Inhibiting Leishmania donovani Sterol Methyltransferase to Identify Lead Compounds Using Molecular Modelling
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Preparation
2.2. Binding Site Prediction
2.3. Ligand-Based Pharmacophore Virtual Screening
2.4. Retrieval and Preparation of Chemical Library for Pharmacophore-Based Screening
2.5. Pharmacophore-Based Virtual Screening of the Libraries
2.6. Validation of Pharmacophore Model and AutoDock Vina
2.6.1. Pharmacophore Model Validation
2.6.2. Validation of AutoDock Vina
2.7. Molecular Docking Studies of Chemical Entities with Good Pharmacophore Fit Scores
2.8. Characterisation of Mechanism of Binding
2.9. ADMET Properties and Drug-Likeness Assessment
2.10. Prediction of Biological Activity of Selected Compounds
2.11. Molecular Dynamics Simulation Analyses of Protein and Protein–Ligand Complexes
2.12. Parasite Culture and In Vitro Effect of Compounds on L. donovani Promastigotes
2.13. In Vitro Culture and Cell Viability Assay for Trypanosoma brucei brucei GUTat 3.1
3. Results and Discussion
3.1. Predictions of LdSMT Binding Site Residues
3.2. Pharmacophore Generation
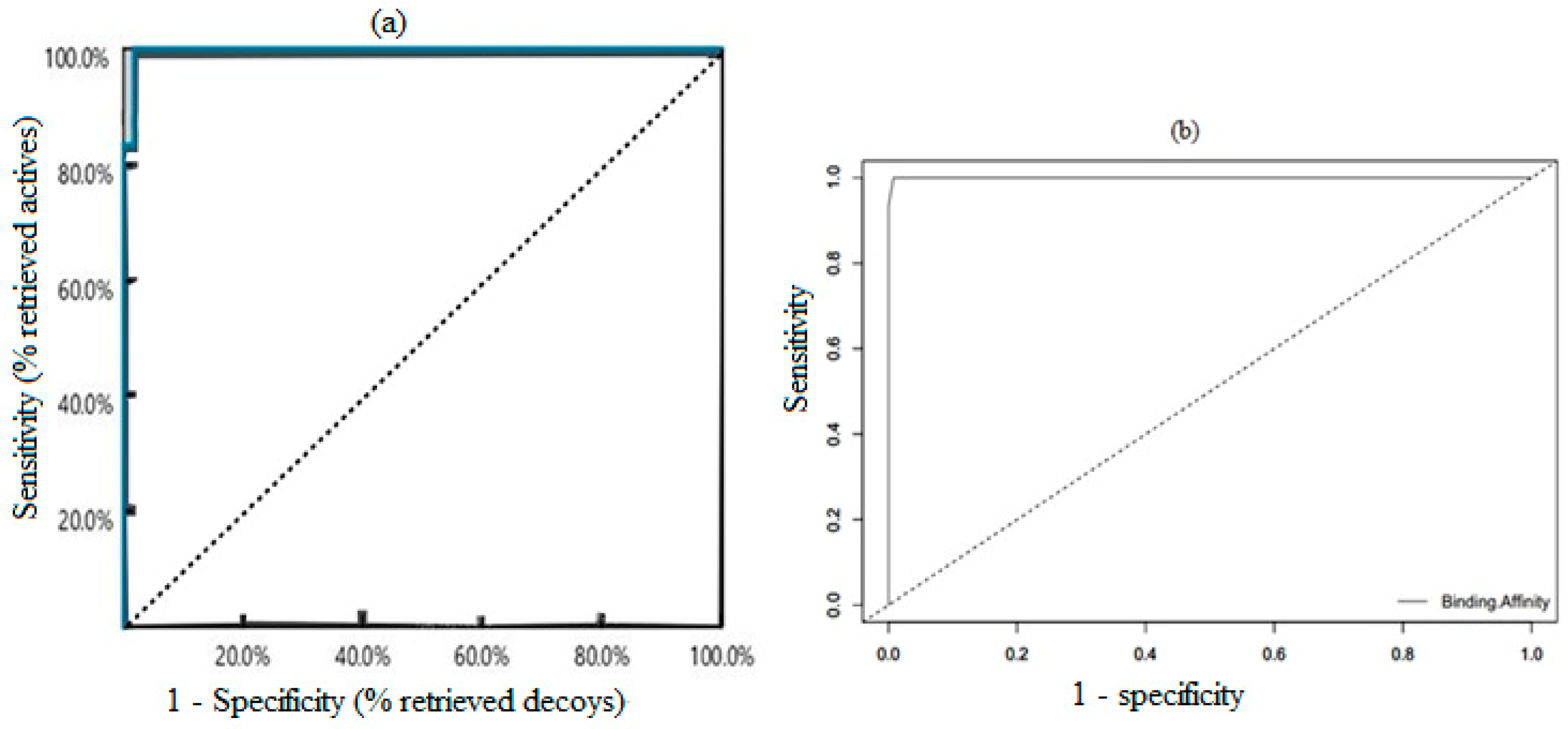
3.3. Validation of Generated Pharmacophore Model
3.4. Validation of Docking Protocol
3.5. Pharmacophore-Based Virtual Screening of Library
3.6. Molecular Docking Analysis
3.7. Protein–Ligand Interaction
3.8. ADMET Profiling
3.9. Biological Activity Predictions of Selected Hit Compounds
3.10. MD Simulations Analyses
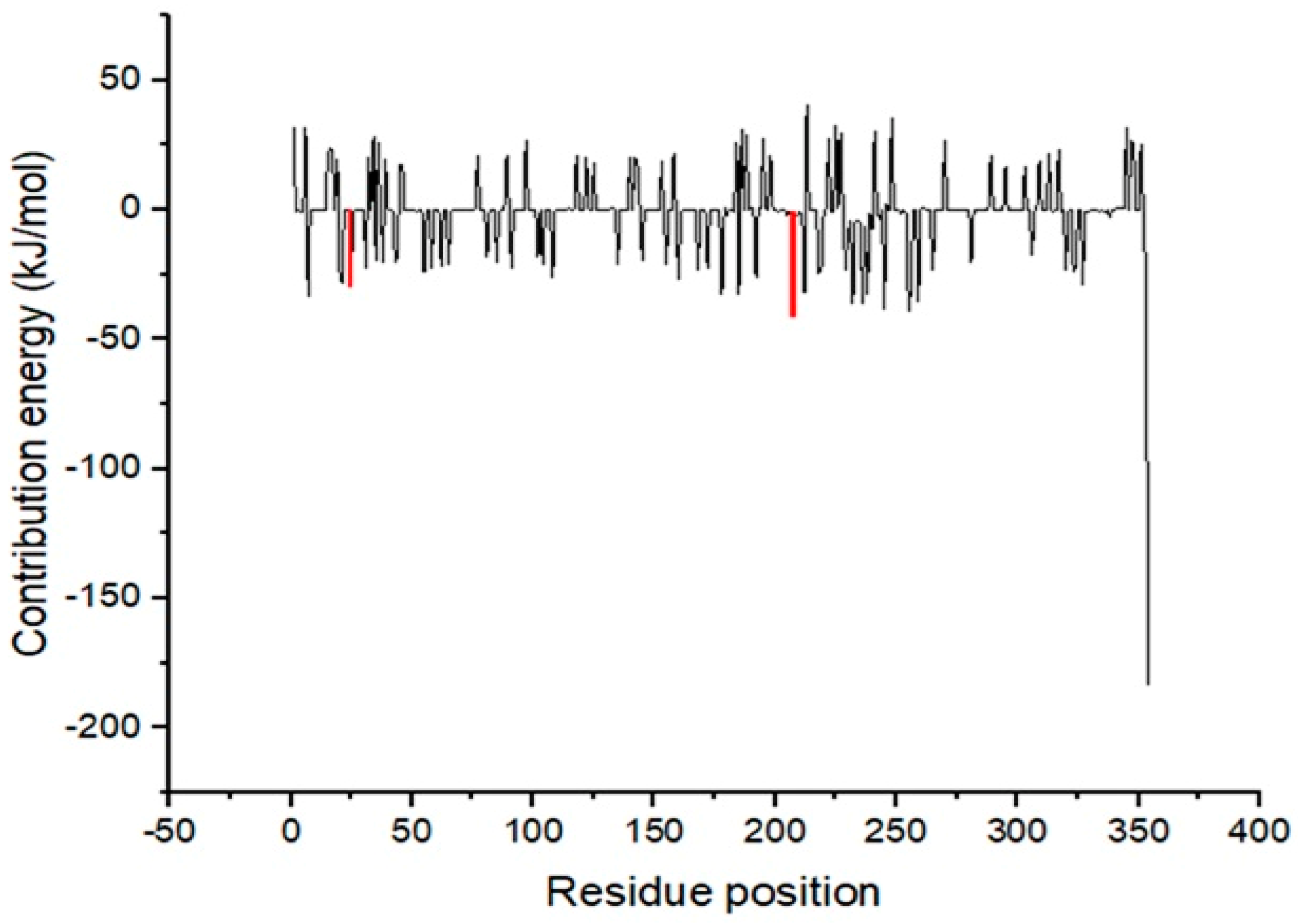
3.11. MM/PBSA Computation of Free Binding Energies
3.12. In Vitro Evaluation of Identified Hits against Leishmania donovani
4. Contribution to the Field
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Camargo, L.B.; Langoni, H. Impact of Leishmaniasis on Public Health. J. Venom. Anim. Toxins incl. Trop. Dis. 2006, 12, 527–548. [Google Scholar] [CrossRef]
- Bamorovat, M.; Sharifi, I.; Tavakoli Oliaee, R.; Jafarzadeh, A.; Khosravi, A. Determinants of Unresponsiveness to Treatment in Cutaneous Leishmaniasis: A Focus on Anthroponotic Form Due to Leishmania Tropica. Front. Microbiol. 2021, 12, 1143–1158. [Google Scholar] [CrossRef]
- Leta, S.; Dao, T.H.T.; Mesele, F.; Alemayehu, G. Visceral Leishmaniasis in Ethiopia: An Evolving Disease. PLoS Negl. Trop. Dis. 2014, 8, e3131. [Google Scholar] [CrossRef]
- Simoben, C.V.; Ntie-Kang, F.; Akone, S.H.; Sippl, W. Compounds from African Medicinal Plants with Activities Against Selected Parasitic Diseases: Schistosomiasis, Trypanosomiasis, and Leishmaniasis. Nat. Prod. Bioprospect. 2018, 8, 151–169. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Broni, E.; Dankwa, B.; Enninful, K.S.; Kwarko, G.B.; Darko, L.; Durvasula, R.; Kempaiah, P.; Rathi, B.; Miller, W.A., III; et al. Outwitting an Old Neglected Nemesis: A Review on Leveraging Integrated Data-Driven Approaches to Aid in Unraveling of Leishmanicides of Therapeutic Potential. Curr. Top. Med. Chem. 2020, 20, 349–366. [Google Scholar] [CrossRef]
- Tiwari, N.; Gedda, M.R.; Tiwari, V.K.; Singh, S.P.; Singh, R.K. Limitations of Current Therapeutic Options, Possible Drug Targets and Scope of Natural Products in Control of Leishmaniasis. Mini-Rev. Med. Chem. 2018, 18, 26–41. [Google Scholar] [CrossRef]
- Singh, O.P.; Singh, B.; Chakravarty, J.; Sundar, S. Current Challenges in Treatment Options for Visceral Leishmaniasis in India: A Public Health Perspective. Infect. Dis. Poverty 2016, 5, 19. [Google Scholar] [CrossRef]
- Sakyi, P.O.; Amewu, R.K.; Devine, R.N.O.A.; Ismaila, E.; Miller, W.A.; Kwofie, S.K. The Search for Putative Hits in Combating Leishmaniasis: The Contributions of Natural Products Over the Last Decade. Nat. Prod. Bioprospect. 2021, 11, 489–544. [Google Scholar] [CrossRef]
- Singh, N.; Mishra, B.B.; Bajpai, S.; Singh, R.K.; Tiwari, V.K. Natural Product Based Leads to Fight against Leishmaniasis. Bioorganic Med. Chem. 2014, 22, 18–45. [Google Scholar] [CrossRef]
