte, A.; Perlis, M.-L.; Spiegelhalder, K. The neurobiology, investigation, and treatment of chronic insomnia. Lancet Neurol. 2015, 14, 547–558. [Google Scholar] [CrossRef]
- Equihua, A.C.; De La Herrán-Arita, A.K.; Edrucker-Colin, R. Orexin receptor antagonists as therapeutic agents for insomnia. Front. Pharmacol. 2013, 4, 163. [Google Scholar] [CrossRef]
- Lader, M. Benzodiazepine harm: How can it be reduced? Br. J. Clin. Pharmacol. 2014, 77, 295–301. [Google Scholar] [CrossRef]
- Gunja, N. In the Zzz Zone: The Effects of Z-Drugs on Human Performance and Driving. J. Med. Toxicol. 2013, 9, 163–171. [Google Scholar] [CrossRef]
- Roch, C.; Bergamini, G.; Steiner, M.A.; Clozel, M. Nonclinical pharmacology of daridorexant: A new dual orexin receptor antagonist for the treatment of insomnia. Psychopharmacology 2021, 238, 2693–2708. [Google Scholar] [CrossRef] [PubMed]
- de Lecea, L.; Kilduff, T.S.; Peyron, C.; Gao, X.-B.; Foye, P.E.; Danielson, P.E.; Fukuhara, C.; Battenberg, E.L.F.; Gautvik, V.T.; Bartlett, F.S., III; et al. The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. USA 1998, 95, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Amemiya, A.; Ishii, M.; Matsuzaki, I.; Chemelli, R.M.; Tanaka, H.; Williams, S.C.; Richardson, J.A.; Kozlowski, G.P.; Wilson, S.; et al. Orexins and Orexin Receptors: A Family of Hypothalamic Neuropeptides and G Protein-Coupled Receptors that Regulate Feeding Behavior. Cell 1998, 92, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yuan, K.; Zheng, Y.; Lu, L. Orexin Receptor Antagonists as Emerging Treatments for Psychiatric Disorders. Neurosci. Bull. 2020, 36, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Pavone, L.M.; Assisi, L.; Langella, E.; Tafuri, S.; Mirabella, N.; Costagliola, A.; Vittoria, A. Expression of orexin B and its receptor 2 in rat testis. Gen. Comp. Endocrinol. 2017, 242, 66–73. [Google Scholar] [CrossRef]
- Huber, M.J.; Chen, Q.-H.; Shan, Z. The Orexin System and Hypertension. Cell. Mol. Neurobiol. 2018, 38, 385–391. [Google Scholar] [CrossRef]
- Barnett, S.; Li, A. Orexin in Respiratory and Autonomic Regulation, Health and Diseases. Compr. Physiol. 2020, 10, 345–363. [Google Scholar] [CrossRef]
- Parekh, R.U.; White, A.; Leffler, K.E.; Biancardi, V.C.; Eells, J.B.; Abdel-Rahman, A.A.; Sriramula, S. Hypothalamic kinin B1 receptor mediates orexin system hyperactivity in neurogenic hypertension. Sci. Rep. 2021, 11, 21050. [Google Scholar] [CrossRef] [PubMed]
- Muehlan, C.; Vaillant, C.; Zenklusen, I.; Kraehenbuehl, S.; Dingemanse, J. Clinical pharmacology, efficacy, and safety of orexin receptor antagonists for the treatment of insomnia disorders. Expert Opin. Drug Metab. Toxicol. 2020, 16, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Vermeeren, A.; Sun, H.; Vuurman, E.F.; Jongen, S.; Van Leeuwen, C.J.; Van Oers, A.C.; Palcza, J.; Li, X.; Laethem, T.; Heirman, I.; et al. On-the-Road Driving Performance the Morning after Bedtime Use of Suvorexant 20 and 40 mg: A Study in Non-Elderly Healthy Volunteers. Sleep 2015, 38, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Palcza, J.; Card, D.; Gipson, A.; Rosenberg, R.; Kryger, M.; Lines, C.; Wagner, J.A.; Troyer, M.D. Effects of Suvorexant, an Orexin Receptor Antagonist, on Respiration during Sleep in Patients with Obstructive Sleep Apnea. J. Clin. Sleep Med. 2016, 12, 9–17. [Google Scholar] [CrossRef]
