Epigenetic Regulation of Macrophage Polarization in Cardiovascular Diseases
Abstract
1. Introduction
2. Macrophage Heterogeneity and Functions
3. Role of Macrophage Polarization in the Pathophysiology of CVDs
4. Epigenetic Regulation of Macrophage Polarization and CVDs
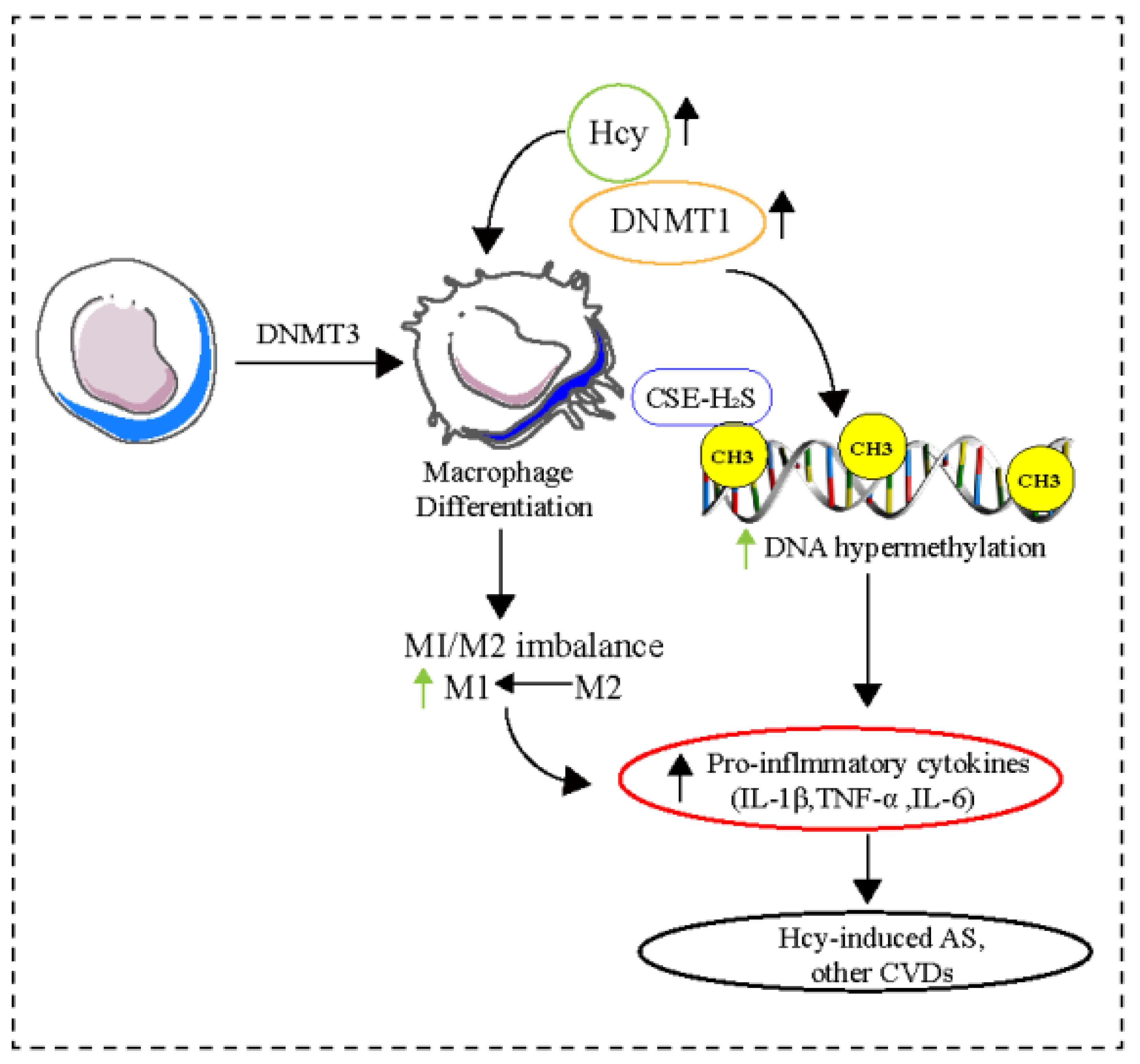
4.1. DNA Methylation
4.2. Histone Modifications
4.3. RNA Regulation
4.4. Noncoding RNA Regulation
4.4.1. miRNA-Mediated Macrophage Polarization and CVDs
4.4.2. lncRNA-Mediated Macrophage Polarization and CVDs
4.4.3. circRNA-Mediated Macrophage Polarization and CVDs
5. Other Mechanisms Underlying Macrophage Activation in CVDs
6. Macrophage Epigenetics as a Potential Therapeutic Target for CVDs
7. Concluding Remarks and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Burden of Cardiovascular Diseases Collaboration; Roth, G.A.; Johnson, C.O.; Abate, K.H.; Abd-Allah, F.; Ahmed, M.; Alam, K.; Alam, T.; Alvis-Guzman, N.; Ansari, H. The Burden of Cardiovascular Diseases Among US States, 1990–2016. JAMA Cardiol. 2018, 3, 375–389. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://www.euro.who.int/en/health-topics/noncommunicablediseases/cardiovascular-diseases (accessed on 21 October 2021).