- Ball, P. Chemistry: Why Synthesize? Nature 2015, 528, 327–329. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug Development from Marine Natural Products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef]
- Berdigaliyev, N.; Aljofan, M. An Overview of Drug Discovery and Development. Future Med. Chem. 2020, 12, 939–947. [Google Scholar] [CrossRef]
- Osorio, Y.; Travi, B.L.; Renslo, A.R.; Peniche, A.G.; Melby, P.C. Identification of Small Molecule Lead Compounds for Visceral Leishmaniasis Using a Novel Ex Vivo Splenic Explant Model System. PLoS Negl. Trop. Dis. 2011, 5, e962. [Google Scholar] [CrossRef]
- Zhang, M.Q.; Wilkinson, B. Drug Discovery beyond the ‘Rule-of-Five’. Curr. Opin. Biotechnol. 2007, 18, 478–488. [Google Scholar] [CrossRef]
- de la Torre, B.G.; Albericio, F. The Pharmaceutical Industry in 2021. An Analysis of FDA Drug Approvals from the Perspective of Molecules. Molecules 2022, 27, 1075. [Google Scholar] [CrossRef]
- Beck, H.; Härter, M.; Haß, B.; Schmeck, C.; Baerfacker, L. Small Molecules and Their Impact in Drug Discovery: A Perspective on the Occasion of the 125th Anniversary of the Bayer Chemical Research Laboratory. Drug Discov. Today 2022, 27, 1560–1574. [Google Scholar] [CrossRef]
- Chawla, B.; Madhubala, R. Drug Targets in Leishmania. J. Parasit. Dis. 2010, 34, 1–13. [Google Scholar] [CrossRef]
- Jones, N.G.; Catta-Preta, C.M.C.; Lima, A.P.C.A.; Mottram, J.C. Genetically Validated Drug Targets in Leishmania: Current Knowledge and Future Prospects. ACS Infect. Dis. 2018, 4, 467–477. [Google Scholar] [CrossRef]
- Le Pape, P. Development of New Antileishmanial Drugs—Current Knowledge and Future Prospects. J. Enzym. Inhib. Med. Chem. 2008, 23, 708–718. [Google Scholar] [CrossRef]
- Mazire, P.; Agarwal, V.; Roy, A. Road-Map of Pre-Clinical Treatment for Visceral Leishmaniasis. Drug Dev. Res. 2022, 83, 317–327. [Google Scholar] [CrossRef]
- Alcazar-Fuoli, L.; Mellado, E. Ergosterol Biosynthesis in Aspergillus Fumigatus: Its Relevance as an Antifungal Target and Role in Antifungal Drug Resistance. Front. Microbiol. 2012, 3, 439–445. [Google Scholar] [CrossRef]
- Mukherjee, S.; Xu, W.; Hsu, F.F.; Patel, J.; Huang, J.; Zhang, K. Sterol Methyltransferase Is Required for Optimal Mitochondrial Function and Virulence in Leishmania Major. Mol. Microbiol. 2019, 111, 65–81. [Google Scholar] [CrossRef]
- Rudmann, D.G. On-Target, and off-Target-Based Toxicologic Effects. Toxicol. Pathol. 2013, 41, 310–314. [Google Scholar] [CrossRef]
- Kidane, M.E.; Vanderloop, B.H.; Zhou, W.; Thomas, C.D.; Ramos, E.; Singha, U.; Chaudhuri, M.; Nes, W.D. Sterol Methyltransferase a Target for Anti-Amoeba Therapy: Towards Transition State Analog and Suicide Substrate Drug Design. J. Lipid Res. 2017, 58, 2310–2323. [Google Scholar] [CrossRef]
- Lorente, S.O.; Rodrigues, J.C.F.; Jiménez, C.J.; Joyce-Menekse, M.; Rodrigues, C.; Croft, S.L.; Yardley, V.; De Luca-Fradley, K.; Ruiz-Pérez, L.M.; Urbina, J.; et al. Novel Azasterols as Potential Agents for Treatment of Leishmaniasis and Trypanosomiasis. Antimicrob. Agents Chemother. 2004, 48, 2937–2950. [Google Scholar] [CrossRef]
- Andrade-Neto, V.V.; Pereira, T.M.; do Canto-Cavalheiro, M.; Torres-Santos, E.C. Imipramine Alters the Sterol Profile in Leishmania Amazonensis and Increases Its Sensitivity to Miconazole. Parasites Vectors 2016, 9, 183. [Google Scholar] [CrossRef]
- Torres-Santos, E.C.; Andrade-Neto, V.V.; Cunha-Júnior, E.F.; Do Canto-Cavalheiro, M.M.; Atella, G.C.; De Almeida Fernandes, T.; Costa, P.R.R. Antileishmanial Activity of Ezetimibe: Inhibition of Sterol Biosynthesis, in Vitro Synergy with Azoles, and Efficacy in Experimental Cutaneous Leishmaniasis. Antimicrob. Agents Chemother. 2016, 60, 6844–6852. [Google Scholar]
- Tabrez, S.; Rahman, F.; Ali, R.; Muhammad, F.; Alshehri, B.M.; Alaidarous, M.A.; Banawas, S.; Dukhyil, A.A.B.; Rub, A. Repurposing of FDA-Approved Drugs as Inhibitors of Sterol C-24 Methyltransferase of Leishmania Donovani to Fight against Leishmaniasis. Drug Dev. Res. 2021, 82, 1154–1161. [Google Scholar] [CrossRef]