- Kong, Y.; Hao, M.; Chen, A.; Yi, T.; Yang, K.; Li, P.; Wang, Y.; Li, P.; Jia, X.; Qin, H.; et al. SymMap database and TMNP algorithm reveal Huanggui Tongqiao granules for Allergic rhinitis through IFN-mediated neuroimmuno-modulation. Pharmacol. Res. 2022, 185, 106483. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Furutani, S.; Itoh, A.; Tanahashi, T.; Nakajima, H.; Oshiro, H.; Sun, S.; Yamada, J. Effects of extracts and neferine from the embryo of Nelumbo nucifera seeds on the central nervous system. Phytomedicine 2008, 15, 1117–1124. [Google Scholar] [CrossRef]
- Jo, K.; Kim, S.; Hong, K.-B.; Suh, H.J. Nelumbo nucifera promotes non-rapid eye movement sleep by regulating GABAergic receptors in rat model. J. Ethnopharmacol. 2021, 267, 113511. [Google Scholar] [CrossRef]
- Wang, T.; Huang, H.; Zhang, Y.; Li, X.; Li, H.; Jiang, Q.; Gao, W. Role of effective composition on antioxidant, anti-inflammatory, sedative-hypnotic capacities of 6 common edible Lilium varieties. J. Food. Sci. 2015, 80, H857–H868. [Google Scholar] [CrossRef]
- Hua, Y.; Guo, S.; Xie, H.; Zhu, Y.; Yan, H.; Tao, W.-W.; Shang, E.-X.; Qian, D.-W.; Duan, J.-A. Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H. F. Chou Seed Ameliorates Insomnia in Rats by Regulating Metabolomics and Intestinal Flora Composition. Front. Pharmacol. 2021, 12, 653767. [Google Scholar] [CrossRef]
- Jiang, J.-G.; Huang, X.-J.; Chen, J. Separation and purification of saponins from Semen Ziziphus jujuba and their sedative and hypnotic effects. J. Pharm. Pharmacol. 2007, 59, 1175–1180. [Google Scholar] [CrossRef]
- Jiang, J.-G.; Huang, X.-J.; Chen, J.; Lin, Q.-S. Comparison of the sedative and hypnotic effects of flavonoids, saponins, and polysaccharides extracted from Semen Ziziphus jujube. Nat. Prod. Res. 2007, 21, 310–320. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, D.; Chen, Y.; Jiang, R.; Xu, H.; Qiu, Z.; Liu, D.; Luo, H. Metabonomics Study of Ginseng Glycoproteins on Improving Sleep Quality in Mice. BioMed. Res. Int. 2019, 2019, 2561828. [Google Scholar] [CrossRef]
- Attele, A.S.; Wu, J.A.; Yuan, C.-S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem. Pharmacol. 1999, 58, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Sachidanandan, D.; Reddy, H.-P.; Mani, A.; Hyde, G.-J.; Bera, A.-K. The Neuropeptide Orexin-A Inhibits the GABA(A) Receptor by PKC and Ca(2+)/CaMKII-Dependent Phosphorylation of Its beta(1) Subunit. J. Mol. Neurosci. 2017, 61, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Asokan, S.M.; Mariappan, R.; Muthusamy, S.; Velmurugan, B.K. Pharmacological benefits of neferine—A comprehensive review. Life Sci. 2018, 199, 60–70. [Google Scholar] [CrossRef]
- Chen, J.; Tang, M.; Liu, M.; Jiang, Y.; Liu, B.; Liu, S. Neferine and lianzixin extracts have protective effects on undifferentiated caffeine-damaged PC12 cells. BMC Complement. Med. Ther. 2020, 20, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, R.; Li, J.; Huang, Q.; Liu, S.; He, J. Pyrrole-2-carbaldehydes with neuroprotective activities from Moringa oleifera seeds. Phytochemistry 2022, 204, 113451. [Google Scholar] [CrossRef]
