Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV
Abstract
1. Introduction
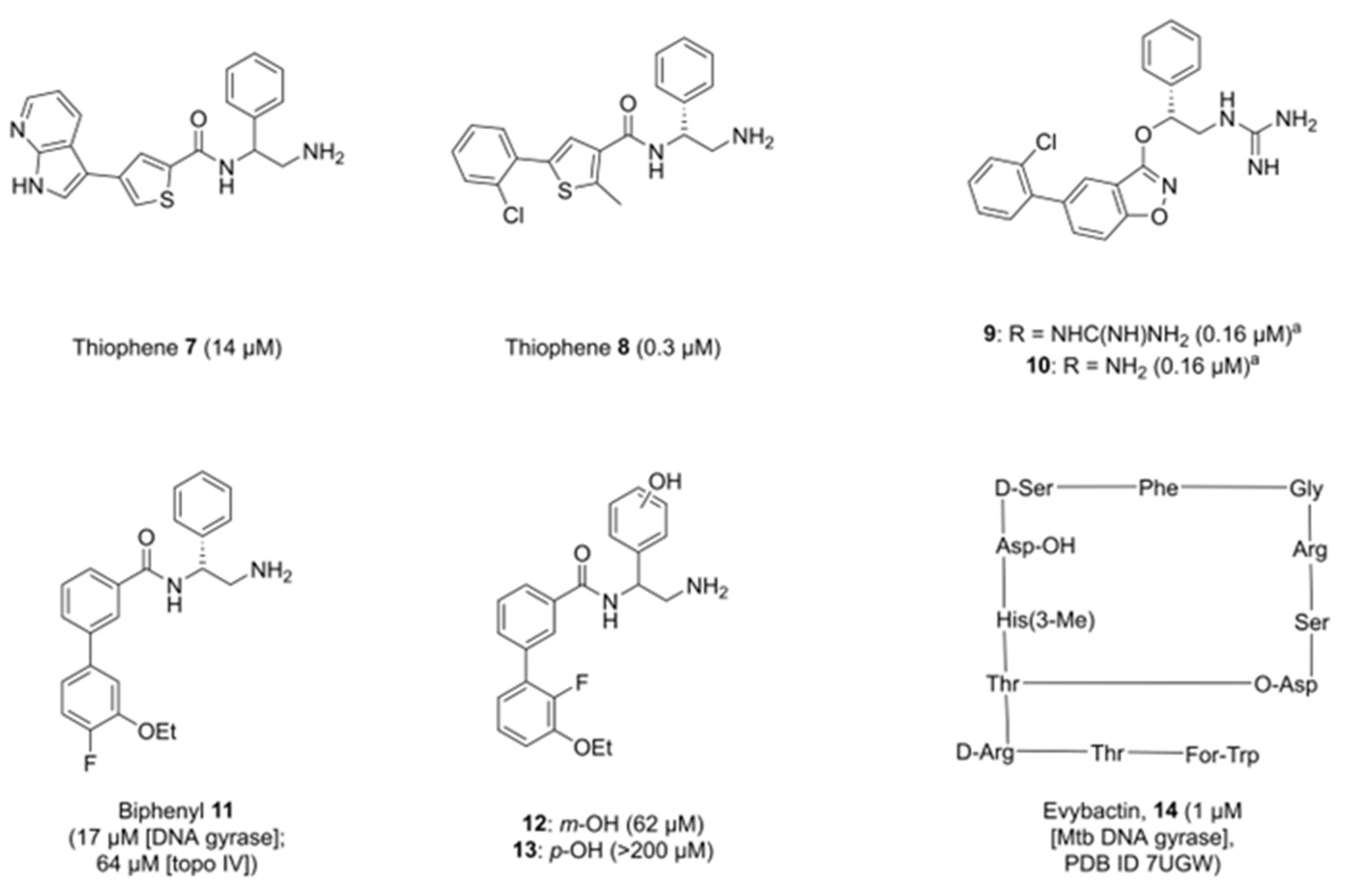
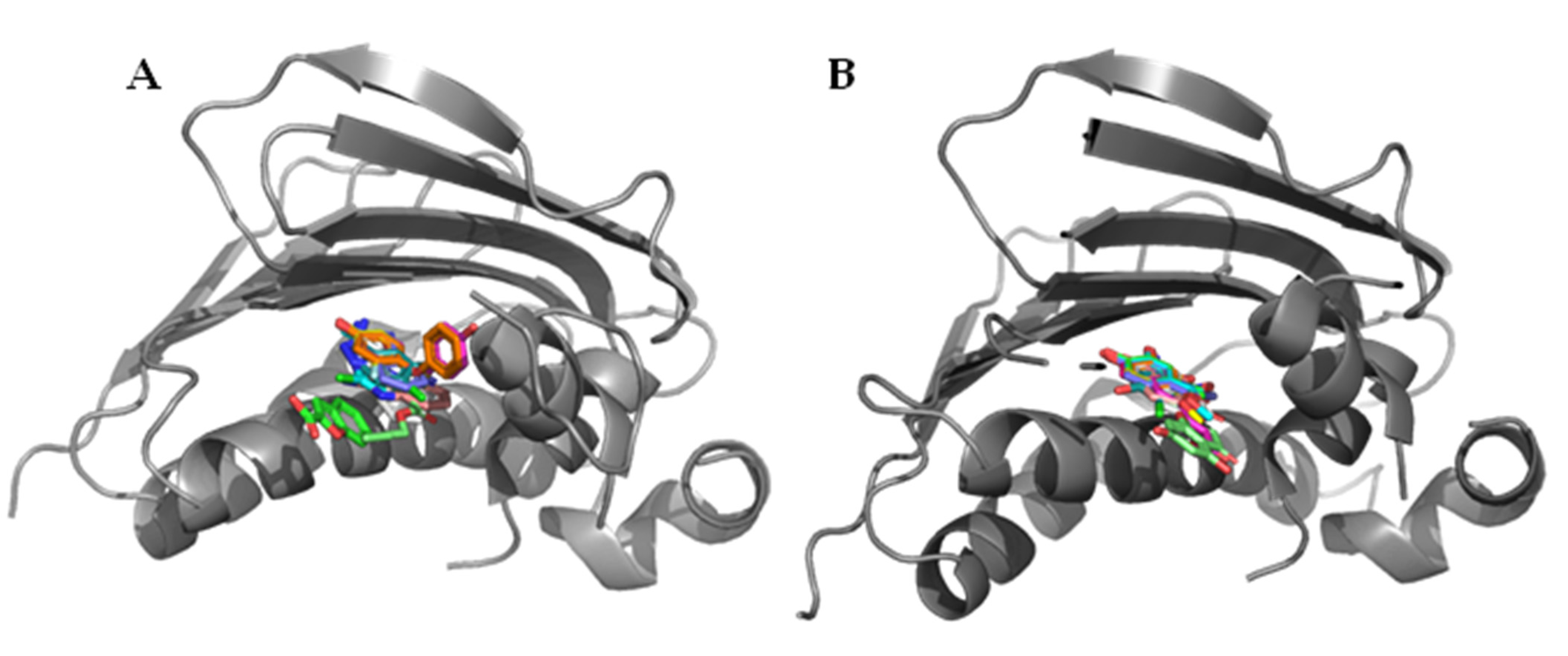
2. Allosteric Inhibitors of GyrA
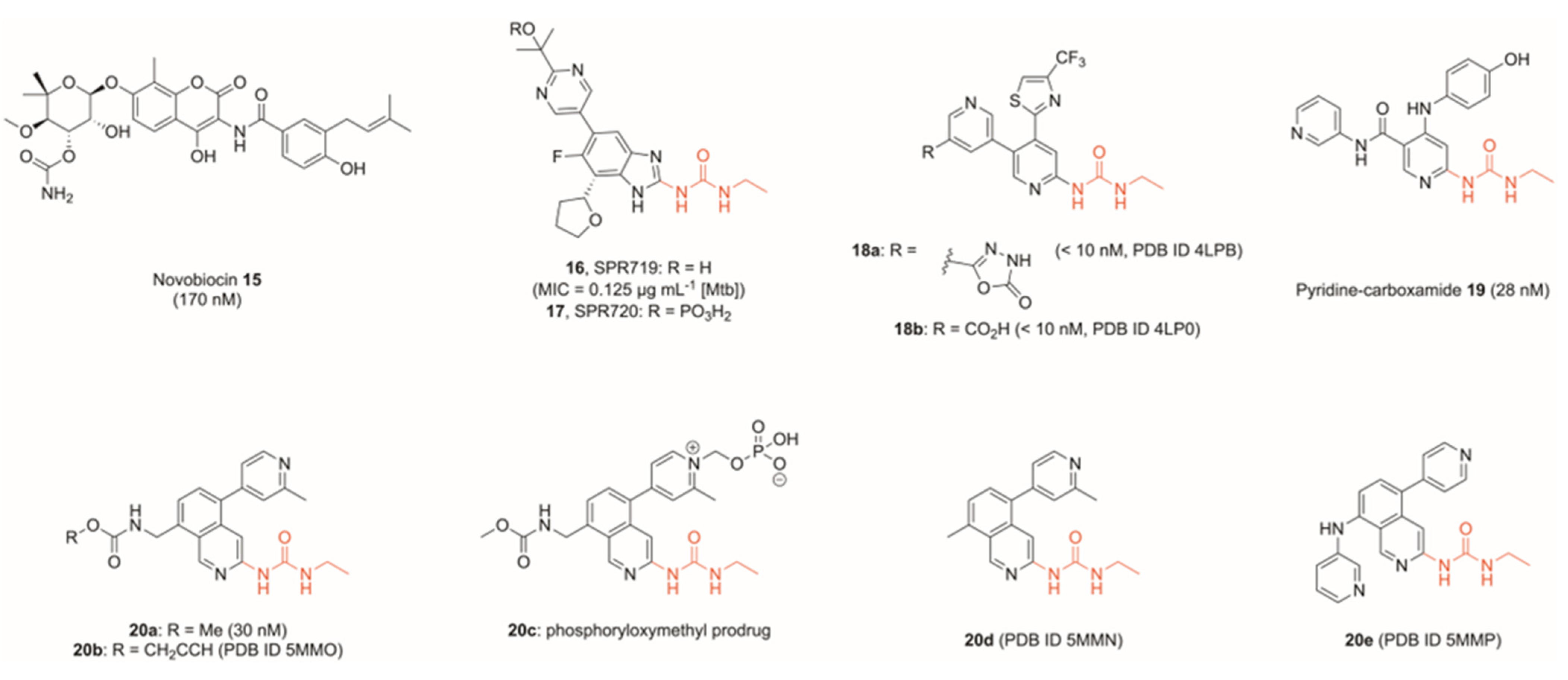
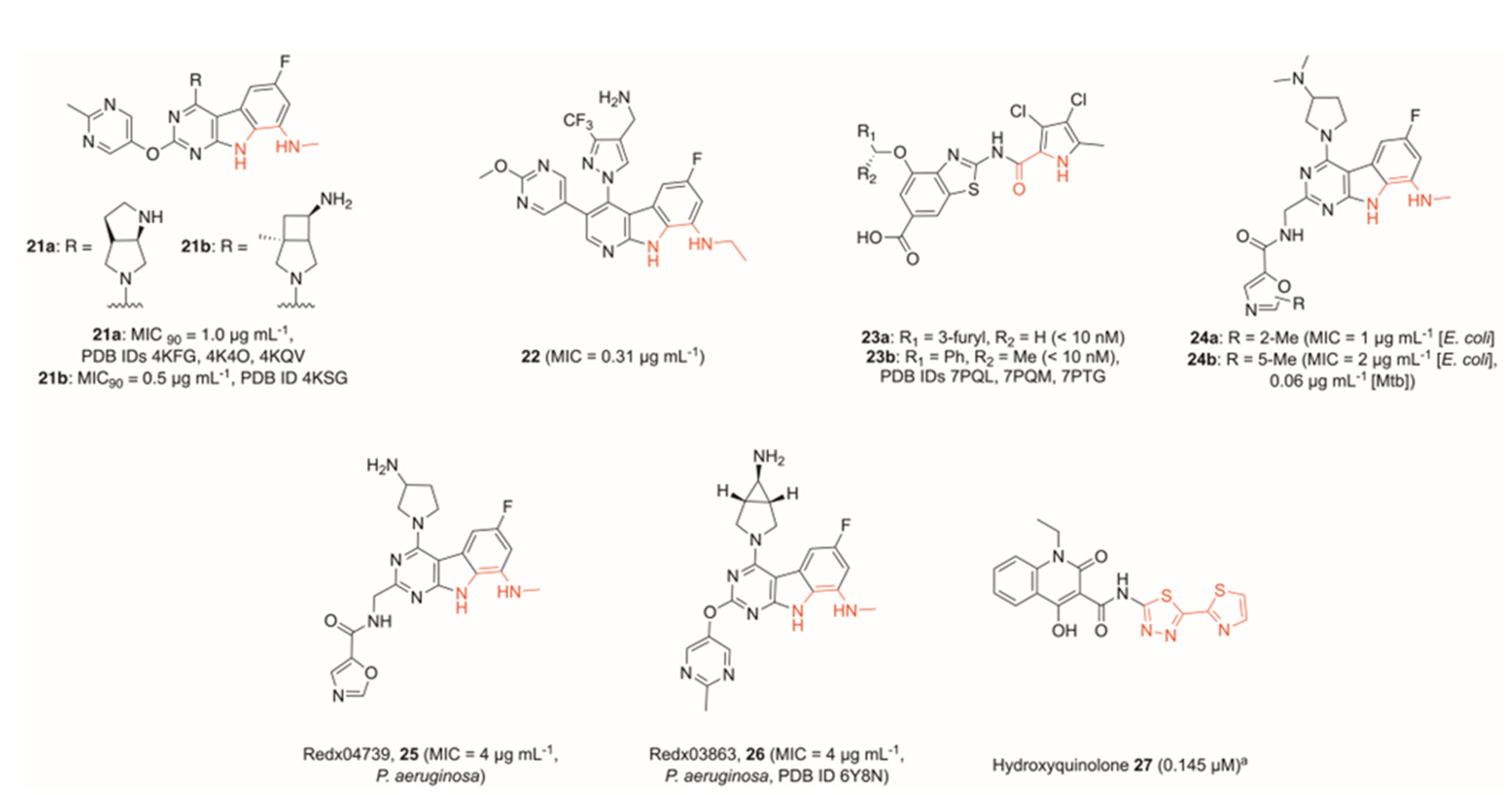
3. Inhibitors of the ATPase Site of GyrB
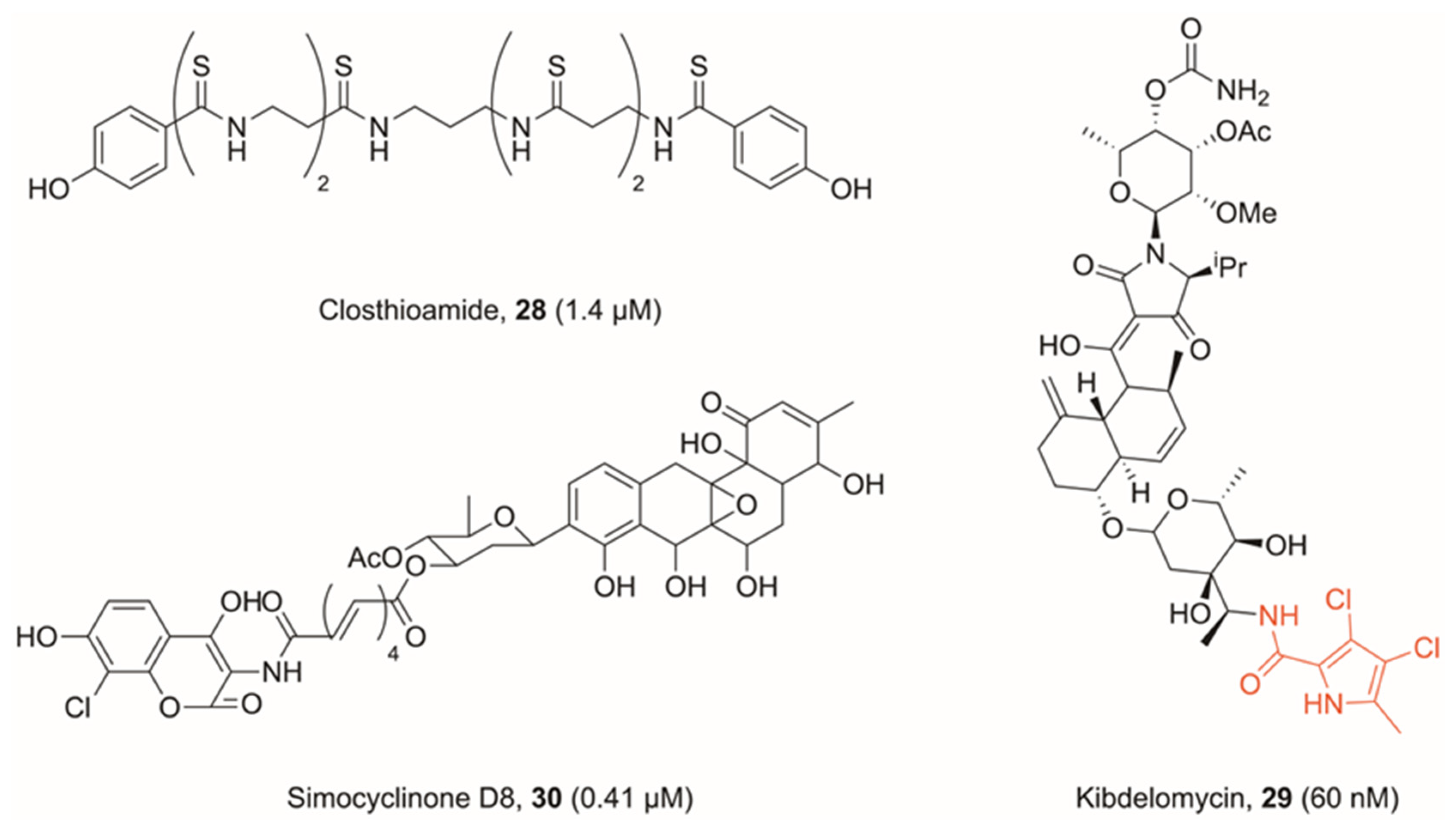
4. Miscellaneous Topoisomerase Inhibitors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2019 Diseases and Injuries Collaborators. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug–Resistant Infections Globally: Final Report and Recommendations; Government of the UK: London, UK, 2016.
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- McCullough, A.R.; Parekh, S.; Rathbone, J.; Del Mar, C.B.; Hoffmann, T.C. A systematic review of the public’s knowledge and beliefs about antibiotic resistance. J. Antimicrob. Chemother. 2016, 71, 27–33. [Google Scholar] [CrossRef]
- Fleming-Dutra, K.E.; Hersh, A.L.; Shapiro, D.J.; Bartoces, M.; Enns, E.A.; File, T.M., Jr.; Finkelstein, J.A.; Gerber, J.S.; Hyun, D.Y.; Linder, J.A.; et al. Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010–2011. JAMA 2016, 315, 1864–1873. [Google Scholar] [CrossRef]
- Forterre, P.; Gribaldo, S.; Gadelle, D.; Serre, M.-C. Origin and evolution of DNA topoisomerases. Biochimie 2007, 89, 427–446. [Google Scholar] [CrossRef]
- Hirsch, J.; Klostermeier, D. What makes a type IIA topoisomerase a gyrase or a Topo IV? Nucleic Acids Res. 2021, 49, 6027–6042. [Google Scholar] [CrossRef]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All tangled up: How cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef]
- Hooper, D.C. Bacterial Topoisomerases, Anti-Topoisomerases, and Anti-Topoisomerase Resistance. Clin. Infect. Dis. 1998, 27, S54–S63. [Google Scholar] [CrossRef]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef]
- Champoux, J.J. DNA Topoisomerases: Structure, Function, and Mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef]
- Thalji, R.K.; Raha, K.; Andreotti, D.; Checchia, A.; Cui, H.; Meneghelli, G.; Profeta, R.; Tonelli, F.; Tommasi, S.; Bakshi, T.; et al. Structure-guided design of antibacterials that allosterically inhibit DNA gyrase. Bioorganic. Med. Chem. Lett. 2019, 29, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.D.; Maxwell, A. Energy Coupling in Type II Topoisomerases: Why Do They Hydrolyze ATP? Biochemistry 2007, 46, 7929–7941. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Bratsman, A.; Mathias, K.; Laubscher, R.; Grigoryan, L.; Rose, S. Outpatient fluoroquinolone prescribing patterns before and after US FDA boxed warning. Pharmacoepidemiol. Drug Saf. 2020, 29, 701–707. [Google Scholar] [CrossRef]