- Haddad, F.; Doyle, R.; Murphy, D.J.; Hunt, S.A. Right Ventricular Function in Cardiovascular Disease, Part II. Circulation 2008, 117, 1717–1731. [Google Scholar] [CrossRef]
- Scholes, S.; Ng Fat, L.; Mindell, J. Trends in Cardiovascular Disease Risk Factors by Body Mass Index Category among Adults in England 2003-18: Analysis of Repeated Cross-Sectional National Health Surveys. medRxiv 2020. [Google Scholar] [CrossRef]
- Duerr, G.D.; Dewald, D.; Schmitz, E.J.; Verfuerth, L.; Keppel, K.; Peigney, C.; Ghanem, A.; Welz, A.; Dewald, O. Metallothioneins 1 and 2 Modulate Inflammation and Support Remodeling in Ischemic Cardiomyopathy in Mice. Mediat. Inflamm. 2016, 2016, 7174127. [Google Scholar] [CrossRef]
- Li, K.; Xu, W.; Guo, Q.; Jiang, Z.; Wang, P.; Yue, Y.; Xiong, S. Differential Macrophage Polarization in Male and Female BALB/c Mice Infected With Coxsackievirus B3 Defines Susceptibility to Viral Myocarditis. Circ. Res. 2009, 105, 353–364. [Google Scholar] [CrossRef]
- Takeda, Y.; Costa, S.; Delamarre, E.; Roncal, C.; de Leite Oliveira, R.; Squadrito, M.L.; Finisguerra, V.; Deschoemaeker, S.; Bruyère, F.; Wenes, M.; et al. Macrophage Skewing by Phd2 Haplodeficiency Prevents Ischaemia by Inducing Arteriogenesis. Nature 2011, 479, 122–126. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Marian, A.J.; Belmont, J.; Ali, J.M.; Hugh, W.; Christine, S. Strategic Approaches to Unraveling Genetic Causes of Cardiovascular Diseases. Circ. Res. 2011, 108, 1252–1269. [Google Scholar] [CrossRef]
- Komal, S.; Zhang, L.-R.; Han, S.-N. Potential Regulatory Role of Epigenetic RNA Methylation in Cardiovascular Diseases. Biomed. Pharmacother. 2021, 137, 111376. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic Modifications. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Lafuse, W.P.; Wozniak, D.J.; Rajaram, M.V.S. Role of Cardiac Macrophages on Cardiac Inflammation, Fibrosis and Tissue Repair. Cells 2020, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Clauss, S.; Xiao, L.; Aguirre, A.D.; King, K.R.; Hanley, A.; Hucker, W.J.; Wülfers, E.M.; Seemann, G.; Courties, G.; et al. Macrophages Facilitate Electrical Conduction in the Heart. Cell 2017, 169, 510–522.e20. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2018, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Laskin, D.L.; Sunil, V.R.; Gardner, C.R.; Laskin, J.D. Macrophages and Tissue Injury: Agents of Defense or Destruction? Annu. Rev. Pharmacol. Toxicol. 2011, 51, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 Macrophages and the Th1/Th2 Paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef]
- Orekhov, A.N.; Orekhova, V.A.; Nikiforov, N.G.; Myasoedova, V.A.; Grechko, A.V.; Romanenko, E.B.; Zhang, D.; Chistiakov, D.A. Monocyte Differentiation and Macrophage Polarization. Vessel Plus 2019, 3, 10. [Google Scholar] [CrossRef]
- Petrova, T.; Zhang, J.; Nanda, S.K.; Figueras-Vadillo, C.; Cohen, P. HOIL-1-Catalysed, Ester-Linked Ubiquitylation Restricts IL-18 Signaling in Cytotoxic T Cells but Promotes TLR Signalling in Macrophages. FEBS J. 2021, 288, 5909–5924. [Google Scholar] [CrossRef]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef]
- Epelman, S.; Lavine, K.J.; Beaudin, A.E.; Sojka, D.K.; Carrero, J.A.; Calderon, B.; Brija, T.; Gautier, E.L.; Ivanov, S.; Satpathy, A.T.; et al. Embryonic and Adult-Derived Resident Cardiac Macrophages Are Maintained through Distinct Mechanisms at Steady State and during Inflammation. Immunity 2014, 40, 91–104. [Google Scholar] [CrossRef]
- Bajpai, G.; Bredemeyer, A.; Li, W.; Zaitsev, K.; Koenig, A.L.; Lokshina, I.; Mohan, J.; Ivey, B.; Hsiao, H.-M.; Weinheimer, C.; et al. Tissue Resident CCR2- and CCR2+ Cardiac Macrophages Differentially Orchestrate Monocyte Recruitment and Fate Specification Following Myocardial Injury. Circ. Res. 2019, 124, 263–278. [Google Scholar] [CrossRef]
- Schultze, J.L.; Schmieder, A.; Goerdt, S. Macrophage Activation in Human Diseases. Semin. Immunol. 2015, 27, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.J.; Sheedy, F.J.; Fisher, E.A. Macrophages in Atherosclerosis: A Dynamic Balance. Nat. Rev. Immunol. 2013, 13, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.; Liu, D.; Yu, L.; Xue, B.; Shi, H. Epigenetic Regulation of Macrophage Polarization by DNA Methyltransferase 3b. Mol. Endocrinol. 2014, 28, 565–574. [Google Scholar] [CrossRef]
