2.1. Defining the Chemical Space Occupied by Terpenes
The authors’ primary source of data was the COCONUT database, created and curated by the Institute for Inorganic and Analytical Chemistry, Friedrich-Schiller University, Germany [
3]. This resource, first made available in 2021, is an online repository of nearly half a million natural products, annotated with several types of physicochemical and topographical information, which compiles the information of over 50 databases of natural molecules. Previous works on terpenes have used commercial databases [
5]; however, the authors believe that a public resource such as the one we describe herein is a positive trend towards the democratization of data and science as a whole. Furthermore, previous works have provided 11 physicochemical properties for their dataset [
5], while we provide over 40 here.
Starting from the COCONUT database (400k+ molecules), the authors initially created a subset comprising solely the chemical SuperClass “Lipids and lipid-like molecules”, which comprises all molecules of lipidic nature, in which terpenes are included. Among this subset (99,696 molecules, distributed according to
Supplementary Table S1), the authors further selected the classes “Monoterpenes, “Sesquiterpenes”, “Diterpenes”, “Sesterterpenes”, “Triterpenes”, “Polyterpenes”, “Sesquaterpenes”, “Terpene glycosides” and “Terpene lactones” in order to include all terpene classes. This yielded the final dataset, in which are included 59,833 molecules. The authors acknowledge that many molecules in the “Terpene glycosides” and “Terpene lactones” subclasses can be, from a phytochemical point of view, part of any of the remaining subclasses. However, for the sake of clarity and access to as many molecules as possible, the authors followed the hierarchical criteria enforced by COCONUT.
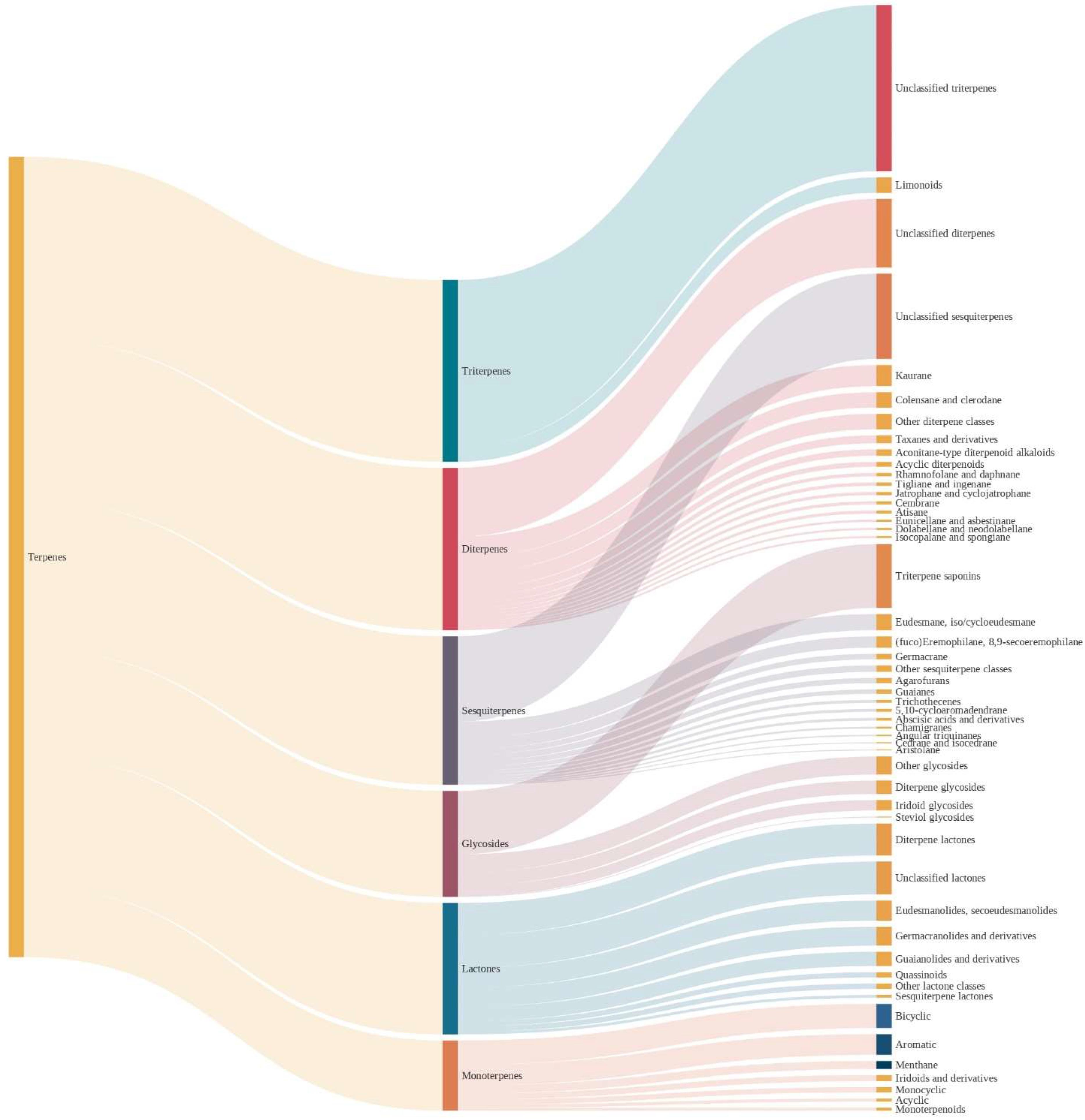
Figure 1 depicts the distribution of the molecules in the dataset across different terpene classes. As shown, the major classes were triterpenes (22.1%, 13,245 molecules), followed by diterpenes (19.7%, 11,814 molecules), and the least expressive class was polyterpenes (0.1%, 48 molecules). Before advancing, it is important to briefly review the organization of COCONUT in terms of chemical ontology.