- Luo, L.-M.; Qian, L.; Pei, G.; Huang, S.-G.; Zhou, X.-J.; Chen, N.-H. Advances in studies on steroidal saponins and their pharmacological activities in genus Lilium. Chin. J. Chin. Mater. Med. 2018, 43, 1416–1426. [Google Scholar]
- Liu, P.; Lin, Z.-J.; Zhang, B. Research progress on chemical constituents and pharmacological effect of Lilii Bulbus. Chin. J. Exp. Trad. Med. Form. 2017, 23, 201–211. [Google Scholar]
- Zhou, J.; An, R.-F.; Huang, X.-F. Genus Lilium: A review on traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2021, 270, 113852. [Google Scholar] [CrossRef]
- Lu Liu, L.; Xu, F.-R.; Wang, Y.-Z. Traditional uses, chemical diversity and biological activities of Panax L. (Araliaceae): A review. J. Ethnopharmacol. 2020, 263, 112792. [Google Scholar] [CrossRef]
- Liu, Y.-G.; Zhang, H.; Dai, X.; Zhu, R.-Y.; Chen, B.-B.; Xia, B.-K.; Ye, Z.-M.-W.; Zhao, D.-D.; Gao, S.-H.; Orekhov, A.-N.; et al. A comprehensive review on the phytochemistry, pharmacokinetics, and antidiabetic effect of Ginseng. Phytomedicine 2021, 92, 153717. [Google Scholar] [CrossRef]
- Shergis, J.-L.; Ni, X.-J.; Sarris, J.; Zhang, A.-L.; Guo, X.-F.; Xue, C.-C.; Lu, C.-J.; Hugel, H. Ziziphus spinosa seeds for insomnia: A review of chemistry and psychopharmacology. Phytomedicine 2017, 34, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Xu, X.-X.; Guo, S.; Xie, H.; Yan, H.; Ma, X.-F.; Niu, Y.; Duan, J.-A. Wild Jujube (Ziziphus jujuba var. spinosa): A review of its phytonutrients, health benefits, metabolism, and applications. J. Agric. Food Chem. 2022, 70, 7871–7886. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Bao, T.; Mo, J.-J.; Ni, J.-D.; Chen, W. Research advances in bioactive components and health benefits of jujube (Ziziphus jujuba Mill.) fruit. J. Zhejiang Univ. Sci. B Biomed. Biotechnol. 2021, 22, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.-B. Study on Triterpenoids from Bamboo Bark; Zhengjiang Unverisity: Hangzhou, China, 2004. [Google Scholar]
- Zhou, B.Q.; Gao, X.M.; Yang, Y.; Wang, L.-C.; Zhang, W.; Zhao., X.-L.; Di, L.-Q. Quality analysis of Caulis Bambusae in taeniam from different origins by HPLC coupled with chemometrics. Chin. Trad. Herb. Drugs 2022, 53, 853–855. [Google Scholar]
- Zhao, S.-M.; Chou, G.-X.; Wang, Z.-T. Chemical constituents from roots and rhizomes of Physochlaina infundibularis. Chin. Trad. Herb. Drugs 2013, 44, 938–941. [Google Scholar]
- Lou, Y.; Xu, R.-Z.; Zhang, L.-P.; Xie, Z.-S.; Zhang, Z.-Q.; Xv, J.-Y.; Li, J.; Wang, P. Two new amides from Physochlaina infundibularis. Chin. Trad. Herb. Drugs 2021, 52, 331–334. [Google Scholar]
- Yang, L.; Li, D.-H. Research progress on chemical composition, pharmacological activity and toxicology of acacia peel. Chin. J. Surg. Integr. Trad. West. Med. 2019, 25, 1061–1064. [Google Scholar]
- Li, W.; Yang, H.-J. Isolation and identification of lignans and other phenolic constituents from the stem bark of Albizia julibrissin Durazz and evaluation of their nitric oxide inhibitory activity. Molecules 2020, 25, 2065. [Google Scholar] [CrossRef]
- Li, R.; Tian, J.-F.; Luo, X.-J.; Li, R.M. Research progress on chemical components and pharmacological effects of the flowers of Albizia julibrissin Durazz. Tianjin Pharm. 2022, 34, 66–71. [Google Scholar]