- Bisacchi, G.S. Origins of the Quinolone Class of Antibacterials: An Expanded “Discovery Story”. J. Med. Chem. 2015, 58, 4874–4882. [Google Scholar] [CrossRef]
- Concia, E.; Bragantini, D.; Mazzaferri, F. Clinical evaluation of guidelines and therapeutic approaches in multi drug-resistant urinary tract infections. J. Chemother. 2017, 29, 19–28. [Google Scholar] [CrossRef]
- Alves, C.; Mendes, D.; Marques, F.B. Fluoroquinolones and the risk of tendon injury: A systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 2019, 75, 1431–1443. [Google Scholar] [CrossRef]
- Sankar, A.; Swanson, K.M.; Zhou, J.; Jena, A.B.; Ross, J.S.; Shah, N.D.; Karaca-Mandic, P. Association of Fluoroquinolone Prescribing Rates with Black Box Warnings from the US Food and Drug Administration. JAMA Netw. Open 2021, 4, e2136662. [Google Scholar] [CrossRef]
- Iacobino, A.; Piccaro, G.; Pardini, M.; Fattorini, L.; Giannoni, F. Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner. Microorganisms 2021, 9, 255. [Google Scholar] [CrossRef]
- Barrett, T.C.; Mok, W.W.K.; Murawski, A.M.; Brynildsen, M.P. Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat. Commun. 2019, 10, 1177. [Google Scholar] [CrossRef]
- Browne, A.J.; Chipeta, M.G.; Haines-Woodhouse, G.; Kumaran, E.P.A.; Hamadani, B.H.K.; Zaraa, S.; Henry, N.J.; Deshpande, A.; Reiner, R.C.; Day, N.P.J.; et al. Global antibiotic consumption and usage in humans, 2000–2018: A spatial modelling study. Lancet Planet. Health 2021, 5, e893–e904. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.M.; Elkhatib, W.F.; El-Mahallawy, H.A.; Helmy, M.M.; Ashour, M.S.; Aboshanab, K.M.A. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci. Rep. 2018, 8, 12268. [Google Scholar] [CrossRef] [PubMed]
- Flamm, R.K.; Vojtko, C.; Chu, D.T.; Li, Q.; Beyer, J.; Hensey, D.; Ramer, N.; Clement, J.J.; Tanaka, S.K. In vitro evaluation of ABT-719, a novel DNA gyrase inhibitor. Antimicrob. Agents Chemother. 1995, 39, 964–970. [Google Scholar] [CrossRef]
- Pan, X.-S.; Gould Katherine, A.; Fisher, L.M. Probing the Differential Interactions of Quinazolinedione PD 0305970 and Quinolones with Gyrase and Topoisomerase IV. Antimicrob. Agents Chemother. 2009, 53, 3822–3831. [Google Scholar] [CrossRef]