- Tan, R.P.; Ryder, I.; Yang, N.; Lam, Y.T.; Santos, M.; Michael, P.L.; Robinson, D.A.; Ng, M.K.; Wise, S.G. Macrophage Polarization as a Novel Therapeutic Target for Endovascular Intervention in Peripheral Artery Disease. JACC Basic Transl. Sci. 2021, 6, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.X.; Gustafson, H.H.; Jackson, D.L.; Pun, S.H.; Trapnell, C. Trajectory Analysis Quantifies Transcriptional Plasticity during Macrophage Polarization. Sci. Rep. 2020, 10, 12273. [Google Scholar] [CrossRef]
- Williams, J.W.; Giannarelli, C.; Rahman, A.; Randolph, G.J.; Kovacic, J.C. Macrophage Biology, Classification, and Phenotype in Cardiovascular Disease. J. Am. Coll. Cardiol. 2018, 72, 2166–2180. [Google Scholar] [CrossRef] [PubMed]
- Verreck, F.A.W.; de Boer, T.; Langenberg, D.M.L.; van der Zanden, L.; Ottenhoff, T.H.M. Phenotypic and Functional Profiling of Human Proinflammatory Type-1 and Anti-Inflammatory Type-2 Macrophages in Response to Microbial Antigens and IFN-γ- and CD40L-Mediated Costimulation. J. Leukoc. Biol. 2006, 79, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Komal, S.; Komal, N.; Mujtaba, A.; Wang, S.-H.; Zhang, L.-R.; Han, S.-N. Potential Therapeutic Strategies for Myocardial Infarction: The Role of Toll-like Receptors. Immunol. Res. 2022, 70, 607–623. [Google Scholar] [CrossRef]
- Marchi, L.F.; Sesti-Costa, R.; Ignacchiti, M.D.C.; Chedraoui-Silva, S.; Mantovani, B. In Vitro Activation of Mouse Neutrophils by Recombinant Human Interferon-Gamma: Increased Phagocytosis and Release of Reactive Oxygen Species and pro-Inflammatory Cytokines. Int. Immunopharmacol. 2014, 18, 228–235. [Google Scholar] [CrossRef]
- Stout, R.D.; Suttles, J. Functional Plasticity of Macrophages: Reversible Adaptation to Changing Microenvironments. J. Leukoc. Biol. 2004, 76, 509–513. [Google Scholar] [CrossRef]
- Bertani, F.R.; Mozetic, P.; Fioramonti, M.; Iuliani, M.; Ribelli, G.; Pantano, F.; Santini, D.; Tonini, G.; Trombetta, M.; Businaro, L.; et al. Classification of M1/M2-Polarized Human Macrophages by Label-Free Hyperspectral Reflectance Confocal Microscopy and Multivariate Analysis. Sci. Rep. 2017, 7, 8965. [Google Scholar] [CrossRef] [PubMed]
- Makita, N.; Hizukuri, Y.; Yamashiro, K.; Murakawa, M.; Hayashi, Y. IL-10 Enhances the Phenotype of M2 Macrophages Induced by IL-4 and Confers the Ability to Increase Eosinophil Migration. Int. Immunol. 2015, 27, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Svedberg, F.R.; Guilliams, M. Cellular Origin of Human Cardiac Macrophage Populations. Nat. Med. 2018, 24, 1091–1092. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.J.; Sun, X.; Ravaud, C.; del Villa Campo, C.; Klaourakis, K.; Lupu, I.-E.; Lord, A.M.; Browne, C.; Jacobsen, S.E.W.; Greaves, D.R.; et al. Tissue-Resident Macrophages Regulate Lymphatic Vessel Growth and Patterning in the Developing Heart. Development 2021, 148, dev194563. [Google Scholar] [CrossRef]
- Bajpai, G.; Schneider, C.; Wong, N.; Bredemeyer, A.; Hulsmans, M.; Nahrendorf, M.; Epelman, S.; Kreisel, D.; Liu, Y.; Itoh, A.; et al. The Human Heart Contains Distinct Macrophage Subsets with Divergent Origins and Functions. Nat. Med. 2018, 24, 1234–1245. [Google Scholar] [CrossRef]
- Moskalik, A.; Niderla-Bielińska, J.; Ratajska, A. Multiple Roles of Cardiac Macrophages in Heart Homeostasis and Failure. Heart Fail. Rev. 2022, 27, 1413–1430. [Google Scholar] [CrossRef]
- Ferrante, C.J.; Leibovich, S.J. Regulation of Macrophage Polarization and Wound Healing. Adv. Wound Care 2012, 1, 10–16. [Google Scholar] [CrossRef]
- Simões, F.C.; Cahill, T.J.; Kenyon, A.; Gavriouchkina, D.; Vieira, J.M.; Sun, X.; Pezzolla, D.; Ravaud, C.; Masmanian, E.; Weinberger, M.; et al. Macrophages Directly Contribute Collagen to Scar Formation during Zebrafish Heart Regeneration and Mouse Heart Repair. Nat. Commun. 2020, 11, 600. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, F.; Chai, R.; Zhou, W.; Hu, M.; Liu, B.; Chen, X.; Liu, M.; Xu, Q.; Liu, N.; et al. Exosomes Derived from Pro-Inflammatory Bone Marrow-Derived Mesenchymal Stem Cells Reduce Inflammation and Myocardial Injury via Mediating Macrophage Polarization. J. Cell. Mol. Med. 2019, 23, 7617–7631. [Google Scholar] [CrossRef]