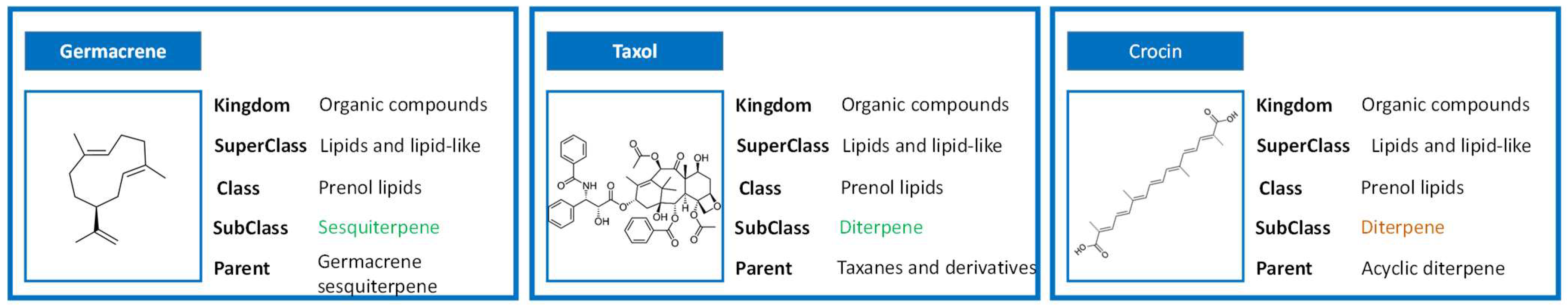
After a registry is created, several parameters are automatically calculated based on well-established algorithms. For example, in the case of the chemical classification, Classyfire is used [
6]. This is a remarkable resource, as this is a purely structure-based chemical taxonomy software that uses chemical structures and structural features as inputs to assign a given molecule to a hierarchical-based taxonomy. This new chemical taxonomy consists of up to 11 different levels (including Kingdom, SuperClass, Class, SubClass, etc.), with each of the categories defined by unambiguous and computable structural rules. This approach is both powerful and dangerous in the case of the natural products chemical space. On the one hand, it is possible to easily classify hundreds of thousands of molecules based on clearly established rules, instead of having to rely in biosynthetic studies. As an example, when we run taxol into Classifyre, we get the classification: Kingdom: Organic compounds > Superclass: Lipids and lipid-like > Class: Prenol Lipids > Subclass: Diterpene > Classification: Taxanes and derivatives (
Figure 2). From a phytochemical point of view, one would not usually include terpenes in the class of lipids (older textbooks would, however), but it is true they are of lipidic nature, so there is no surprise here. The most important levels of taxonomical classification for a natural products chemist are Subclass: Diterpenes and Parent: Taxanes. This works fine, as they can get both the natural product family and its nucleus/immediate family. To be able to have this degree of information, even in cases where biosynthetic studies are not yet available, is remarkable.
On the other hand, the disadvantage of this rule-based classification is that natural products are a wonderful source of rule breakers and deviants. Take, for instance, crocin, from
Crocus sativus. From a phytochemical point of view, crocin is a (di)apocarotenoid, as its loss of isoprene units from both ends renders it a chemical entity with C
20. Naturally, a rule-based algorithm has no way to know that from a biosynthetically point of view, this molecule is a carotenoid (typically C
40) that has lost carbons. As so, it is classified as “acyclic diterpenoid” (compounds comprising four consecutive isoprene units that do not contain a cycle,
Figure 2).