- Yao, X.-M.; Zhou, X.-J.; Zhou, Y.-Y.; Yu, M.; Guan, H.-B. Research progress on chemical composition and pharmacological effects of Yuanzhi. Acta Chin. Med. Pharmacol. 2022, 50, 103–107. [Google Scholar]
- Li, X.-R.; Chen, S.-M.; Chen, W.-Y.; Song, J.-M.; Zhang, Y.-Y. Research progress on the chemical composition of polygala tenuifolia and prevention and treatment of Alzheimer’s disease. Chin. Pharmaceut. J. 2022, 57, 15–23. [Google Scholar]
- Wang, Y.; Zhang, X. Polygalae Radix: A review of its traditional uses, phytochemistry, pharmacology, toxicology, and pharmacokinetics. Fitoterapia 2020, 147, 104759. [Google Scholar]
- Feng, N.; Yue, Y.-W.; Chen, C.-L.; Yang, M.; Wang, C.; Zhang, J.-S. Research progress of triterpenes from mycelia of Ganoderma lingzhi and its pharmacological effects. Mycosystema 2022, 41, 1341–1353. [Google Scholar]
- Huang, J.; Wang, H.-J.; Wu, Q.; Wang, X.-L.; Liu, G.-Q. Research progress and prospect of active proteins and peptides of Ganoderma. J. Fungal Res. 2022, 20, 79–86. [Google Scholar]
- Luo, Y.; Chen, L.; Zhang, X.-L.; Lan, J.; Wei, F.; Sun, X.B. Research progress on pharmacological activity of Ganoderma lucidum triterpenoids. Chin. Pharmacol. Bull. 2021, 37, 1185–1188. [Google Scholar]
- Gong, T.; Yan, R.; Kang, J.; Chen, R. Chemical components of ganoderma. Adv. Exp. Med. Biol. 2019, 1181, 59–106. [Google Scholar]
- Wang, L.; Li, J.-Q.; Zhang, J.; Li, Z.-M.; Liu, H.-G.; Wang, Y.-Z. Traditional uses, chemical components and pharmacological activities of the genus Ganoderma P. Karst.: A review. RSC Adv. 2020, 10, 42084–42097. [Google Scholar] [CrossRef]
- Gao, L.-Y.; Li, W.-H.; Kou, N.; Shen, F.-L.; Li, J. Research progress on chemical composition and pharmacological effects of eleuthero. Inform. Trad. Chin. Med. 2019, 36, 113–116. [Google Scholar]
- Jia, A.; Zhang, Y.; Gao, H.; Zhang, Z.; Zhang, Y.; Wang, Z.; Zhang, J.; Deng, B.; Qiu, Z.; Fu, C. A review of acanthopanax senticosus (Rupr and Maxim.) harms: From ethnopharmacological use to modern application. J. Ethnopharmacol. 2021, 268, 113586. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-L.; Liu, Q.-T.; Zhang, J.-S.; Li, Q.-S. Research progress on chemical composition and pharmacological effects of Apocynum venetum L. leaves. Lishizhen Med. Mater. Med. Res. 2022, 33, 2739–2742. [Google Scholar]
- Chou, C.-L.; Kong, L.-F. GC-MS analyzes the chemical composition of the volatile oil of Apocynum venetum L. leaves. Chin. Trad. Patent Med. 2011, 33, 338–340. [Google Scholar]
- Xie, W.; Zhang, X.; Wang, T.; Hu, J. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): A review. J. Ethnopharmacol. 2012, 141, 1–8. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, Y.-F. Advances in chemical constituents and pharmacological activities of arboruitae seed. Chem. Res. 2019, 30, 429–433. [Google Scholar]
- Ren, J.; Liao, L.; Shang, S.; Zheng, Y.; Sha, W.; Yuan, E. Purification, characterization, and bioactivities of polyphenols from Platycladus orientalis (L.) Franco. J. Food Sci. 2019, 84, 667–677. [Google Scholar] [CrossRef]
- Ren, X.-Y.; Ye, Y. Labdane diterpenes from the seeds of Platycladus orientalis. J. Asian Nat. Prod. Res. 2006, 8, 677–682. [Google Scholar] [CrossRef]