- Pucci Michael, J.; Podos Steven, D.; Thanassi Jane, A.; Leggio Melissa, J.; Bradbury Barton, J.; Deshpande, M. In Vitro and In Vivo Profiles of ACH-702, an Isothiazoloquinolone, against Bacterial Pathogens. Antimicrob. Agents Chemother. 2011, 55, 2860–2871. [Google Scholar] [CrossRef]
- Savage, V.J.; Charrier, C.; Salisbury, A.-M.; Moyo, E.; Forward, H.; Chaffer-Malam, N.; Metzger, R.; Huxley, A.; Kirk, R.; Uosis-Martin, M.; et al. Biological profiling of novel tricyclic inhibitors of bacterial DNA gyrase and topoisomerase IV. J. Antimicrob. Chemother. 2016, 71, 1905–1913. [Google Scholar] [CrossRef]
- Desai, J.; Sachchidanand, S.; Kumar, S.; Sharma, R. Novel Bacterial Topoisomerase inhibitors (NBTIs)—A comprehensive review. Eur. J. Med. Chem. Rep. 2021, 3, 100017. [Google Scholar] [CrossRef]
- Kokot, M.; Anderluh, M.; Hrast, M.; Minovski, N. The Structural Features of Novel Bacterial Topoisomerase Inhibitors That Define Their Activity on Topoisomerase IV. J. Med. Chem. 2022, 65, 6431–6440. [Google Scholar] [CrossRef]
- Kolarič, A.; Anderluh, M.; Minovski, N. Two Decades of Successful SAR-Grounded Stories of the Novel Bacterial Topoisomerase Inhibitors (NBTIs). J. Med. Chem. 2020, 63, 5664–5674. [Google Scholar] [CrossRef]
- Basarab, G.S.; Kern, G.H.; McNulty, J.; Mueller, J.P.; Lawrence, K.; Vishwanathan, K.; Alm, R.A.; Barvian, K.; Doig, P.; Galullo, V.; et al. Responding to the challenge of untreatable gonorrhea: ETX0914, a first-in-class agent with a distinct mechanism-of-action against bacterial Type II topoisomerases. Sci. Rep. 2015, 5, 11827. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A.; Miller, A.A.; O’Donnell, J.; Mueller, J.P. Zoliflodacin: An Oral Spiropyrimidinetrione Antibiotic for the Treatment of Neisseria gonorrheae, Including Multi-Drug-Resistant Isolates. ACS Infect. Dis. 2020, 6, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Damião Gouveia, A.C.; Unemo, M.; Jensen, J.S. In vitro activity of zoliflodacin (ETX0914) against macrolide-resistant, fluoroquinolone-resistant and antimicrobial-susceptible Mycoplasma genitalium strains. J. Antimicrob. Chemother. 2018, 73, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, R.A.; Huband, M.D.; de Jonge, B.L.M.; Bradford, P.A. Effect of Susceptibility Testing Conditions on the In Vitro Antibacterial Activity of ETX0914. Diagn. Microbiol. Infect. Dis. 2017, 87, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Basarab, G.S.; Doig, P.; Galullo, V.; Kern, G.; Kimzey, A.; Kutschke, A.; Newman, J.P.; Morningstar, M.; Mueller, J.; Otterson, L.; et al. Discovery of Novel DNA Gyrase Inhibiting Spiropyrimidinetriones: Benzisoxazole Fusion with N-Linked Oxazolidinone Substituents Leading to a Clinical Candidate (ETX0914). J. Med. Chem. 2015, 58, 6264–6282. [Google Scholar] [CrossRef]