Nevertheless, despite these limitations, it is helpful to have a tool to quickly classify millions of molecules, with the advances in technology allowing to fine-tune the rules towards increasingly accurate performances. However, some caution must be taken for structures that may be incorrectly classified. In
Figure 3, the authors present the different levels of chemical taxonomy for the dataset used in this work.
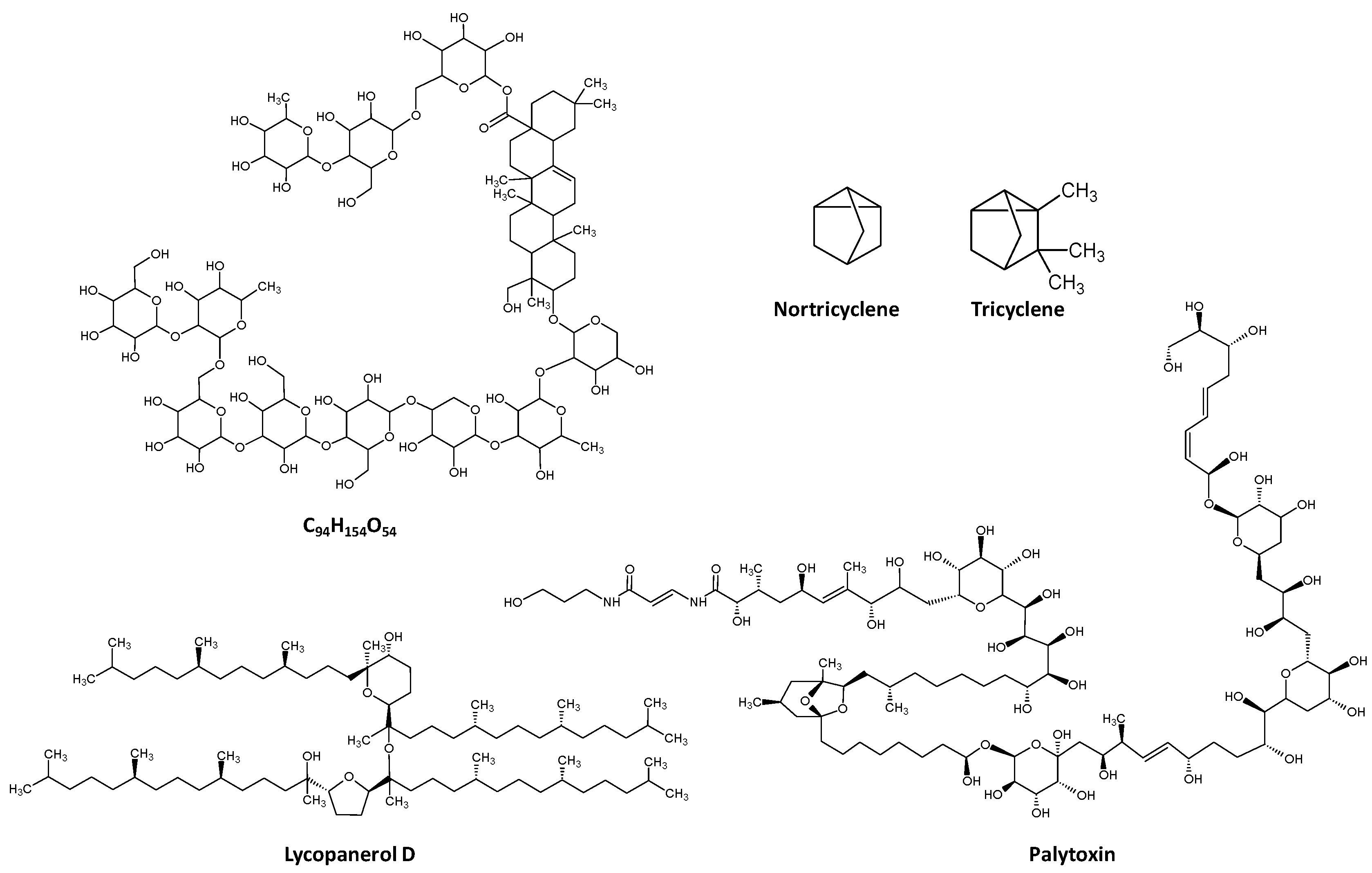
Considering all terpenes studied here, the average molecular weight was 534.9 Da (min: 94.2, nortricyclene and max 2680.1, palytoxin,
Figure 4).