- Sakyi, P.O.; Broni, E.; Amewu, R.K.; Miller, W.A.I.; Wilson, M.D.; Kwofie, S.K. Homology Modeling, de Novo Design of Ligands, and Molecular Docking Identify Potential Inhibitors of Leishmania Donovani 24-Sterol Methyltransferase. Front. Cell. Infect. Microbiol. 2022, 12, 859981. [Google Scholar] [CrossRef]
- Perez, J.; Fuertes, M.; Nguewa, P.; Castilla, J.; Alonso, C. Anticancer Compounds as Leishmanicidal Drugs: Challenges in Chemotherapy and Future Perspectives. Curr. Med. Chem. 2008, 15, 433–439. [Google Scholar] [CrossRef]
- Sanderson, L.; Yardley, V.; Croft, S.L. Activity of Anti-Cancer Protein Kinase Inhibitors against Leishmania spp. J. Antimicrob. Chemother. 2014, 69, 1888–1891. [Google Scholar] [CrossRef]
- Broni, E.; Kwofie, S.K.; Asiedu, S.O.; Miller, W.A.; Wilson, M.D. A Molecular Modeling Approach to Identify Potential Antileishmanial Compounds Against the Cell Division Cycle (Cdc)-2-Related Kinase 12 (CRK12) Receptor of Leishmania Donovani. Biomolecules 2021, 11, 458. [Google Scholar] [CrossRef]
- Kalmi, G.; Vignon-Pennamen, M.D.; Ram-Wolff, C.; Battistella, M.; Lafaurie, M.; Bouaziz, J.D.; Hamane, S.; Bernard, S.; Bretagne, S.; Thiéblemont, C.; et al. Visceral Leishmaniasis in Patients with Lymphoma: Case Reports and Review of the Literature. Medicine 2020, 99, e22787. [Google Scholar] [CrossRef]
- Machado, P.R.L.; Ribeiro, C.S.; França-Costa, J.; Dourado, M.E.F.; Trinconi, C.T.; Yokoyama-Yasunaka, J.K.U.; Malta-Santos, H.; Borges, V.M.; Carvalho, E.M.; Uliana, S.R.B. Tamoxifen and Meglumine Antimoniate Combined Therapy in Cutaneous Leishmaniasis Patients: A Randomised Trial. Trop. Med. Int. Health 2018, 23, 936–942. [Google Scholar] [CrossRef]
- Mak, K.K.; Pichika, M.R. Artificial Intelligence in Drug Development: Present Status and Future Prospects. Drug Discov. Today 2019, 24, 773–780. [Google Scholar] [CrossRef]
- Pathania, S.; Singh, P.K.; Narang, R.K.; Rawal, R.K. Identifying Novel Putative ERK1/2 Inhibitors via Hybrid Scaffold Hopping –FBDD Approach. J. Biomol. Struct. Dyn. 2021, 39, 1–16. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein Structure Modeling with MODELLER. Methods Mol. Biol. 2008, 426, 145–159. [Google Scholar]
- Siu, S.W.I.; Pluhackova, K.; Böckmann, R.A. Optimization of the OPLS-AA Force Field for Long Hydrocarbons. J. Chem. Theory Comput. 2012, 8, 1459–1470. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High-Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. Softwares 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Šudomová, M.; Hassan, S.T.S.; Khan, H.; Rasekhian, M.; Nabavi, S.M. A Multi-Biochemical and In silico Study on Anti-Enzymatic Actions of Pyroglutamic Acid against PDE-5, ACE, and Urease Using Various Analytical Techniques: Unexplored Pharmacological Properties and Cytotoxicity Evaluation. Biomolecules 2019, 9, 392. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Sorokina, M.; Steinbeck, C. Review on Natural Products Databases: Where to Find Data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef]
- Magaraci, F.; Jimenez, C.J.; Rodrigues, C.; Rodrigues, J.C.F.; Vianna Braga, M.; Yardley, V.; De Luca-Fradley, K.; Croft, S.L.; De Souza, W.; Ruiz-Perez, L.M.; et al. Azasterols as Inhibitors of Sterol 24-Methyltransferase in Leishmania Species and Trypanosoma Cruzi. J. Med. Chem. 2003, 46, 4714–4727. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, A.E. EasyROC: An Interactive Web Tool for ROC Curve Analysis Using R Language Environment. R J. 2016, 8, 213–230. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization and Analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of Activity Spectra for Biologically Active Substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. Potential Energy Functions for Atomic-Level Simulations of Water and Organic and Biomolecular Systems. Proc. Natl. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef]