- Yang, P.; Liu, L.; Sun, L.; Fang, P.; Snyder, N.; Saredy, J.; Ji, Y.; Shen, W.; Qin, X.; Wu, Q.; et al. Immunological Feature and Transcriptional Signaling of Ly6C Monocyte Subsets From Transcriptome Analysis in Control and Hyperhomocysteinemic Mice. Front. Immunol. 2021, 12, 632333. [Google Scholar] [CrossRef]
- Liu, G.; Liu, Q.; Shen, Y.; Kong, D.; Gong, Y.; Tao, B.; Chen, G.; Guo, S.; Li, J.; Zuo, S. Early Treatment with Resolvin E1 Facilitates Myocardial Recovery from Ischaemia in Mice. Br. J. Pharmacol. 2018, 175, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, M.J.; Harmsen, M.C.; van Rooijen, N.; Petersen, A.H.; van Luyn, M.J.A. Macrophage Depletion Impairs Wound Healing and Increases Left Ventricular Remodeling after Myocardial Injury in Mice. Am. J. Pathol. 2007, 170, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Raedschelders, K.; Ansley, D.M.; Chen, D.D.Y. The Cellular and Molecular Origin of Reactive Oxygen Species Generation during Myocardial Ischemia and Reperfusion. Pharmacol. Ther. 2012, 133, 230–255. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Tao, R.; Zhang, H.; Xie, H.; Lu, L.; Wang, T.; Su, M.; Hu, J.; Zhang, Q.; Chen, Q.; et al. Dectin-1 Contributes to Myocardial Ischemia/Reperfusion Injury by Regulating Macrophage Polarization and Neutrophil Infiltration. Circulation 2019, 139, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Tian, Y.; Luo, Y.; Xu, X.; Ge, W.; Sun, G.; Sun, X. Iminostilbene, a Novel Small-Molecule Modulator of PKM2, Suppresses Macrophage Inflammation in Myocardial Ischemia–Reperfusion Injury. J. Adv. Res. 2021, 29, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Monnerat, G.; Alarcón, M.L.; Vasconcellos, L.R.; Hochman-Mendez, C.; Brasil, G.; Bassani, R.A.; Casis, O.; Malan, D.; Travassos, L.H.; Sepúlveda, M. Macrophage-Dependent IL-1β Production Induces Cardiac Arrhythmias in Diabetic Mice. Nat. Commun. 2016, 7, 13344. [Google Scholar] [CrossRef]
- Glezeva, N.; Voon, V.; Watson, C.; Horgan, S.; McDonald, K.; Ledwidge, M.; Baugh, J. Exaggerated Inflammation and Monocytosis Associate with Diastolic Dysfunction in Heart Failure with Preserved Ejection Fraction: Evidence of M2 Macrophage Activation in Disease Pathogenesis. J. Card. Fail. 2015, 21, 167–177. [Google Scholar] [CrossRef]
- Jaenisch, R.; Bird, A. Epigenetic Regulation of Gene Expression: How the Genome Integrates Intrinsic and Environmental Signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef]
- Amit, I.; Winter, D.R.; Jung, S. The Role of the Local Environment and Epigenetics in Shaping Macrophage Identity and Their Effect on Tissue Homeostasis. Nat. Immunol. 2016, 17, 18–25. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Jin, J.; Lian, T.; Gu, C.; Yu, K.; Gao, Y.Q.; Su, X.-D. The Effects of Cytosine Methylation on General Transcription Factors. Sci. Rep. 2016, 6, 29119. [Google Scholar] [CrossRef] [PubMed]
- Duthie, S.J. Epigenetic Modifications and Human Pathologies: Cancer and CVD. Proc. Nutr. Soc. 2011, 70, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Song, M.; Qu, J.; Liu, G.-H. Epigenetic Modifications in Cardiovascular Aging and Diseases. Circ. Res. 2018, 123, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.A.; Lalla, E.; Lu, Y.; Gleason, M.R.; Wolf, B.M.; Tanji, N.; Ferran, L.J.; Kohl, B.; Rao, V.; Kisiel, W.; et al. Hyperhomocysteinemia Enhances Vascular Inflammation and Accelerates Atherosclerosis in a Murine Model. J. Clin. Investig. 2001, 107, 675–683. [Google Scholar] [CrossRef]
- Gao, S.; Wang, L.; Liu, W.; Wu, Y.; Yuan, Z. The Synergistic Effect of Homocysteine and Lipopolysaccharide on the Differentiation and Conversion of Raw264.7 Macrophages. J. Inflamm. 2014, 11, 13. [Google Scholar] [CrossRef]
- Wang, X.-H.; Wang, F.; You, S.-J.; Cao, Y.-J.; Cao, L.-D.; Han, Q.; Liu, C.-F.; Hu, L.-F. Dysregulation of Cystathionine γ-Lyase (CSE)/Hydrogen Sulfide Pathway Contributes to Ox-LDL-Induced Inflammation in Macrophage. Cell. Signal. 2013, 25, 2255–2262. [Google Scholar] [CrossRef]
- Li, J.-J.; Li, Q.; Du, H.-P.; Wang, Y.-L.; You, S.-J.; Wang, F.; Xu, X.-S.; Cheng, J.; Cao, Y.-J.; Liu, C.-F.; et al. Homocysteine Triggers Inflammatory Responses in Macrophages through Inhibiting CSE-H2S Signaling via DNA Hypermethylation of CSE Promoter. Int. J. Mol. Sci. 2015, 16, 12560–12577. [Google Scholar] [CrossRef]