- Li, M.-C.; Fu, Y.-X.; Zhang, X.-Y.; Liu, Y.; Xv, T.-R.; Hao, Q. Research progress on chemical composition and pharmacological activity of Tuber Fleece flower Stem. Yunnan J. Trad. Chin. Med. Mater. Med. 2018, 39, 81–84. [Google Scholar]
- Liang, Y.; Tian, W.-X.; Ma, X.-F. The chemical composition of Tuber Fleece flower Stem. J. Shenyang Pharm. Univ. 2009, 26, 536–538. [Google Scholar]
- Lin, L.; Ni, B.; Lin, H.; Zhang, M.; Li, X.; Yin, X.; Qu, C.; Ni, J. Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: A review. J. Ethnopharmacol. 2015, 159, 158–183. [Google Scholar] [CrossRef]
- Jiang, Y.-P.; Liu, R.; Liu, M.; Yi, L.-Z.; Liu, S. An integrated strategy to rapidly characterize non-targeted benzylisoquinoline alkaloids from Plumula nelumbinis ethanol extract using UHPLC/Q-orbitrap HRMS. Inter. J. Mass Spectrom. 2018, 432, 26–35. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Wu, J.; Li, J.; Xiao, M.; Yang, Y.; Liu, Z.; Cheng, Y. Plumula Nelumbinis: A review of traditional uses, phytochemistry, pharmacology, pharmacokinetics and safety. J. Ethnopharmacol. 2021, 266, 113429. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, Z.-Z.; Yao, Y.-X. Research progress on the determination method of chemical composition of lotus seed. Chin. Trad. Patent Med. 2020, 42, 446–451. [Google Scholar]
- Zhao, X.-L.; Dang, Y.-L. Recent advances in chemical components, extraction and pharmacological effects of Nelumbinis Plumula. Food Sci. 2018, 39, 329–336. [Google Scholar]

| No. | Compound Name | Binding Affinity (S Value) b | |
|---|---|---|---|
| Orexin 1 | Orexin 2 | ||
| Positive control | Suvorexant | −8.2193 | −8.7622 |
| 1 | Liensinine | −11.0564 | −11.3679 |
| 2 | 6-hydroxynorisoliensinine | −10.3332 | −11.0539 |
| 3 | N-norisoliensinine | −10.4403 | −11.3397 |
| 4 | Norisoliensinine | −10.5099 | −11.3855 |
| 5 | Isoliensinine | −11.2011 | −11.7965 |
| 6 | Neferine | −10.9392 | −11.6055 |
| 7 | 3‴-hydroxylisoliensine | −10.9323 | −11.2588 |
| 8 | 3‴-neferine | −11.6650 | −11.0768 |
| 9 | Nelumborine A | −10.0248 | −10.6367 |
| 10 | Nelumborine B | −10.0467 | −10.3183 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Fang, J.; Wang, Y.; Ge, C.; Liu, S.; Jiang, Y. Discovery of Small-Molecule Antagonists of Orexin 1/2 Receptors from Traditional Chinese Medicinal Plants with a Hypnotic Effect. Pharmaceuticals 2023, 16, 542. https://doi.org/10.3390/ph16040542
He J, Fang J, Wang Y, Ge C, Liu S, Jiang Y. Discovery of Small-Molecule Antagonists of Orexin 1/2 Receptors from Traditional Chinese Medicinal Plants with a Hypnotic Effect. Pharmaceuticals. 2023; 16(4):542. https://doi.org/10.3390/ph16040542
Chicago/Turabian StyleHe, Jia, Jing Fang, Yuxin Wang, Chengyu Ge, Shao Liu, and Yueping Jiang. 2023. "Discovery of Small-Molecule Antagonists of Orexin 1/2 Receptors from Traditional Chinese Medicinal Plants with a Hypnotic Effect" Pharmaceuticals 16, no. 4: 542. https://doi.org/10.3390/ph16040542
APA StyleHe, J., Fang, J., Wang, Y., Ge, C., Liu, S., & Jiang, Y. (2023). Discovery of Small-Molecule Antagonists of Orexin 1/2 Receptors from Traditional Chinese Medicinal Plants with a Hypnotic Effect. Pharmaceuticals, 16(4), 542. https://doi.org/10.3390/ph16040542