- Lemkul, J.A. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2019, 1, 5068. [Google Scholar] [CrossRef]
- Dahiya, R.; Mohammad, T.; Gupta, P.; Haque, A.; Alajmi, M.F.; Hussain, A.; Hassan, M.I. Molecular Interaction Studies on Ellagic Acid for Its Anticancer Potential Targeting Pyruvate Dehydrogenase Kinase 3. RSC Adv. 2019, 9, 23302–23315. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa —A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Antwi, C.A.; Amisigo, C.M.; Adjimani, J.P.; Gwira, T.M. In Vitro Activity and Mode of Action of Phenolic Compounds on Leishmania Donovani. PLoS Negl. Trop. Dis. 2019, 13, e0007206. [Google Scholar] [CrossRef]
- Amisigo, C.M.; Antwi, C.A.; Adjimani, J.P.; Gwira, T.M. In Vitro Anti-Trypanosomal Effects of Selected Phenolic Acids on Trypanosoma Brucei. PLoS ONE 2019, 14, e0216078. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Dolling, N.N.O.; Donkoh, E.; Laryea, G.M.; Mosi, L.; Miller, W.A.; Adinortey, M.B.; Wilson, M.D. Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium Ulcerans Cystathionine γ-Synthase Metb. Computation 2021, 9, 32. [Google Scholar] [CrossRef]
- Nahm, F.S. Receiver Operating Characteristic Curve: Overview and Practical Use for Clinicians. Korean J. Anesthesiol. 2022, 75, 25. [Google Scholar] [CrossRef]
- Jung, S.W.; Kim, M.; Ramsey, S.; Kurtzman, T.; Cho, A.E. Water Pharmacophore: Designing Ligands Using Molecular Dynamics Simulations with Water. Sci. Rep. 2018, 8, 10400. [Google Scholar] [CrossRef]
- Kchouk, S.; Hegazy, L. Pharmacophore Modeling for Biological Targets with High Flexibility: LXRβ Case Study. Med. Drug Discov. 2022, 15, 100135. [Google Scholar] [CrossRef]
- Pascual, R.; Almansa, C.; Plata-Salamán, C.; Vela, J.M. A New Pharmacophore Model for the Design of Sigma-1 Ligands Validated on a Large Experimental Dataset. Front. Pharmacol. 2019, 10, 519. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Taylor, R.D.; Chessari, G.; Murray, C.W. Illustration of Current Challenges in Molecular Docking. In Structure-Based Drug Discovery; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Hsin, K.Y.; Matsuoka, Y.; Asai, Y.; Kamiyoshi, K.; Watanabe, T.; Kawaoka, Y.; Kitano, H. SystemsDock: A Web Server for Network Pharmacology-Based Prediction and Analysis. Nucleic Acids Res. 2016, 44, W507–W513. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef]
- Kaserer, T.; Beck, K.R.; Akram, M.; Odermatt, A.; Schuster, D.; Willett, P. Pharmacophore Models, and Pharmacophore-Based Virtual Screening: Concepts and Applications Exemplified on Hydroxysteroid Dehydrogenases. Molecules 2015, 20, 22799–22832. [Google Scholar] [CrossRef]
- Chen, Z.; Li, H.L.; Zhang, Q.J.; Bao, X.G.; Yu, K.Q.; Luo, X.M.; Zhu, W.L.; Jiang, H.L. Pharmacophore-Based Virtual Screening versus Docking-Based Virtual Screening: A Benchmark Comparison against Eight Targets. Acta Pharmacol. Sin. 2009, 30, 1694–1708. [Google Scholar] [CrossRef]
- Sharma, V.; Panwar, A.; Gupta, G.K.; Sharma, A.K. Molecular Docking and MD: Mimicking the Real Biological Process. Silico Chem. Biol. Curr. Futur. Prospect. 2022, 7, 133–144. [Google Scholar] [CrossRef]
- Sun, Z.; Zheng, L.; Wang, K.; Huai, Z.; Liu, Z. Primary vs Secondary: Directionalized Guest Coordination in β-Cyclodextrin Derivatives. Carbohydr. Polym. 2022, 297, 120050. [Google Scholar] [CrossRef]
- Duan, L.L.; Feng, G.Q.; Zhang, Q.G. Large-Scale Molecular Dynamics Simulation: Effect of Polarization on Thrombin-Ligand Binding Energy. Sci. Rep. 2016, 6, 31488. [Google Scholar] [CrossRef]
- Swegat, W.; Schlitter, J.; Krüger, P.; Wollmer, A. MD Simulation of Protein-Ligand Interaction: Formation and Dissociation of an Insulin-Phenol Complex. Biophys. J. 2003, 84, 1493. [Google Scholar] [CrossRef]
- Mady, F.M.; Aly, U.F. Experimental, Molecular Docking Investigations and Bioavailability Study on the Inclusion Complexes of Finasteride and Cyclodextrins. Drug Des. Devel. Ther. 2017, 11, 1681. [Google Scholar] [CrossRef]