- Chan, P.F.; Srikannathasan, V.; Huang, J.; Cui, H.; Fosberry, A.P.; Gu, M.; Hann, M.M.; Hibbs, M.; Homes, P.; Ingraham, K.; et al. Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin. Nat. Commun. 2015, 6, 10048. [Google Scholar] [CrossRef]
- Morgan, H.; Lipka-Lloyd, M.; Warren, A.; Hughes, N.; Holmes, J.; Burton, N.; Mahenthiralingam, E.; Bax, B. A 2.8 Å structure of zoliflodacin in a DNA-cleavage complex with Staphylococcus aureus DNA gyrase. bioRxiv, 2022; preprint. [Google Scholar] [CrossRef]
- Jones, J.A.; Virga, K.G.; Gumina, G.; Hevener, K.E. Recent advances in the rational design and optimization of antibacterial agents. MedChemComm 2016, 7, 1694–1715. [Google Scholar] [CrossRef]
- Badshah, S.L.; Ullah, A. New developments in non-quinolone-based antibiotics for the inhibiton of bacterial gyrase and topoisomerase IV. Eur. J. Med. Chem. 2018, 152, 393–400. [Google Scholar] [CrossRef]
- Chan, P.F.; Germe, T.; Bax, B.D.; Huang, J.; Thalji, R.K.; Bacqué, E.; Checchia, A.; Chen, D.; Cui, H.; Ding, X.; et al. Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase. Proc. Natl. Acad. Sci. 2017, 114, E4492–E4500. [Google Scholar] [CrossRef]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef]
- Imai, Y.; Hauk, G.; Quigley, J.; Liang, L.; Son, S.; Ghiglieri, M.; Gates, M.F.; Morrissette, M.; Shahsavari, N.; Niles, S.; et al. Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis. Nat. Chem. Biol. 2022, 18, 1236–1244. [Google Scholar] [CrossRef]
- Asha, M.K.; Debraj, D.; Prashanth, D.s.; Edwin, J.R.; Srikanth, H.S.; Muruganantham, N.; Dethe, S.M.; Anirban, B.; Jaya, B.; Deepak, M.; et al. In vitro anti-Helicobacter pylori activity of a flavonoid rich extract of Glycyrrhiza glabra and its probable mechanisms of action. J. Ethnopharmacol. 2013, 145, 581–586. [Google Scholar] [CrossRef]
- Orritt, K.M.; Feng, L.; Newell, J.F.; Sutton, J.N.; Grossman, S.; Germe, T.; Abbott, L.R.; Jackson, H.L.; Bury, B.K.L.; Maxwell, A.; et al. De novo design of type II topoisomerase inhibitors as potential antimicrobial agents targeting a novel binding region. RSC Med. Chem. 2022, 13, 831–839. [Google Scholar] [CrossRef]