- Yang, A.-N.; Zhang, H.-P.; Sun, Y.; Yang, X.-L.; Wang, N.; Zhu, G.; Zhang, H.; Xu, H.; Ma, S.-C.; Zhang, Y.; et al. High-Methionine Diets Accelerate Atherosclerosis by HHcy-Mediated FABP4 Gene Demethylation Pathway via DNMT1 in ApoE−/− Mice. FEBS Lett. 2015, 589, 3998–4009. [Google Scholar] [CrossRef]
- Greco, C.M.; Kunderfranco, P.; Rubino, M.; Larcher, V.; Carullo, P.; Anselmo, A.; Kurz, K.; Carell, T.; Angius, A.; Latronico, M.V.G.; et al. DNA Hydroxymethylation Controls Cardiomyocyte Gene Expression in Development and Hypertrophy. Nat. Commun. 2016, 7, 12418. [Google Scholar] [CrossRef]
- Bakshi, C.; Vijayvergiya, R.; Dhawan, V. Aberrant DNA Methylation of M1-Macrophage Genes in Coronary Artery Disease. Sci. Rep. 2019, 9, 1429. [Google Scholar] [CrossRef]
- Luttmer, R.; Spijkerman, A.M.; Kok, R.M.; Jakobs, C.; Blom, H.J.; Serne, E.H.; Dekker, J.M.; Smulders, Y.M. Metabolic Syndrome Components Are Associated with DNA Hypomethylation. Obes. Res. Clin. Pract. 2013, 7, e106–e115. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-Y.; Zhao, D.; Li, J.; Su, D.; Hochstrasser, M. Histone Sumoylation Promotes Set3 Histone-Deacetylase Complex-Mediated Transcriptional Regulation. Nucleic Acids Res. 2020, 48, 12151–12168. [Google Scholar] [CrossRef] [PubMed]
- Fulton, M.D.; Zhang, J.; He, M.; Ho, M.-C.; Zheng, Y.G. Intricate Effects of α-Amino and Lysine Modifications on Arginine Methylation of the N-Terminal Tail of Histone H4. Biochemistry 2017, 56, 3539–3548. [Google Scholar] [CrossRef] [PubMed]
- Daskalaki, M.G.; Tsatsanis, C.; Kampranis, S.C. Histone Methylation and Acetylation in Macrophages as a Mechanism for Regulation of Inflammatory Responses. J. Cell. Physiol. 2018, 233, 6495–6507. [Google Scholar] [CrossRef] [PubMed]
- Mathiyalagan, P.; Chang, L.; Du, X.-J.; El-Osta, A. Cardiac Ventricular Chambers Are Epigenetically Distinguishable. Cell Cycle 2010, 9, 612–617. [Google Scholar] [CrossRef]
- Greißel, A.; Culmes, M.; Burgkart, R.; Zimmermann, A.; Eckstein, H.-H.; Zernecke, A.; Pelisek, J. Histone Acetylation and Methylation Significantly Change with Severity of Atherosclerosis in Human Carotid Plaques. Cardiovasc. Pathol. 2016, 25, 79–86. [Google Scholar] [CrossRef]
- Lv, Y.-C.; Tang, Y.-Y.; Zhang, P.; Wan, W.; Yao, F.; He, P.-P.; Xie, W.; Mo, Z.-C.; Shi, J.-F.; Wu, J.-F.; et al. Histone Methyltransferase Enhancer of Zeste Homolog 2-Mediated ABCA1 Promoter DNA Methylation Contributes to the Progression of Atherosclerosis. PLoS ONE 2016, 11, e0157265. [Google Scholar] [CrossRef]
- Davis, F.M.; Gallagher, K.A. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 623–634. [Google Scholar] [CrossRef]
- Neele, A.E.; Prange, K.H.; Hoeksema, M.A.; van der Velden, S.; Lucas, T.; Dimmeler, S.; Lutgens, E.; Van den Bossche, J.; de Winther, M.P. Macrophage Kdm6b Controls the Pro-Fibrotic Transcriptome Signature of Foam Cells. Epigenomics 2017, 9, 383–391. [Google Scholar] [CrossRef]
- Targeting Macrophage Histone Deacetylase 3 Stabilizes Atherosclerotic Lesions. EMBO Mol. Med. 2014, 6, 1124–1132. [CrossRef]
- Vlad, M.-L.; Manea, S.-A.; Lazar, A.-G.; Raicu, M.; Muresian, H.; Simionescu, M.; Manea, A. Histone Acetyltransferase-Dependent Pathways Mediate Upregulation of NADPH Oxidase 5 in Human Macrophages under Inflammatory Conditions: A Potential Mechanism of Reactive Oxygen Species Overproduction in Atherosclerosis. Oxidative Med. Cell Longev. 2019, 2019, 3201062. [Google Scholar] [CrossRef]
- Oksala, N.K.J.; Seppälä, I.; Rahikainen, R.; Mäkelä, K.-M.; Raitoharju, E.; Illig, T.; Klopp, N.; Kholova, I.; Laaksonen, R.; Karhunen, P.J.; et al. Synergistic Expression of Histone Deacetylase 9 and Matrix Metalloproteinase 12 in M4 Macrophages in Advanced Carotid Plaques. Eur. J. Vasc. Endovasc. Surg. 2017, 53, 632–640. [Google Scholar] [CrossRef]
- Wang, Y.; Miao, X.; Liu, Y.; Li, F.; Liu, Q.; Sun, J.; Cai, L. Dysregulation of Histone Acetyltransferases and Deacetylases in Cardiovascular Diseases. Oxidative Med. Cell. Longev. 2014, 2014, e641979. [Google Scholar] [CrossRef] [PubMed]
- Garikipati, V.N.S.; Uchida, S. Elucidating the Functions of Non-Coding RNAs from the Perspective of RNA Modifications. Non-Coding RNA 2021, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mohapatra, T. Deciphering Epitranscriptome: Modification of MRNA Bases Provides a New Perspective for Post-Transcriptional Regulation of Gene Expression. Front. Cell Dev. Biol. 2021, 9, 628415. [Google Scholar] [CrossRef] [PubMed]