Nortricyclene does not comply with the standard monoterpene definition given its odd number of carbons, 7; however, this classification arises from its structural relation to tricyclene (C
10H
16,
Figure 4). Palytoxin, from Palythoa corals, is one of the largest non-polymeric natural products described and one of the most poisonous non-protein molecules known, second only to maitotoxin in terms of mice toxicity [
7]. It is a terpene glycoside and displays 8 double bonds, 40 hydroxyl groups and 64 chiral centres, which gives rise to over 1021 possible stereoisomers.
The distribution of MW across all terpenes is presented in
Figure 1. From a phytochemical point of view, the different terpene subclasses are grouped based on their biosynthesis and, consequently, the number of isoprene units in their structure. For this reason, it is obvious that marked differences in molecular weight can be found across distinct subclasses. For the sake of compiling the molecular weight intervals in each terpene class,
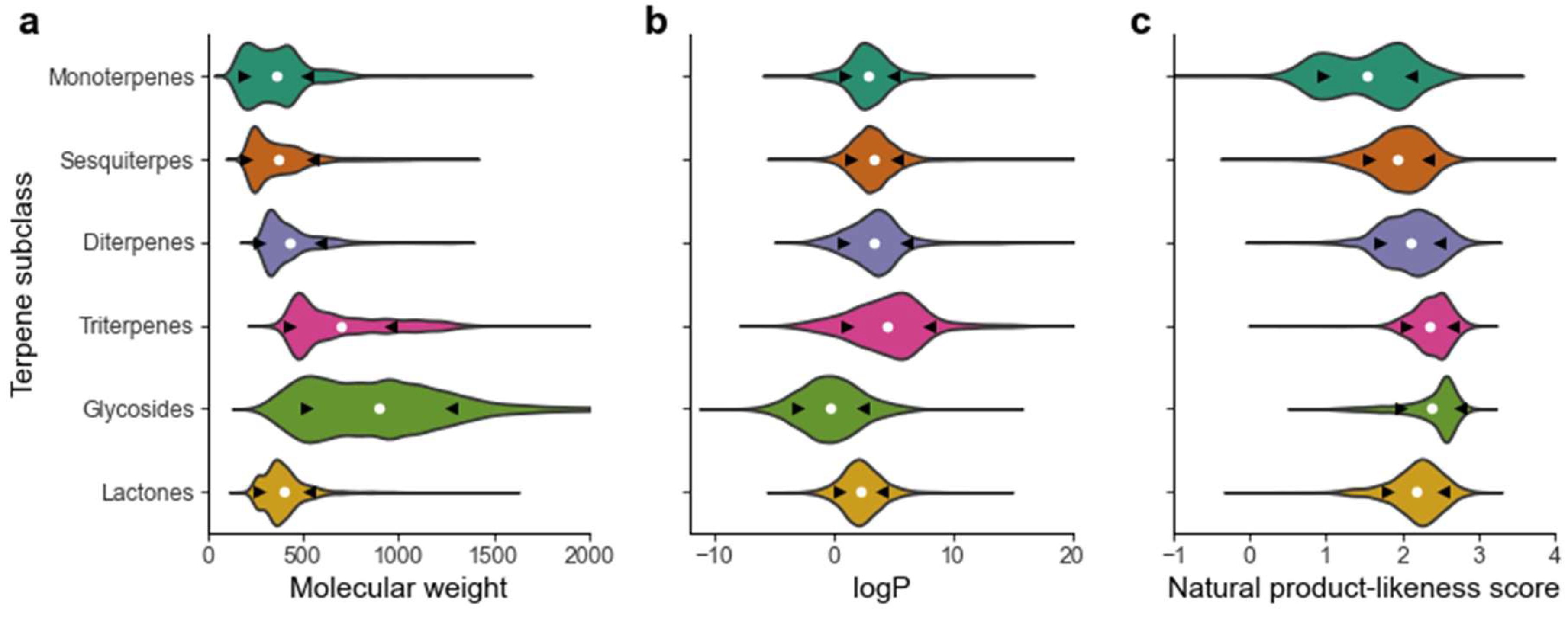
Figure 5a shows the molecular weight distribution as a function of the subclass.
The average molecular weight increases with the increasing number of carbons in the monoterpene < sesquiterpene < diterpene < triterpene order. Terpene glycosides present the widest range of molecular weight, an expected consequence of the occurrence of a different number of sugars and their diverse identity.