- Domenech, P.; Kobayashi, H.; LeVier, K.; Walker Graham, C.; Barry Clifton, E. BacA, an ABC Transporter Involved in Maintenance of Chronic Murine Infections with Mycobacterium tuberculosis. J. Bacteriol. 2009, 191, 477–485. [Google Scholar] [CrossRef]
- Zhou, J.; Gelot, C.; Pantelidou, C.; Li, A.; Yücel, H.; Davis, R.E.; Färkkilä, A.; Kochupurakkal, B.; Syed, A.; Shapiro, G.I.; et al. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2021, 2, 598–610. [Google Scholar] [CrossRef]
- Bisacchi, G.S.; Manchester, J.I. A New-Class Antibacterial—Almost. Lessons in Drug Discovery and Development: A Critical Analysis of More than 50 Years of Effort toward ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. ACS Infect. Dis. 2015, 1, 4–41. [Google Scholar] [CrossRef]
- Grillot, A.-L.; Charifson, P.; Stamos, D.; Liao, Y.; Badia, M.; Trudeau, M. Bacterial Gyrase Inhibitors and Uses Thereof. WIPO Patent WO2002060879A3, 27 March 2003. [Google Scholar]
- Grillot, A.-L.; Tiran, A.L.; Shannon, D.; Krueger, E.; Liao, Y.; O’Dowd, H.; Tang, Q.; Ronkin, S.; Wang, T.; Waal, N.; et al. Second-Generation Antibacterial Benzimidazole Ureas: Discovery of a Preclinical Candidate with Reduced Metabolic Liability. J. Med. Chem. 2014, 57, 8792–8816. [Google Scholar] [CrossRef]
- Locher Christopher, P.; Jones Steven, M.; Hanzelka Brian, L.; Perola, E.; Shoen Carolyn, M.; Cynamon Michael, H.; Ngwane Andile, H.; Wiid Ian, J.; van Helden Paul, D.; Betoudji, F.; et al. A Novel Inhibitor of Gyrase B Is a Potent Drug Candidate for Treatment of Tuberculosis and Nontuberculosis Mycobacterial Infections. Antimicrob. Agents Chemother. 2015, 59, 1455–1465. [Google Scholar] [CrossRef]
- Talley Angela, K.; Thurston, A.; Moore, G.; Gupta Vipul, K.; Satterfield, M.; Manyak, E.; Stokes, S.; Dane, A.; Melnick, D. First-in-Human Evaluation of the Safety, Tolerability, and Pharmacokinetics of SPR720, a Novel Oral Bacterial DNA Gyrase (GyrB) Inhibitor for Mycobacterial Infections. Antimicrob. Agents Chemother. 2021, 65, e01208–e01221. [Google Scholar] [CrossRef]
- Basarab, G.S.; Manchester, J.I.; Bist, S.; Boriack-Sjodin, P.A.; Dangel, B.; Illingworth, R.; Sherer, B.A.; Sriram, S.; Uria-Nickelsen, M.; Eakin, A.E. Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents. J. Med. Chem. 2013, 56, 8712–8735. [Google Scholar] [CrossRef]