- Gupta, Y.; Maciorowski, D.; Zak, S.E.; Jones, K.A.; Kathayat, R.S.; Azizi, S.A.; Mathur, R.; Pearce, C.M.; Ilc, D.J.; Husein, H.; et al. Bisindolylmaleimide IX: A Novel Anti-SARS-CoV-2 Agent Targeting Viral Main Protease 3CLpro Demonstrated by Virtual Screening Pipeline and in-Vitro Validation Assays. Methods 2021, 195, 57–71. [Google Scholar] [CrossRef]
- Li, J.; McKay, K.T.; Remington, J.M.; Schneebeli, S.T. A Computational Study of Cooperative Binding to Multiple SARS-CoV-2 Proteins. Sci. Rep. 2021, 11, 16307. [Google Scholar] [CrossRef]
- Azam, S.S.; Abro, A.; Raza, S.; Saroosh, A. Structure and Dynamics Studies of Sterol 24-C-Methyltransferase with Mechanism Based Inactivators for the Disruption of Ergosterol Biosynthesis. Mol. Biol. Rep. 2014, 41, 4279–4293. [Google Scholar] [CrossRef]
- Rahman, F.; Tabrez, S.; Ali, R.; Akand, S.K.; Zahid, M.; Alaidarous, M.A.; Alsaweed, M.; Alshehri, B.M.; Banawas, S.; Dukhyil, A.A.B.; et al. Virtual Screening of Natural Compounds for Potential Inhibitors of Sterol C-24 Methyltransferase of Leishmania Donovani to Overcome Leishmaniasis. J. Cell. Biochem. 2021, 122, 1216–1228. [Google Scholar] [CrossRef]
- Kwofie, S.; Broni, E.; Yunus, F.; Nsoh, J.; Adoboe, D.; Miller, W.; Wilson, M. Molecular Docking Simulation Studies Identifies Potential Natural Product Derived-Antiwolbachial Compounds as Filaricides against Onchocerciasis. Biomedicines 2021, 9, 1682. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and Drug-like Compounds: The Rule-of-Five Revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Kwarko, G.B.; Broni, E.; Adinortey, M.B.; Wilson, M.D. Molecular Informatics of Trypanothione Reductase of Leishmania Major Reveals Novel Chromen-2-One Analogues as Potential Leishmanicides; IntechOpen: London, UK, 2021; p. 100594. [Google Scholar]
- Kwofie, S.K.; Broni, E.; Asiedu, S.O.; Kwarko, G.B.; Dankwa, B.; Enninful, K.S.; Tiburu, E.K.; Wilson, M.D. Cheminformatics-Based Identification of Potential Novel Anti-SARS-CoV-2 Natural Compounds of African Origin. Molecules 2021, 26, 406. [Google Scholar] [CrossRef]
- Islam, M.A.; Pillay, T.S. Identification of Promising Anti-DNA Gyrase Antibacterial Compounds Using de Novo Design, Molecular Docking, and Molecular Dynamics Studies. J. Biomol. Struct. Dyn. 2019, 38, 1798–1809. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Sawale, R.T.; Kalyankar, T.M.; George, R.; Deosarkar, S.D. Molar Refraction and Polarizability of Antiemetic Drug 4-Amino-5-Chloro-N-(2-(Diethylamino)Ethyl)-2 Methoxybenzamide Hydrochloride Monohydrate in {Aqueous-Sodium or Lithium Chloride} Solutions at 30 o C. J. Appl. Pharm. Sci. 2016, 6, 120–124. [Google Scholar] [CrossRef]
- van den Anker, J.; Reed, M.D.; Allegaert, K.; Kearns, G.L. Developmental Changes in Pharmacokinetics and Pharmacodynamics. J. Clin. Pharmacol. 2018, 58, S10–S25. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Annan, D.G.; Adinortey, C.A.; Boison, D.; Kwarko, G.B.; Abban, R.A.; Adinortey, M.B. Identification of Novel Potential Inhibitors of Varicella-Zoster Virus Thymidine Kinase from Ethnopharmacologic Relevant Plants through an in-Silico Approach. J. Biomol. Struct. Dyn. 2021, 40, 12932–12947. [Google Scholar] [CrossRef]
- Banks, W.A. Characteristics of Compounds That Cross the Blood-Brain Barrier. BMC Neurol. 2009, 9, S3. [Google Scholar] [CrossRef]
- Melo, G.D.; Goyard, S.; Fiette, L.; Boissonnas, A.; Combadiere, C.; Machado, G.F.; Minoprio, P.; Lang, T. Unveiling Cerebral Leishmaniasis: Parasites and Brain Inflammation in Leishmania Donovani Infected Mice. Sci. Rep. 2017, 7, 8454. [Google Scholar] [CrossRef]
- Abreu-Silva, A.L.; Calabrese, K.S.; Tedesco, R.C.; Mortara, R.A.; Gonçalves Da Costa, S.C. Central Nervous System Involvement in Experimental Infection with Leishmania (Leishmania) Amazonensis. Am. J. Trop. Med. Hyg. 2003, 68, 661–665. [Google Scholar] [CrossRef]