The partition coefficient (P), which describes the suitability of a neutral molecule to dissolve in immiscible biphasic systems comprised of lipids and water, is a pivotal characteristic that has a marked impact on the “druggability” of a compound, as extreme values, either too negative or too positive, can hinder its adequate pharmacokinetics in the human body. For example, a study comprising a dataset of 4 leading pharmaceutical companies showed that among 812 molecules studied as drug candidates, the average logP was 3.2. As is shown in
Figure 1, the average logP of terpenes is approximately 3, with a molecule with formula C
94H
154O
54 (no common name) and the tetraterpene ether lycopanerol H being the molecules in the lower and upper limit (−10.3 and 50.2, respectively,
Figure 4).
As shown in the class-by-class analysis (
Figure 5b), the average value increased in the monoterpene < sesquiterpene < diterpene < triterpene order, the latter being the subclass of terpenes with the highest average logP, 4.6. Terpene glycosides was the only class presenting a negative average logP, as expected given their heterosidic nature that greatly increases their polarity.
The natural product-likeness score is a descriptor that takes into account several structural and topographical features and tries to quantify the “degree of naturalness” of a given molecule [
8]. Naturally occurring molecules, owing to their stereochemical complexity and diversified ring systems, usually afford higher values, whereas compounds arising from synthetic chemistry display lower values. As it is shown in
Figure 5c, the average value for terpenes was 2.1. Monoterpenes, the simplest class from a structural point of view, had the lowest average natural product-likeness score, 1.5, owing to their simpler chemical structures. This value increases with the chemical complexity of the subclasses in the monoterpene > sesquiterpene > diterpene > triterpene order.
Several terpenes are currently being used in the drugs’ arsenal against a number of diseases. This includes, for example, paclitaxel for cancer, artemisinin for malaria and manoalide for inflammatory processes.
In the process of pharmaceutical development, it is important to take into account the “drug-likeness” of a given molecule in order to identify unlikely candidates as upstream in the development process as possible. One of the most widely spread set of “rules of thumb” are the Lipinski’s rule of five [
9], which postulate that ideal drugs would have a number of hydrogen bond donors ≤ 5, number of hydrogen bond acceptors ≤ 10, molecular mass ≤ 500 Da and logP ≤ 5. Although this set of anecdotal rules was postulated a long time ago, most of its basis is still widely used today: in a study with over 800 drug candidates, only 24% had a molecular weight > 500 Da and only 15% of the candidates had a logP > 5. Notably, only 7.6% of all the molecules violated both rules simultaneously [
10].
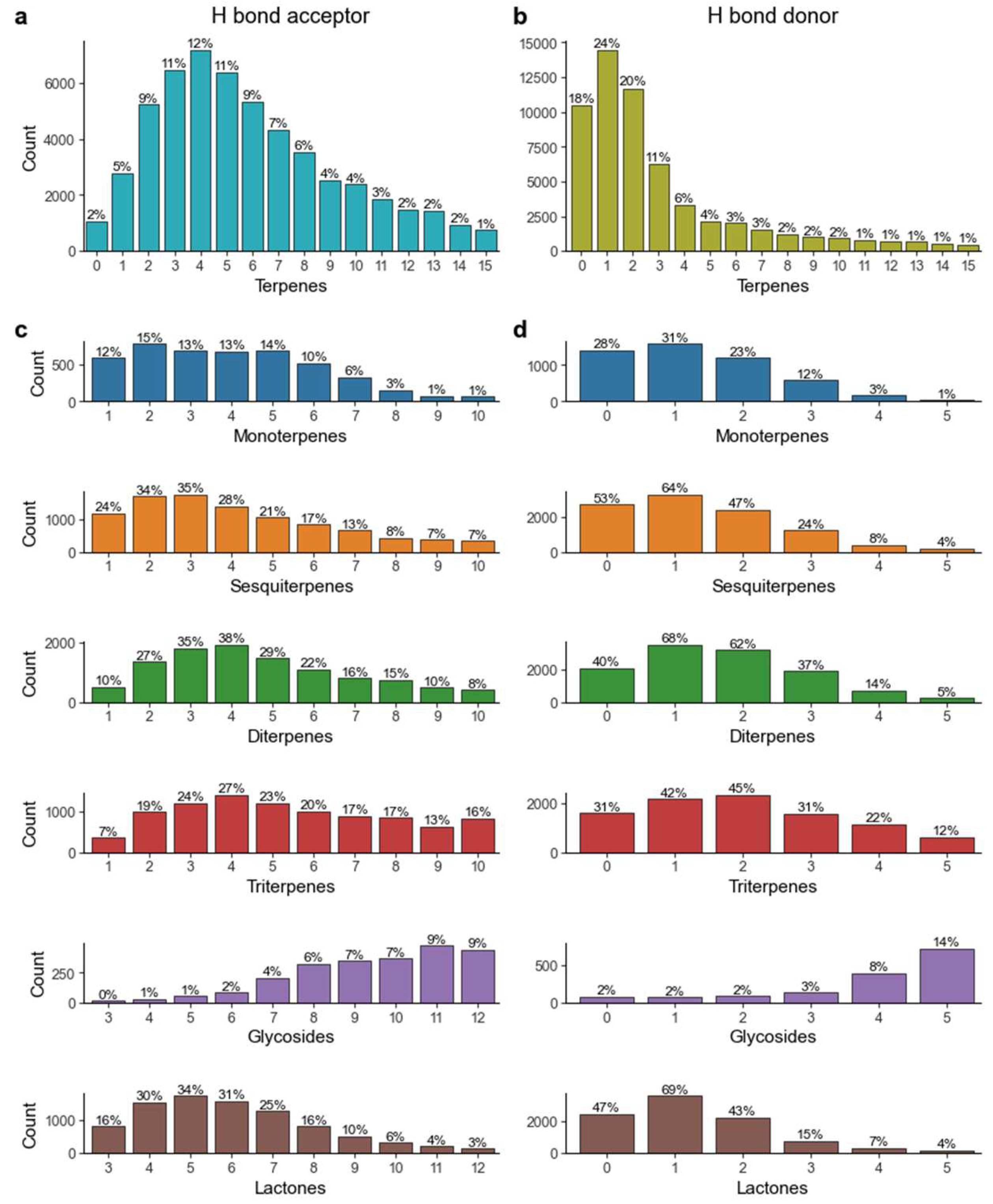
We also investigated the rate of violation of the terpenes chemical space in terms of the number of hydrogen bond donors and acceptors. As is shown in