- Yule, I.A.; Czaplewski, L.G.; Pommier, S.; Davies, D.T.; Narramore, S.K.; Fishwick, C.W.G. Pyridine-3-carboxamide-6-yl-ureas as novel inhibitors of bacterial DNA gyrase: Structure based design, synthesis, SAR and antimicrobial activity. Eur. J. Med. Chem. 2014, 86, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, P.; Bruyère, T.; Blumstein, A.-C.; Bur, D.; Chambovey, A.; Ertel, E.A.; Gude, M.; Hubschwerlen, C.; Jacob, L.; Kimmerlin, T.; et al. Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents. J. Med. Chem. 2017, 60, 3755–3775. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.W.; Li, X.; Trzoss, M.; Bensen, D.C.; Chen, Z.; Lam, T.; Zhang, J.; Lee, S.J.; Hough, G.; Phillipson, D.; et al. Tricyclic GyrB/ParE (TriBE) Inhibitors: A New Class of Broad-Spectrum Dual-Targeting Antibacterial Agents. PLoS ONE 2013, 8, e84409. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Shi, H.; Zhou, M.; Ren, Q.; Zhu, W.; Zhang, W.; Zhang, Z.; Zhou, C.; Liu, Y.; Ding, X.; et al. Discovery of Pyrido [2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV. J. Med. Chem. 2020, 63, 9623–9649. [Google Scholar] [CrossRef] [PubMed]
- Nyerges, A.; Tomašič, T.; Durcik, M.; Revesz, T.; Szili, P.; Draskovits, G.; Bogar, F.; Skok, Ž.; Zidar, N.; Ilaš, J.; et al. Rational design of balanced dual-targeting antibiotics with limited resistance. PLoS Biol. 2020, 18, e3000819. [Google Scholar] [CrossRef]
- Durcik, M.; Nyerges, Á.; Skok, Ž.; Skledar, D.G.; Trontelj, J.; Zidar, N.; Ilaš, J.; Zega, A.; Cruz, C.D.; Tammela, P.; et al. New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens. Eur. J. Med. Chem. 2021, 213, 113200. [Google Scholar] [CrossRef]
- Cotman, A.E.; Durcik, M.; Benedetto Tiz, D.; Fulgheri, F.; Secci, D.; Sterle, M.; Možina, Š.; Skok, Ž.; Zidar, N.; Zega, A.; et al. Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Med. Chem. 2023, 66, 1380–1425. [Google Scholar] [CrossRef]
- McGarry, D.H.; Cooper, I.R.; Walker, R.; Warrilow, C.E.; Pichowicz, M.; Ratcliffe, A.J.; Salisbury, A.-M.; Savage, V.J.; Moyo, E.; Maclean, J.; et al. Design, synthesis and antibacterial properties of pyrimido [4,5-b]indol-8-amine inhibitors of DNA gyrase. Bioorganic Med. Chem. Lett. 2018, 28, 2998–3003. [Google Scholar] [CrossRef]
- Cooper, I.; Pichowicz, M.; Stokes, N. Compounds with Activity against Bacteria and Mycobacteria. WIPO Patent WO2016067009A1, 6 May 2016. [Google Scholar]
- Henderson, S.R.; Stevenson, C.E.M.; Malone, B.; Zholnerovych, Y.; Mitchenall, L.A.; Pichowicz, M.; McGarry, D.H.; Cooper, I.R.; Charrier, C.; Salisbury, A.-M.; et al. Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase. J. Antimicrob. Chemother. 2020, 75, 2835–2842. [Google Scholar] [CrossRef]