- Maia, C.S.F.; Monteiro, M.C.; Gavioli, E.C.; Oliveira, F.R.; Oliveira, G.B.; Romão, P.R.T. Neurological Disease in Human and Canine Leishmaniasis—Clinical Features and Immunopathogenesis. Parasite Immunol. 2015, 37, 385–393. [Google Scholar] [CrossRef]
- Ali, N.; Ali, N.M.; Fariba, J.; Elaheh, H. The Efficacy of 5% Trichloroacetic Acid Cream in the Treatment of Cutaneous Leishmaniasis Lesions. J. Dermatolog. Treat. 2012, 23, 136–139. [Google Scholar] [CrossRef]
- Momeni, A.Z.; Aminjavaheri, M.; Omidghaemi, M.R. Treatment of Cutaneous Leishmaniasis with Ketoconazole Cream. J. Dermatolog. Treat. 2003, 14, 26–29. [Google Scholar] [CrossRef]
- Sakyi, P.O.; Amewu, R.K.; Devine, R.N.O.A.; Bienibuor, A.K.; Miller, W.A.; Kwofie, S.K. Unravelling the Myth Surrounding Sterol Biosynthesis as Plausible Target for Drug Design against Leishmaniasis. J. Parasit. Dis. 2021, 45, 1152–1171. [Google Scholar] [CrossRef]
- Galvão, E.L.; Rabello, A.; Cota, G.F. Efficacy of Azole Therapy for Tegumentary Leishmaniasis: A Systematic Review and Meta-Analysis. PLoS ONE 2017, 12, e0186117. [Google Scholar] [CrossRef]
- Weidner, S.; Kittelmann, M.; Goeke, K.; Ghisalba, O.; Zähner, H. 3’-Demethoxy-3’-Hydroxystaurosporine-O-Methyltransferase from Streptomyces Longisporoflavus Catalyzing the Last Step in the Biosynthesis of Staurosporine. J. Antibiot. 1998, 51, 679–682. [Google Scholar] [CrossRef]
- Axelrod, J.; Daly, J. Phenol-O-Methyltransferase. BBA-Enzymol. 1968, 159, 472–478. [Google Scholar] [CrossRef]
- Coque, J.J.R.; Alvarez-Rodríguez, M.L.; Larriba, G. Characterization of an Inducible Chlorophenol O-Methyltransferase from Trichoderma longibrachiatum Involved in the Formation of Chloroanisoles and Determination of Its Role in Cork Taint of Wines. Appl. Environ. Microbiol. 2003, 69, 5089–5095. [Google Scholar] [CrossRef]
- Bustamante, C.; Ochoa, R.; Asela, C.; Muskus, C. Repurposing of Known Drugs for Leishmaniasis Treatment Using Bioinformatic Predictions, in Vitro Validations and Pharmacokinetic Simulations. J. Comput. Aided. Mol. Des. 2019, 33, 845–854. [Google Scholar] [CrossRef]
- Cavaliere, F.; Fornarelli, A.; Bertan, F.; Russo, R.; Marsal-Cots, A.; Morrone, L.A.; Adornetto, A.; Corasaniti, M.T.; Bano, D.; Bagetta, G.; et al. The Tricyclic Antidepressant Clomipramine Inhibits Neuronal Autophagic Flux. Sci. Rep. 2019, 9, 4881. [Google Scholar] [CrossRef]
- da Silva Rodrigues, J.H.; Miranda, N.; Volpato, H.; Ueda-Nakamura, T.; Nakamura, C.V. The Antidepressant Clomipramine Induces Programmed Cell Death in Leishmania Amazonensis through a Mitochondrial Pathway. Parasitol. Res. 2019, 118, 977–989. [Google Scholar] [CrossRef]
- Manandhar, S.; Sankhe, R.; Priya, K.; Hari, G.; Kumar, B.H.; Mehta, C.H.; Nayak, U.Y.; Pai, K.S.R. Molecular Dynamics and Structure-Based Virtual Screening and Identification of Natural Compounds as Wnt Signaling Modulators: Possible Therapeutics for Alzheimer’s Disease. Mol. Divers. 2022, 26, 2793–2811. [Google Scholar] [CrossRef]
- Ahamad, S.; Kanipakam, H.; Birla, S.; Ali, M.S.; Gupta, D. Screening Malaria-Box Compounds to Identify Potential Inhibitors against SARS-CoV-2 Mpro, Using Molecular Docking and Dynamics Simulation Studies. Eur. J. Pharmacol. 2021, 890, 173664. [Google Scholar] [CrossRef]
- Kant, V.; Ranjan, R.; Kumar, P.; Mandal, D.; Vijayakumar, S. In silico Screening, Molecular Dynamic Simulations, and in Vitro Activity of Selected Natural Compounds as an Inhibitor of Leishmania Donovani 3 - Mercaptopyruvate Sulfurtransferase. Parasitol. Res. 2022, 121, 2093–2109. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jha, S.; Rathore, A.S.; Gupta, K. Identification of Small Molecule Modulators of Class II Transactivator-I Using Computational Approaches. J. Biomol. Struct. Dyn. 2022, 1–13. [Google Scholar] [CrossRef]