- Suñer, C.; Sibilio, A.; Martín, J.; Castellazzi, C.L.; Reina, O.; Dotu, I.; Caballé, A.; Rivas, E.; Calderone, V.; Díez, J.; et al. Macrophage Inflammation Resolution Requires CPEB4-Directed Offsetting of MRNA Degradation. eLife 2022, 11, e75873. [Google Scholar] [CrossRef]
- Wardowska, A. M6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy? Pharmaceuticals 2021, 14, 218. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; He, C. RNA N6-Methyladenosine Methylation in Post-Transcriptional Gene Expression Regulation. Genes Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Z.; Tang, H.; Shen, Y.; Gong, Z.; Xie, N.; Zhang, X.; Wang, W.; Kong, W.; Zhou, Y.; et al. The N6-Methyladenosine (M6A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization through the Methylation of STAT1 MRNA. Am. J. Physiol.-Cell Physiol. 2019, 317, C762–C775. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Y.; Ran, X.; Wang, D.; Zheng, X.; Zhang, M.; Yu, B.; Sun, Y.; Wu, J. Mettl14 Mediates the Inflammatory Response of Macrophages in Atherosclerosis through the NF-ΚB/IL-6 Signaling Pathway. Cell. Mol. Life Sci. 2022, 79, 311. [Google Scholar] [CrossRef]
- Gu, X.; Zhang, Y.; Li, D.; Cai, H.; Cai, L.; Xu, Q. N6-Methyladenosine Demethylase FTO Promotes M1 and M2 Macrophage Activation. Cell. Signal. 2020, 69, 109553. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Yang, M.; Han, X.; Li, J.; Gao, G.; Tai, H.; Huang, N.; Xiao, H. Fat Mass and Obesity-Associated Protein Attenuates Lipid Accumulation in Macrophage Foam Cells and Alleviates Atherosclerosis in Apolipoprotein E-Deficient Mice. J. Hypertens. 2017, 35, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological Roles of RNA M5C Modification and Its Implications in Cancer Immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Feng, J.; Xu, Q.; Wang, W.; Wang, X. NSun2 Deficiency Protects Endothelium From Inflammation via MRNA Methylation of ICAM-1. Circ. Res. 2016, 118, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tang, H.; Wang, X.; Wang, W.; Feng, J. Homocysteine Upregulates Interleukin-17A Expression via NSun2-Mediated RNA Methylation in T Lymphocytes. Biochem. Biophys. Res. Commun. 2017, 493, 94–99. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, D.; Zhang, H.; Yin, F.; Guo, P.; Zhang, X.; Bian, C.; Chen, C.; Li, S.; Yin, Y.; et al. N1-Methyladenosine (M1A) Regulation Associated With the Pathogenesis of Abdominal Aortic Aneurysm Through YTHDF3 Modulating Macrophage Polarization. Front. Cardiovasc. Med. 2022, 9, 883155. [Google Scholar] [CrossRef]
- Ecker, S.; Chen, L.; Pancaldi, V.; Bagger, F.O.; Fernández, J.M.; de Carrillo Santa Pau, E.; Juan, D.; Mann, A.L.; Watt, S.; Casale, F.P.; et al. Genome-Wide Analysis of Differential Transcriptional and Epigenetic Variability across Human Immune Cell Types. Genome Biol. 2017, 18, 18. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.-M.; Jakob, P.; Nakagawa, S.; Blankenberg, S. Non-Coding RNAs in Cardiovascular Diseases: Diagnostic and Therapeutic Perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.-C. MiRNA-Mediated Macrophage Polarization and Its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, M.; Tian, J.; Li, X.; Yang, M.; Zhang, K.; Tan, S.; Luo, L.; Luo, C.; Peng, L.; et al. Long Non-Coding RNA Expressed in Macrophage Co-Varies with the Inflammatory Phenotype during Macrophage Development and Polarization. J. Cell. Mol. Med. 2019, 23, 6530–6542. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Li, X.; Zhang, M.; Lv, K. Microarray Analysis of Circular RNA Expression Patterns in Polarized Macrophages. Int. J. Mol. Med. 2017, 39, 373–379. [Google Scholar] [CrossRef]
- Khan, A.W.; Paneni, F.; Jandeleit-Dahm, K.A.M. Cell-Specific Epigenetic Changes in Atherosclerosis. Clin. Sci. 2021, 135, 1165–1187. [Google Scholar] [CrossRef]
- Sugita, J.; Fujiu, K.; Nakayama, Y.; Matsubara, T.; Matsuda, J.; Oshima, T.; Liu, Y.; Maru, Y.; Hasumi, E.; Kojima, T.; et al. Cardiac Macrophages Prevent Sudden Death during Heart Stress. Nat. Commun. 2021, 12, 1910. [Google Scholar] [CrossRef]
- Komal, S.; Yin, J.-J.; Wang, S.-H.; Huang, C.-Z.; Tao, H.-L.; Dong, J.-Z.; Han, S.-N.; Zhang, L.-R. MicroRNAs: Emerging Biomarkers for Atrial Fibrillation. J. Cardiol. 2019, 74, 475–482. [Google Scholar] [CrossRef]