- Huang, X.; Guo, J.; Liu, Q.; Gu, Q.; Xu, J.; Zhou, H. Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes. MedChemComm 2018, 9, 1619–1629. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, J.; Cai, Z.; Ju, Y.; Xu, J.; Gu, Q.; Zhou, H. Identification of new building blocks by fragment screening for discovering GyrB inhibitors. Bioorganic Chem. 2021, 114, 105040. [Google Scholar] [CrossRef]
- Xue, W.; Li, X.; Ma, G.; Zhang, H.; Chen, Y.; Kirchmair, J.; Xia, J.; Wu, S. N-thiadiazole-4-hydroxy-2-quinolone-3-carboxamides bearing heteroaromatic rings as novel antibacterial agents: Design, synthesis, biological evaluation and target identification. Eur. J. Med. Chem. 2020, 188, 112022. [Google Scholar] [CrossRef]
- Xue, W.; Wang, Y.; Lian, X.; Li, X.; Pang, J.; Kirchmair, J.; Wu, K.; Han, Z.; You, X.; Zhang, H.; et al. Discovery of N-quinazolinone-4-hydroxy-2-quinolone-3-carboxamides as DNA gyrase B-targeted antibacterial agents. J. Enzym. Inhib. Med. Chem. 2022, 37, 1620–1631. [Google Scholar] [CrossRef]
- Chiriac, A.I.; Kloss, F.; Krämer, J.; Vuong, C.; Hertweck, C.; Sahl, H.-G. Mode of action of closthioamide: The first member of the polythioamide class of bacterial DNA gyrase inhibitors. J. Antimicrob. Chemother. 2015, 70, 2576–2588. [Google Scholar] [CrossRef]
- Lincke, T.; Behnken, S.; Ishida, K.; Roth, M.; Hertweck, C. Closthioamide: An Unprecedented Polythioamide Antibiotic from the Strictly Anaerobic Bacterium Clostridium cellulolyticum. Angew. Chem. 2010, 122, 2055–2057. [Google Scholar] [CrossRef]
- Phillips, J.W.; Goetz, M.A.; Smith, S.K.; Zink, D.L.; Polishook, J.; Onishi, R.; Salowe, S.; Wiltsie, J.; Allocco, J.; Sigmund, J.; et al. Discovery of Kibdelomycin, A Potent New Class of Bacterial Type II Topoisomerase Inhibitor by Chemical-Genetic Profiling in Staphylococcus aureus. Chem. Biol. 2011, 18, 955–965. [Google Scholar] [CrossRef]
- Lu, J.; Patel, S.; Sharma, N.; Soisson, S.M.; Kishii, R.; Takei, M.; Fukuda, Y.; Lumb, K.J.; Singh, S.B. Structures of Kibdelomycin Bound to Staphylococcus aureus GyrB and ParE Showed a Novel U-Shaped Binding Mode. ACS Chem. Biol. 2014, 9, 2023–2031. [Google Scholar] [CrossRef]
- Eakin Ann, E.; Green, O.; Hales, N.; Walkup Grant, K.; Bist, S.; Singh, A.; Mullen, G.; Bryant, J.; Embrey, K.; Gao, N.; et al. Pyrrolamide DNA Gyrase Inhibitors: Fragment-Based Nuclear Magnetic Resonance Screening to Identify Antibacterial Agents. Antimicrob. Agents Chemother. 2012, 56, 1240–1246. [Google Scholar] [CrossRef]