Figure 6, 78.7% of all terpenes had 10 hydrogen bond acceptors or less, while 81% had 5 hydrogen bond donors or less, thus complying with Lipinski’s rule of five. When studying these parameters on a subclass level, we see that in a general way, monoterpenes, sesquiterpenes, diterpenes and terpene lactones all present over 85% of molecules with 10 or less hydrogen bond acceptors and 70% of molecules with 5 or less hydrogen bond donors. Triterpenes are slightly different, as they exhibit 70% and 59% for the same parameters, respectively. The most distinct set of molecules are terpene glycosides, in which only 18.7% of molecules observe these rules.
After this, the authors were interested in quantifying how many molecules violated any of the postulated Lipinski rules. As shown in
Figure 7, 41.6% of terpenes do not present any violation. While the most common deviation is 1 violation (22.8%), it is of note that there is an almost equivalent count of molecules with 2, 3 and 4 violations, which cumulatively account for 35% of all molecules. The authors further investigated the number of violations across different terpenes subclasses. As seen in
Figure 7, there are remarkable differences.
Monoterpenes and sesquiterpenes present a similar and nearly superimposable profile and distribution of number of violations. Triterpenes are unique in the way they present the lowest percentage of compounds with 0 violations, only 5.2%, while nearly 60% present 1 or 2 violations. Equally unique are terpene glycosides, which include over 50% of molecules with 4 violations, a unique trait, considering that the second class in this category is sesquiterpenes, with only 1.9%. Likewise, terpene lactones are a rather particular class, as nearly 80% of the molecules in this class present no violations of Lipinski’s rule of five. The results presented so far provide a valuable source of information that can be used to select the terpene subclasses that present a chemical profile more compatible to future drug development frameworks.
2.2. Clustering the Terpene Chemical Space
In high dimensional data, clustering can be an effective method to make sense of heterogeneous data. In the case of chemical information, it can be used to identify the most important physicochemical parameters that allow grouping different families of molecules. In the specific case of terpenes, this can be useful where a new chemical entity is found, and its sorting in a given class is challenging owing to inexistent biosynthetic data, mostly in the case of molecules with an odd number of carbons.
In the case of terpenes, the authors were interested in studying if their physicochemical parameters could per se help position a given molecule in a particular subclass. To this end, and given the high dimensionality of the data, the authors employed several dimensionality reduction methods, namely PCA, UMAP, t-SNE, FastICA and Kernel PCA. After this, the authors tried to cluster the data, both the original data along with the reduced form, by k-means and hierarchical clustering and benchmarked the results.