- Zaki, A.A.; Ashour, A.; Elhady, S.S.; Darwish, K.M.; Al-Karmalawy, A.A. Calendulaglycoside A Showing Potential Activity against SARS-CoV-2 Main Protease: Molecular Docking, Molecular Dynamics, and SAR Studies. J. Tradit. Complement. Med. 2022, 12, 16. [Google Scholar] [CrossRef]
- Steinbrecher, T.; Labahn, A. Towards Accurate Free Energy Calculations in Ligand Protein-Binding Studies. Curr. Med. Chem. 2010, 17, 767–785. [Google Scholar] [CrossRef]
- Sun, Z.; Yan, Y.N.; Yang, M.; Zhang, J.Z.H. Interaction Entropy for Protein-Protein Binding. J. Chem. Phys. 2017, 146, 124124. [Google Scholar] [CrossRef]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Stuart, K.; Brun, R.; Croft, S.; Fairlamb, A.; Gürtler, R.E.; McKerrow, J.; Reed, S.; Tarleton, R. Kinetoplastids: Related Protozoan Pathogens, Different Diseases. J. Clin. Investig. 2008, 118, 1301–1310. [Google Scholar] [CrossRef]
- Phan, T.N.; Baek, K.H.; Lee, N.; Byun, S.Y.; Shum, D.; No, J.H. In Vitro and in Vivo Activity of MTOR Kinase and PI3K Inhibitors Against Leishmania Donovani and Trypanosoma Brucei. Molecules 2020, 25, 1980. [Google Scholar] [CrossRef]
- Rojas Vargas, J.A.; López, A.G.; Pérez, Y.; Cos, P.; Froeyen, M. In Vitro Evaluation of Arylsubstituted Imidazoles Derivatives as Antiprotozoal Agents and Docking Studies on Sterol 14α-Demethylase (CYP51) from Trypanosoma Cruzi, Leishmania Infantum, and Trypanosoma Brucei. Parasitol. Res. 2019, 118, 1533–1548. [Google Scholar] [CrossRef]
- Baek, K.H.; Phan, T.N.; Malwal, S.R.; Lee, H.; Li, Z.H.; Moreno, S.N.J.; Oldfield, E.; No, J.H. In Vivo Efficacy of SQ109 against Leishmania Donovani, Trypanosoma spp., and Toxoplasma gondii and In Vitro Activity of SQ109 Metabolites. Biomedicines 2022, 10, 670. [Google Scholar] [CrossRef]
- Alves, F.; Bilbe, G.; Blesson, S.; Goyal, V.; Monnerat, S.; Mowbray, C.; Ouattara, G.M.; Pécoul, B.; Rijal, S.; Rode, J.; et al. Recent Development of Visceral Leishmaniasis Treatments: Successes, Pitfalls, and Perspectives. Clin. Microbiol. Rev. 2018, 31, e00048-18. [Google Scholar] [CrossRef]


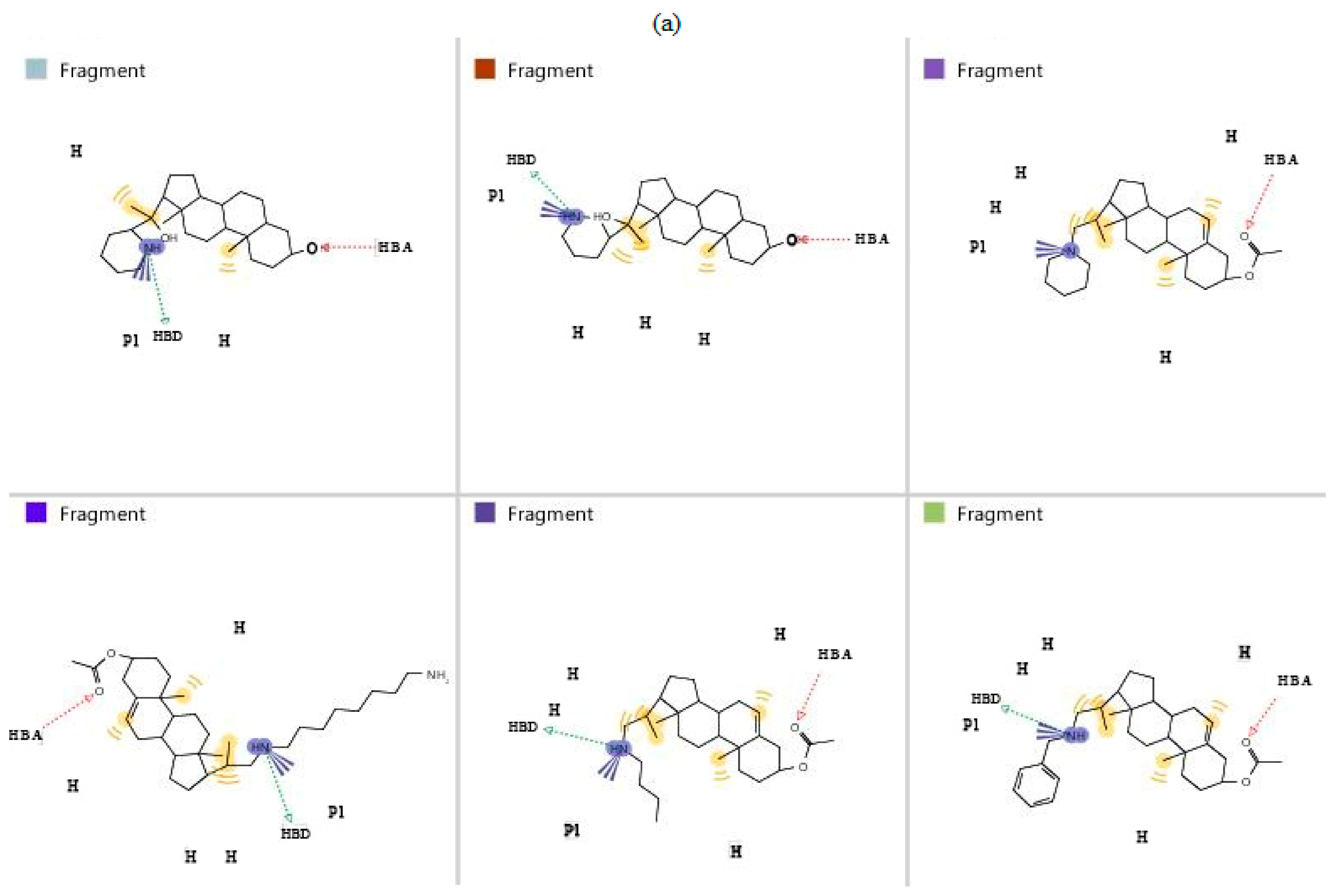


| Compound ID | Pharmacophore Fit Score | Compound ID | Pharmacophore Fit Score |
|---|---|---|---|
| STOCK6S-64941 | 59.39 | STOCK6S-14893 | 58.37 |
| STOCK6S-43563 | 59.23 | STOCK6S-06707 | 58.24 |
| STOCK6S-07535 | 59.19 | STOCK7S-11482 | 58.14 |
| STOCK6S-33909 | 59.19 | STOCK6S-16994 | 58.05 |
| STOCK6S-39547 | 59.18 | STOCK6S-64616 | 57.53 |
| STOCK6S-19430 | 59.00 | STOCK6S-65229 | 57.44 |
| STOCK7S-75883 | 58.89 | STOCK6S-63483 | 57.37 |
| STOCK7S-14941 | 58.87 | STOCK6S-55084 | 57.33 |
| STOCK6S-84928 | 58.85 | STOCK6S-65920 | 57.3 |
| STOCK6S-47549 | 58.47 | STOCK6S-47366 | 57.15 |
| Ligands | Binding Energy/kcal/mol | Binding Residues | |
|---|---|---|---|
| Hydrogen Bonds | Hydrophobic Bonds | ||
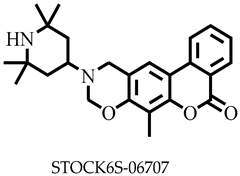 | −8.7 | Asp25, Ala28, Asp31, Arg32, Phe307, Ala311 | |
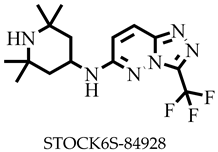 | −8.2 | Arg15, Asp28, Arg227 | Leu13, Lys313 |
 | −8.0 | Ser350 | Glu192, Phe194, Arg195, Lys351 |
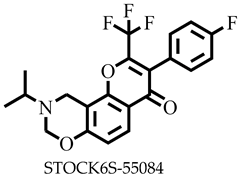 | −7.9 | Glu178, Trp208, His226, Ile228 | |
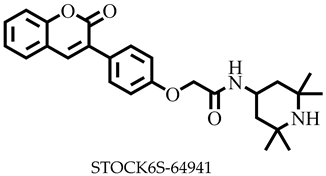 | −7.8 | Asp281, Ser284 | Leu322 |
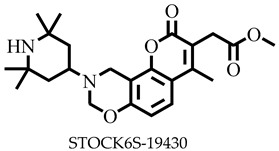 | −7.7 | Arg289, Arg295 | Tyr316, Glu320 |
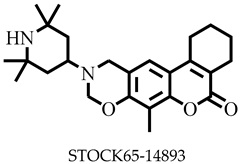 | −7.6 | Ala28, Phe307, Val308, Ala311 | |
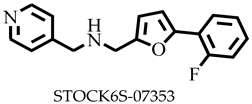 | −7.5 | Glu219, Glu229, Tyr275 | Trp208, Ile224, Lys225, Ile228, Ile272 |
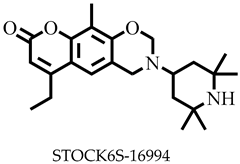 | −7.5 | Thr338 | Phe93, Ile258 |
 | −7.6 | Glu102, Gly200 | Phe100, Lys198, Pro199 |
 | −5.3 | Arg309, Asn299, Gly294, Leu288 | |
 | −5.0 | Asp31, Arg32, Val308, Arg309, Leu310 | Glu306 |
 | −4.0 | Cys202 | Phe100, Met101, Asp104, Asp172, Pro199, Gly200, Thr201, Tyr343, Ile344 |
| Compound ID | MW (g/mol) | NRB | MR | TPSA (Å2) | LogS | SC | BS | GI | BBB | vRoF |
|---|---|---|---|---|---|---|---|---|---|---|
| STOCK6S-06707 | 406.52 | 1 | 129.05 | 54.71 | −5.31 | Moderate | 0.55 | High | Yes | 0 |