- Liu, S.; Chen, J.; Shi, J.; Zhou, W.; Wang, L.; Fang, W.; Zhong, Y.; Chen, X.; Chen, Y.; Sabri, A.; et al. M1-like Macrophage-Derived Exosomes Suppress Angiogenesis and Exacerbate Cardiac Dysfunction in a Myocardial Infarction Microenvironment. Basic Res. Cardiol. 2020, 115, 22. [Google Scholar] [CrossRef]
- Lu, Y.; Thavarajah, T.; Gu, W.; Cai, J.; Xu, Q. Impact of MiRNA in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e159–e170. [Google Scholar] [CrossRef]
- Zheng, C.-G.; Chen, B.-Y.; Sun, R.-H.; Mou, X.-Z.; Han, F.; Li, Q.; Huang, H.-J.; Liu, J.-Q.; Tu, Y.-X. MiR-133b Downregulation Reduces Vulnerable Plaque Formation in Mice with AS through Inhibiting Macrophage Immune Responses. Mol. Nucleic Acids 2019, 16, 745–757. [Google Scholar] [CrossRef]
- Tan, L.; Liu, L.; Jiang, Z.; Hao, X. Inhibition of MicroRNA-17-5p Reduces the Inflammation and Lipid Accumulation, and up-Regulates ATP-Binding Cassette TransporterA1 in Atherosclerosis. J. Pharmacol. Sci. 2019, 139, 280–288. [Google Scholar] [CrossRef]
- Chen, W.; Li, X.; Wang, J.; Song, N.; Zhu, A.; Jia, L. MiR-378a Modulates Macrophage Phagocytosis and Differentiation through Targeting CD47-SIRPα Axis in Atherosclerosis. Scand. J. Immunol. 2019, 90, e12766. [Google Scholar] [CrossRef]
- Li, J.; Xue, H.; Li, T.; Chu, X.; Xin, D.; Xiong, Y.; Qiu, W.; Gao, X.; Qian, M.; Xu, J.; et al. Exosomes Derived from Mesenchymal Stem Cells Attenuate the Progression of Atherosclerosis in ApoE−/− Mice via MiR-Let7 Mediated Infiltration and Polarization of M2 Macrophage. Biochem. Biophys. Res. Commun. 2019, 510, 565–572. [Google Scholar] [CrossRef]
- Meng, X.; Yin, J.; Yu, X.; Guo, Y. MicroRNA-205-5p Promotes Unstable Atherosclerotic Plaque Formation In Vivo. Cardiovasc. Drugs Ther. 2020, 34, 25–39. [Google Scholar] [CrossRef]
- Zhao, Z.; Hao, J.; Li, X.; Chen, Y.; Qi, X. MiR-21-5p Regulates Mycobacterial Survival and Inflammatory Responses by Targeting Bcl-2 and TLR4 in Mycobacterium Tuberculosis-Infected Macrophages. FEBS Lett. 2019, 593, 1326–1335. [Google Scholar] [CrossRef]
- Garikipati, V.N.S.; Verma, S.K.; Jolardarashi, D.; Cheng, Z.; Ibetti, J.; Cimini, M.; Tang, Y.; Khan, M.; Yue, Y.; Benedict, C.; et al. Therapeutic Inhibition of MiR-375 Attenuates Post-Myocardial Infarction Inflammatory Response and Left Ventricular Dysfunction via PDK-1-AKT Signalling Axis. Cardiovasc. Res. 2017, 113, 938–949. [Google Scholar] [CrossRef]
- Yang, J.; Brown, M.E.; Zhang, H.; Martinez, M.; Zhao, Z.; Bhutani, S.; Yin, S.; Trac, D.; Xi, J.J.; Davis, M.E. High-Throughput Screening Identifies MicroRNAs That Target Nox2 and Improve Function after Acute Myocardial Infarction. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H1002–H1012. [Google Scholar] [CrossRef]
- Zhao, J.; Li, X.; Hu, J.; Chen, F.; Qiao, S.; Sun, X.; Gao, L.; Xie, J.; Xu, B. Mesenchymal Stromal Cell-Derived Exosomes Attenuate Myocardial Ischaemia-Reperfusion Injury through MiR-182-Regulated Macrophage Polarization. Cardiovasc. Res. 2019, 115, 1205–1216. [Google Scholar] [CrossRef]
- Corsten, M.F.; Papageorgiou, A.; Verhesen, W.; Carai, P.; Lindow, M.; Obad, S.; Summer, G.; Coort, S.L.M.; Hazebroek, M.; van Leeuwen, R.; et al. MicroRNA Profiling Identifies MicroRNA-155 as an Adverse Mediator of Cardiac Injury and Dysfunction During Acute Viral Myocarditis. Circ. Res. 2012, 111, 415–425. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Li, X.; Tang, Z.; Wang, X.; Zhong, M.; Suo, Q.; Zhang, Y.; Lv, K. Silencing MicroRNA-155 Attenuates Cardiac Injury and Dysfunction in Viral Myocarditis via Promotion of M2 Phenotype Polarization of Macrophages. Sci. Rep. 2016, 6, 22613. [Google Scholar] [CrossRef]
- Heymans, S.; Corsten, M.F.; Verhesen, W.; Carai, P.; van Leeuwen, R.E.W.; Custers, K.; Peters, T.; Hazebroek, M.; Stöger, L.; Wijnands, E.; et al. Macrophage MicroRNA-155 Promotes Cardiac Hypertrophy and Failure. Circulation 2013, 128, 1420–1432. [Google Scholar] [CrossRef]
- Gou, W.; Zhang, Z.; Yang, C.; Li, Y. MiR-223/Pknox1 Axis Protects Mice from CVB3-Induced Viral Myocarditis by Modulating Macrophage Polarization. Exp. Cell Res. 2018, 366, 41–48. [Google Scholar] [CrossRef]
- Unique Features of Long Non-Coding RNA Biogenesis and Function|Nature Reviews Genetics. Available online: https://www.nature.com/articles/nrg.2015.10 (accessed on 5 December 2021).