To evaluate the quality of the clustering, the authors considered the following metrics. The homogeneity score (shorthand: Homo) measures if each cluster includes only the datapoints that are members of a single class. The completeness score (shorthand: Compl) shows if all datapoints of a given class are members of the same cluster. The harmonic mean between the homogeneity and completeness scores is computed by v-measure (shorthand: V-meas) [
11]. The Rand index [
12] shows the similarity between two label assignments, and the adjusted Rand index (shorthand: ARI) is a version of the Rand index that corrects for chance. The agreement between two label assignments, ignoring permutations, is computed by mutual information. Here, we used adjusted mutual information (shorthand: AMI), which is normalized against the chance version of it. The above-mentioned measures fall within the range 0 to 1, with score 1 meaning perfectly labelling.
The silhouette coefficient [
13], however, ranges from –1 to 1 and measures how similar a datapoint is to its own cluster rather than other clusters. Higher silhouette values show that a datapoint is highly similar to its own cluster and poorly similar to neighbouring clusters.
In addition to the mentioned metrics, the authors measured the execution time of different methods running on Apple M1 CPU.
Supplementary Tables S4 and S5 demonstrate, in detail, the results of carrying out k-means and agglomerative clustering on imbalanced and balanced datasets, with and without applying dimensionality reduction methods with the parameters shown in
Supplementary Table S3.
Of those results, the best ones are shown in
Table 1.
In the case of k-means on imbalanced data, PCA provides metric values ≤ 0.33 with 11 principal components, which are low-performing values, as they should be as close to 1 as possible. When compared to PCA, UMAP (n_neighbors = 45, min_dist = 0.1,
Supplementary Table S3) did not provide any substantial increase in performance, except for silhouette, which yielded an increase of 0.21. t-SNE did not provide any improvement over UMAP, despite the multiple settings tried; the same was true for FastICA and Kernal PCA, except for the silhouette metric. Kernal PCA afforded the best silhouette, 0.50, which is still low and, hence, not suitable for our clustering purposes. Considering these results, the authors wondered if the reason for this poor performance could be related to some degree of imbalance in the data.
To account for this, the authors applied random oversampling (ROS) for data balancing, after which all algorithms were run once more. Note that since it was important for the authors not to generate synthetic data, they did not employ synthetic sampling approaches. As shown in the table, data balancing did not improve the results; the three subclasses we fed to the clustering algorithm, i.e., “Triterpenes”, “Diterpenes” and “Monoterpenes”, had the occurrences of 13245, 11814 and 5115, respectively.
The same procedure as above was repeated by hierarchical (agglomerative) clustering.
Table 1 shows that applying UMAP (UMAP7), before clustering, provides the best results when the authors cluster on imbalanced data. These results are almost the same as the ones obtained by UMAP7 plus k-means, with the difference that the latter is 70% faster. Similar to before, data balancing could not help improve the results. Therefore, the best results are obtained by first applying UMAP7 to reduce the dimensionality of the data (the original imbalanced one) and then k-means to cluster it. Note that even these methods could not provide a satisfactory result in clustering the three target subclasses.
2.3. Classification
The authors were also interested in assessing the suitability of this data for classification purposes. This is an important task, as it may help classify novel molecules to a given terpene subclass in cases where biosynthetic studies are not yet available.
Several classification methods were fitted, namely LightGBM, kNN, random forests, Gaussian naïve Bayes and MLP, with their default parameters in scikit-learn on the terpenes training data, which was obtained after the data cleansing described in the “Methods” section; then, we validated them by the 5-fold cross-validation technique.
To evaluate the classification methods, we measured the following metrics, which range from 0 to 1, with the values closer to 1 showing better performance. Considering the fact that the target feature, i.e., “chemicalSubClass”, is imbalanced, the authors used the weighted version of the metrics.
Precision shows if the classifier is able to not label negative samples as positive. Recall shows if the classifier can find all the positive samples. The F1 score is calculated by taking the harmonic mean of the precision and recall. Balanced accuracy is the average of recall scores for each class. Area under curve of the receiver operating characteristic (shorthand: ROC-AUC) measures the performance, as the discrimination threshold of the classifier varies. The authors set the “multi_class” parameter in the implementation as “ovo”, which stands for one-vs-one and calculates the average area under curve (AUC) for all pairs of classes.
Supplementary Table S6 shows the results of cross-validation on the terpenes training data. Since LightGBM and random forest performed similarly in producing the best results, we optimized hyperparameters of both methods, employing randomized search, and cross-validated them again on the training data. Carrying out with the optimized hyperparameters (
Supplementary Table S7), it is shown in
Supplementary Table S8 that LightGBM performed better, however slightly, while running 4.7 times faster. It reaches to 91% balanced accuracy and 99% ROC-AUC on the validation data. Based on what we described, LightGBM was chosen to classify the terpenes subclasses.