- Flatman Ruth, H.; Howells Alison, J.; Heide, L.; Fiedler, H.-P.; Maxwell, A. Simocyclinone D8, an Inhibitor of DNA Gyrase with a Novel Mode of Action. Antimicrob. Agents Chemother. 2005, 49, 1093–1100. [Google Scholar] [CrossRef]
- Sissi, C.; Vazquez, E.; Chemello, A.; Mitchenall, L.A.; Maxwell, A.; Palumbo, M. Mapping Simocyclinone D8 Interaction with DNA Gyrase: Evidence for a New Binding Site on GyrB. Antimicrob. Agents Chemother. 2010, 54, 213–220. [Google Scholar] [CrossRef]
- Edwards, M.J.; Williams, M.A.; Maxwell, A.; McKay, A.R. Mass Spectrometry Reveals That the Antibiotic Simocyclinone D8 Binds to DNA Gyrase in a “Bent-Over” Conformation: Evidence of Positive Cooperativity in Binding. Biochemistry 2011, 50, 3432–3440. [Google Scholar] [CrossRef] [PubMed]
- Hearnshaw, S.J.; Edwards, M.J.; Stevenson, C.E.; Lawson, D.M.; Maxwell, A. A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight into the Mechanism of Inhibition. J. Mol. Biol. 2014, 426, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Oppegard, L.M.; Hamann, B.L.; Streck, K.R.; Ellis, K.C.; Fiedler, H.-P.; Khodursky, A.B.; Hiasa, H. In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob. Agents Chemother. 2009, 53, 2110–2119. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Nissink, J.W.M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol. 2018, 13, 36–44. [Google Scholar] [CrossRef]
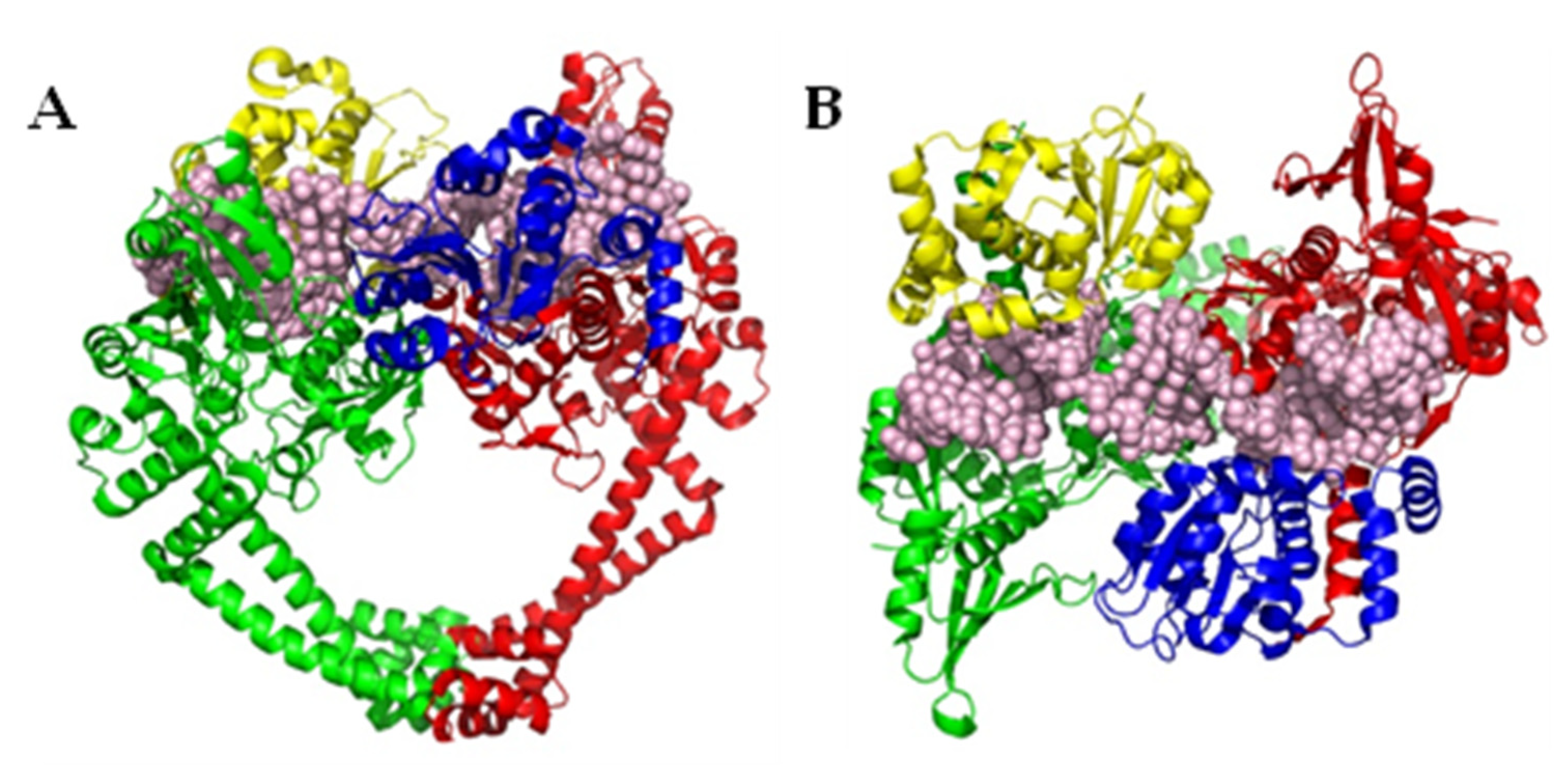
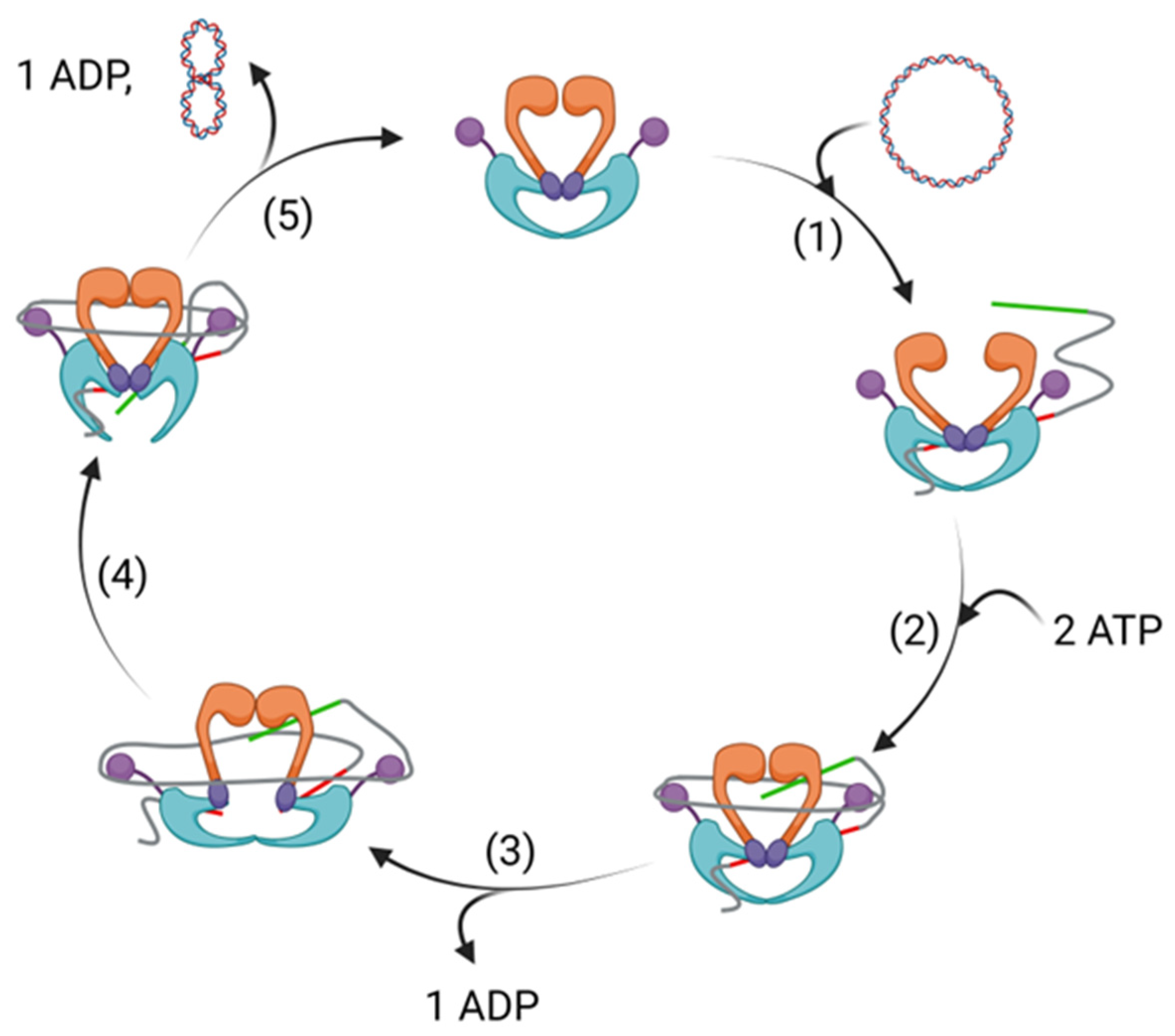
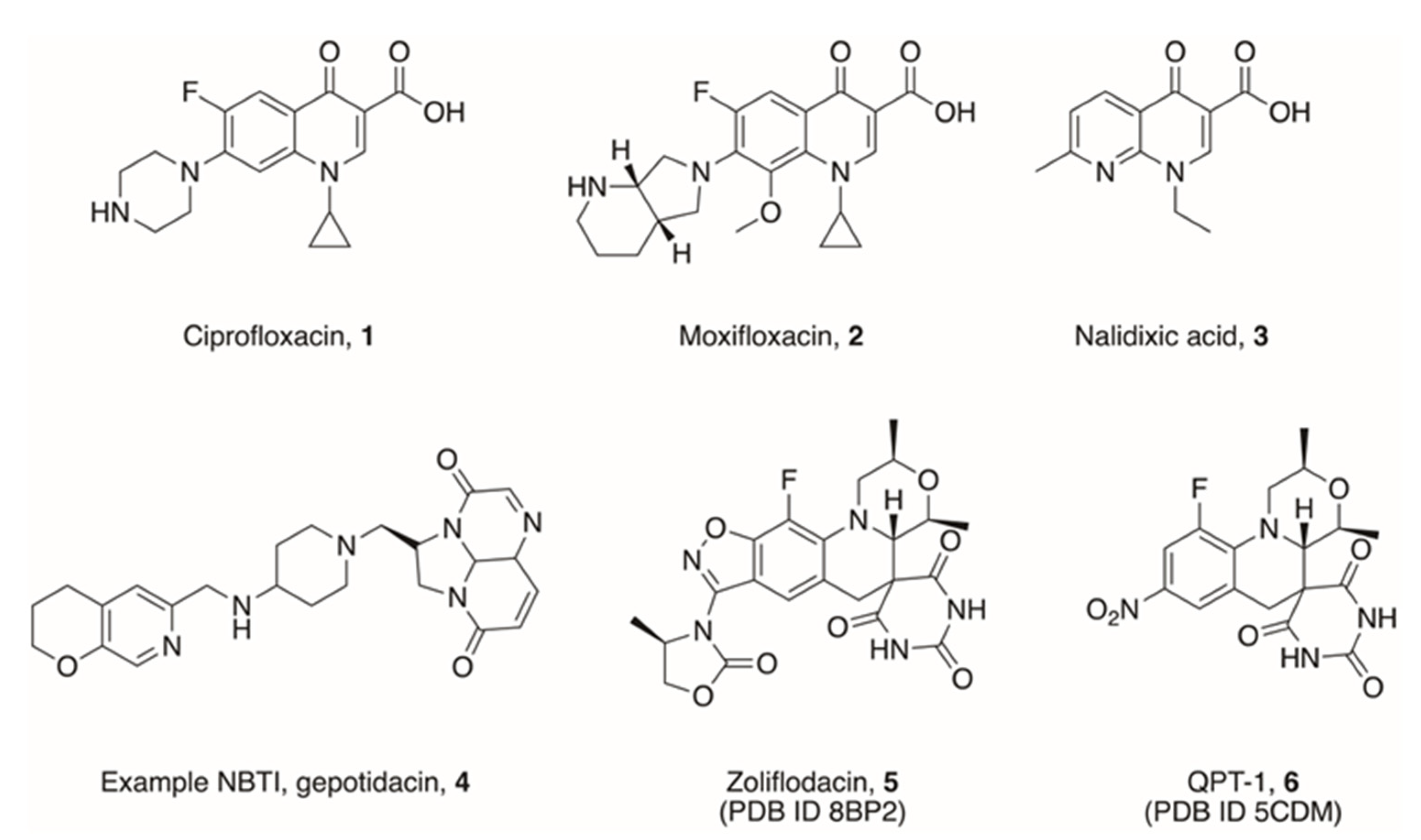
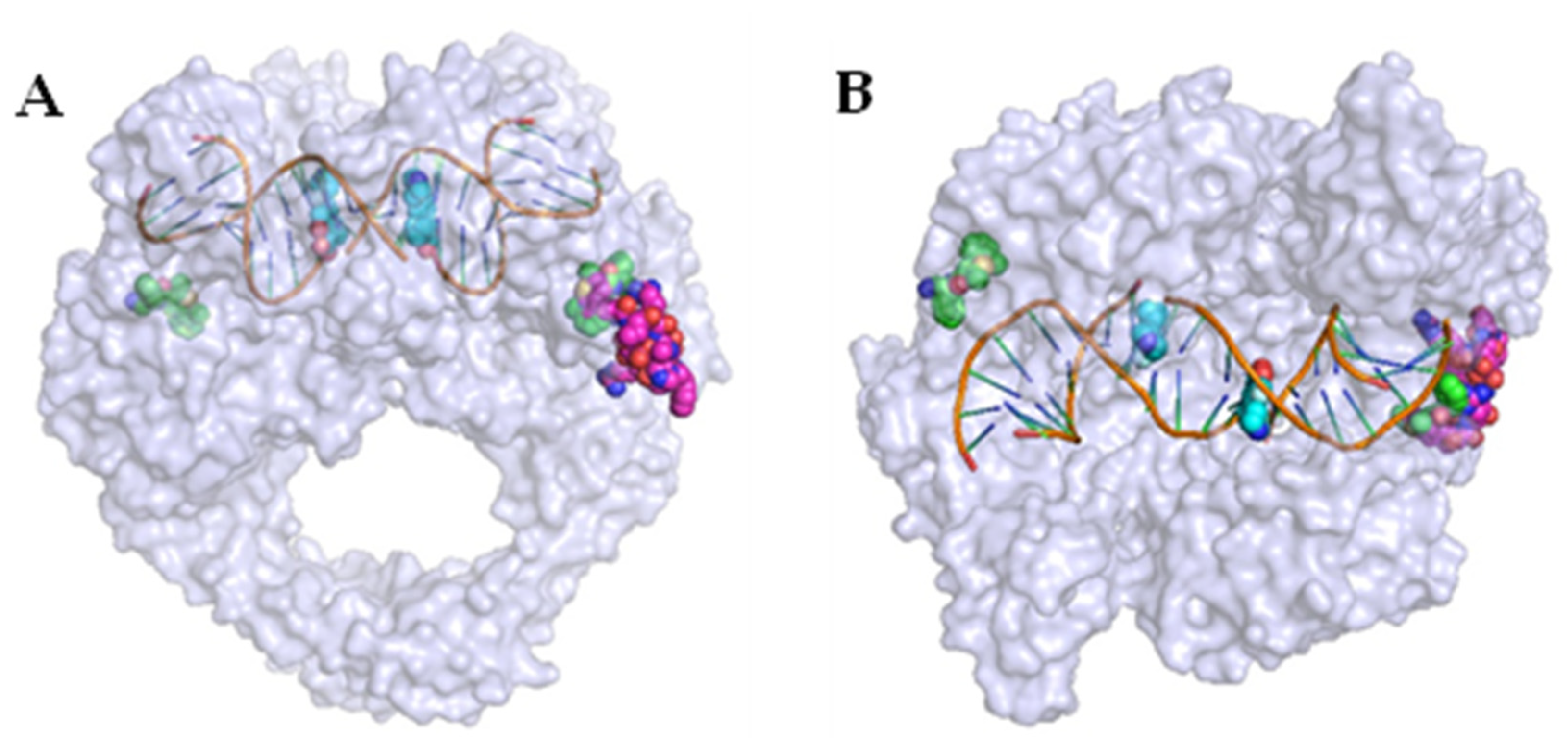
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grossman, S.; Fishwick, C.W.G.; McPhillie, M.J. Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. Pharmaceuticals 2023, 16, 261. https://doi.org/10.3390/ph16020261
Grossman S, Fishwick CWG, McPhillie MJ. Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. Pharmaceuticals. 2023; 16(2):261. https://doi.org/10.3390/ph16020261
Chicago/Turabian StyleGrossman, Scott, Colin W. G. Fishwick, and Martin J. McPhillie. 2023. "Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV" Pharmaceuticals 16, no. 2: 261. https://doi.org/10.3390/ph16020261
APA StyleGrossman, S., Fishwick, C. W. G., & McPhillie, M. J. (2023). Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. Pharmaceuticals, 16(2), 261. https://doi.org/10.3390/ph16020261