| STOCK6S-84928 | 342.36 | 3 | 88.22 | 67.14 | −3.39 | Soluble | 0.55 | High | Yes | 0 |
| STOCK6S-65920 | 353.39 | 2 | 103.42 | 42.68 | −4.73 | Moderate | 0.55 | High | Yes | 0 |
| STOCK6S-55084 | 407.36 | 3 | 103.46 | 42.68 | −5.27 | Moderate | 0.55 | High | No | 0 |
| STOCK6S-64941 | 434.53 | 6 | 130.16 | 80.57 | −5.62 | Moderate | 0.55 | High | No | 0 |
| STOCK6S-19430 | 428.52 | 4 | 127.41 | 81.01 | −4.01 | Moderate | 0.55 | High | No | 0 |
| STOCK6S-14893 | 410.55 | 1 | 128.98 | 54.71 | −5.11 | Moderate | 0.55 | High | Yes | 0 |
| STOCK6S-07353 | 282.31 | 5 | 78.96 | 38.06 | −3.03 | Soluble | 0.55 | High | Yes | 0 |
| STOCK6S-16994 | 384.51 | 2 | 121.32 | 54.71 | −4.5 | Soluble | 0.55 | High | Yes | 0 |
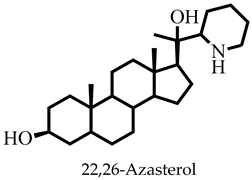
| 22,26-Azasterol | 403.64 | 2 | 125.08 | 52.49 | −5.66 | Moderate | 0.55 | High | Yes | 1 |
| Amphotericin B | 924.08 | 3 | 239.06 | 319.61 | −5.37 | Moderate | 0.55 | Low | No | 3 |
| Miltefosine | 407.57 | 20 | 115.9 | 68.40 | −5.32 | Moderate | 0.55 | Low | No | 0 |
| Paromomycin | 615.63 | 9 | 133.56 | 347.32 | −2.44 | Soluble | 0.55 | Low | No | 3 |
| Ligand | DataWarrior Predictions | |||
|---|---|---|---|---|
| Tumorigenic | Mutagenic | Irritant | Reproductive Effect | |
| STOCK6S-06707 | None | None | High | None |
| STOCK6S-84928 | None | None | None | None |
| STOCK6S-65920 | None | None | None | Low |
| STOCK6S-55084 | None | None | None | Low |
| STOCK6S-64941 | None | None | None | High |
| STOCK6S-19430 | None | None | High | high |
| STOCK6S-14893 | None | None | High | High |
| STOCK6S-07353 | None | None | None | None |
| STOCK6S-16994 | None | None | High | High |
| 22,26-Azasterol | None | None | None | None |
| Amphotericin B | None | None | None | None |
| Miltefosine | None | None | None | None |
| Paromomycin | None | None | None | None |
| Complex | ΔGvdW (kJ/mol) | ΔGele (kJ/mol) | ΔGpol,sol (kJ/mol) | ΔGSASA (kJ/mol) | ΔGbind (kJ/mol) |
|---|---|---|---|---|---|
| STOCKIN6S-O6707 | −232.978 ± 1.954 | −983.173 ± 4.875 | 863.728 ± 3.447 | −18.722 ± 2.602 | −371.146 ± 2.105 |
| STOCKIN6S−84928 | −142.275 ± 5.679 | −3.390 ± 0.417 | 29.163 ± 2.101 | −13.223 ± 4.385 | −129.725 ± 4.799 |
| STOCKIN6S-65920 | −217.243 ± 3.589 | −6.439 ± 1.576 | 90.570 ± 4.465 | −16.786 ± 1.646 | −149.899 ± 3.600 |
| 22,26-Azasterol | −186.874 ± 2.143 | −217.654± 0.546 | 199.212± 4.701 | −16.211 ± 1.632 | −221.527 ± 3.716 |
| Compound | Leishmania donovani IC50 (μM) ± SD | Trypanosoma brucei (IC50) (μM) ± SD |
|---|---|---|
| STOCK6S-65920 | 23.5 ± 1.1 | 18.1 ± 1.4 |
| STOCK6S-06707 | 21.9 ± 1.5 | NA |
| STOCK6S-84928 | 118.3 ± 5.8 | 14.3 ± 2.0 |
| Amphotericin B | 6.56 ± 0.06 | - |
| Diminazene | - | 0.1 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakyi, P.O.; Kwofie, S.K.; Tuekpe, J.K.; Gwira, T.M.; Broni, E.; Miller, W.A., III; Wilson, M.D.; Amewu, R.K. Inhibiting Leishmania donovani Sterol Methyltransferase to Identify Lead Compounds Using Molecular Modelling. Pharmaceuticals 2023, 16, 330. https://doi.org/10.3390/ph16030330
Sakyi PO, Kwofie SK, Tuekpe JK, Gwira TM, Broni E, Miller WA III, Wilson MD, Amewu RK. Inhibiting Leishmania donovani Sterol Methyltransferase to Identify Lead Compounds Using Molecular Modelling. Pharmaceuticals. 2023; 16(3):330. https://doi.org/10.3390/ph16030330
Chicago/Turabian StyleSakyi, Patrick O., Samuel K. Kwofie, Julius K. Tuekpe, Theresa M. Gwira, Emmanuel Broni, Whelton A. Miller, III, Michael D. Wilson, and Richard K. Amewu. 2023. "Inhibiting Leishmania donovani Sterol Methyltransferase to Identify Lead Compounds Using Molecular Modelling" Pharmaceuticals 16, no. 3: 330. https://doi.org/10.3390/ph16030330
APA StyleSakyi, P. O., Kwofie, S. K., Tuekpe, J. K., Gwira, T. M., Broni, E., Miller, W. A., III, Wilson, M. D., & Amewu, R. K. (2023). Inhibiting Leishmania donovani Sterol Methyltransferase to Identify Lead Compounds Using Molecular Modelling. Pharmaceuticals, 16(3), 330. https://doi.org/10.3390/ph16030330