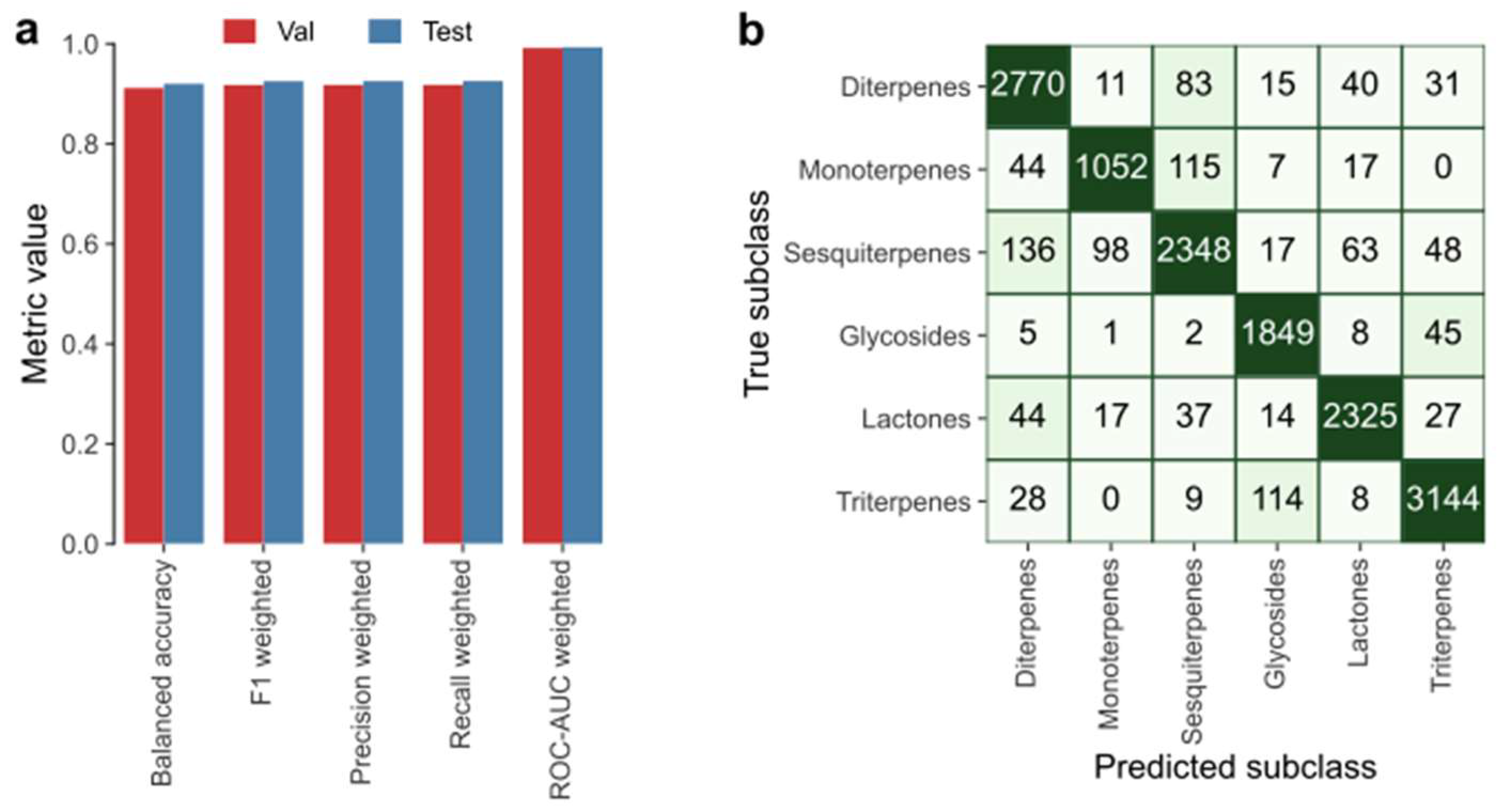
In
Figure 8, the evaluation results of applying LightGBM on the terpenes data is depicted.
Figure 8a shows that at least 0.91 was approached (out of 1.00) by all the considered metrics; for instance, balanced accuracy and weighted F1 score on the test data are 0.92 and 0.93, respectively.
Figure 8b shows the confusion matrix obtained by applying LightGBM on the test data, from which we could calculate the weighted metrics shown in
Figure 8a, since it denotes true positives, false positives, true negatives and false negatives for each subclass. Given the positive results obtained, the authors succeeded in applying a classifier capable of classification of terpene molecules on the grounds of their physicochemical properties.