- Margulies, K.B.; Matiwala, S.; Cornejo, C.; Olsen, H.; Craven, W.A.; Bednarik, D. Transcription Patterns in Failing and Recovering Human Myocardium. Circ. Res. 2005, 96, 592–599. [Google Scholar] [CrossRef]
- Huangfu, N.; Xu, Z.; Zheng, W.; Wang, Y.; Cheng, J.; Chen, X. LncRNA MALAT1 Regulates OxLDL-Induced CD36 Expression via Activating β-Catenin. Biochem. Biophys. Res. Commun. 2018, 495, 2111–2117. [Google Scholar] [CrossRef]
- Identification of Differentially Expressed Long Non-Coding RNAs in Polarized Macrophages|Scientific Reports. Available online: https://www.nature.com/articles/srep19705 (accessed on 5 December 2021).
- The LPS-Inducible LncRNA Mirt2 Is a Negative Regulator of Inflammation|Nature Communications. Available online: https://www.nature.com/articles/s41467-017-02229-1 (accessed on 5 December 2021).
- Meng, X.-D.; Yao, H.-H.; Wang, L.-M.; Yu, M.; Shi, S.; Yuan, Z.-X.; Liu, J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE−/− Mice. Mol. Ther. Nucleic Acids 2020, 19, 84–96. [Google Scholar] [CrossRef]
- Lu, W.; He, X.; Su, L.; Miao, J. Long Noncoding RNA-CERNA1 Stabilized Atherosclerotic Plaques in Apolipoprotein E−/− Mice. J. Cardiovasc. Trans. Res. 2019, 12, 425–434. [Google Scholar] [CrossRef]
- Macrophage-Enriched LncRNA RAPIA|Arteriosclerosis, Thrombosis, and Vascular Biology. Available online: https://www.ahajournals.org/doi/full/10.1161/ATVBAHA.119.313749 (accessed on 5 December 2021).
- Sun, F.; Guo, Z.; Zhang, C.; Che, H.; Gong, W.; Shen, Z.; Shi, Y.; Ge, S. LncRNA NRON Alleviates Atrial Fibrosis through Suppression of M1 Macrophages Activated by Atrial Myocytes. Biosci. Rep. 2019, 39, BSR20192215. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Kong, X.; Zhang, M.; Wang, D.; Liu, Y.; Lv, K. Long Non-Coding RNA AK085865 Ablation Confers Susceptibility to Viral Myocarditis by Regulating Macrophage Polarization. J. Cell. Mol. Med. 2020, 24, 5542–5554. [Google Scholar] [CrossRef]
- Xue, Y.-L.; Zhang, S.-X.; Zheng, C.-F.; Li, Y.-F.; Zhang, L.-H.; Su, Q.-Y.; Hao, Y.-F.; Wang, S.; Li, X.-W. Long Non-Coding RNA MEG3 Inhibits M2 Macrophage Polarization by Activating TRAF6 via MicroRNA-223 down-Regulation in Viral Myocarditis. J. Cell. Mol. Med. 2020, 24, 12341–12354. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Kessler, T.; Vilne, B.; Schunkert, H. The Impact of Genome-wide Association Studies on the Pathophysiology and Therapy of Cardiovascular Disease. EMBO Mol. Med. 2016, 8, 688–701. [Google Scholar] [CrossRef]
- Song, H.; Yang, Y.; Sun, Y.; Wei, G.; Zheng, H.; Chen, Y.; Cai, D.; Li, C.; Ma, Y.; Lin, Z.; et al. Circular RNA Cdyl Promotes Abdominal Aortic Aneurysm Formation by Inducing M1 Macrophage Polarization and M1-Type Inflammation. Mol. Ther. 2022, 30, 915–931. [Google Scholar] [CrossRef]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-Based Network Analysis Reveals a Spectrum Model of Human Macrophage Activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef]
- Moore, K.J.; Tabas, I. Macrophages in the Pathogenesis of Atherosclerosis. Cell 2011, 145, 341–355. [Google Scholar] [CrossRef]
- Tugal, D.; Liao, X.; Jain, M.K. Transcriptional Control of Macrophage Polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1135–1144. [Google Scholar] [CrossRef]
- Yang, M.; Li, F.; Wang, L.; Yukht, A.; Arias, A.; Tian, F.; Williamson, I.; Shah, P.K.; Sharifi, B.G. Abstract 13424: GATA3 Regulates Macrophage Polarization and Phenotype. Circulation 2012, 126, A13424. [Google Scholar] [CrossRef]
- Sweet, D.R.; Fan, L.; Hsieh, P.N.; Jain, M.K. Krüppel-Like Factors in Vascular Inflammation: Mechanistic Insights and Therapeutic Potential. Front. Cardiovasc. Med. 2018, 5, 6. [Google Scholar] [CrossRef]
- Peterson, J.M.; Wang, D.J.; Shettigar, V.; Roof, S.R.; Canan, B.D.; Bakkar, N.; Shintaku, J.; Gu, J.-M.; Little, S.C.; Ratnam, N.M.; et al. NF-ΚB Inhibition Rescues Cardiac Function by Remodeling Calcium Genes in a Duchenne Muscular Dystrophy Model. Nat. Commun. 2018, 9, 3431. [Google Scholar] [CrossRef]
- Schiano, C.; Benincasa, G.; Franzese, M.; Della Mura, N.; Pane, K.; Salvatore, M.; Napoli, C. Epigenetic-Sensitive Pathways in Personalized Therapy of Major Cardiovascular Diseases. Pharmacol. Ther. 2020, 210, 107514. [Google Scholar] [CrossRef]
- Ye, F.; Huang, J.; Wang, H.; Luo, C.; Zhao, K. Targeting Epigenetic Machinery: Emerging Novel Allosteric Inhibitors. Pharmacol. Ther. 2019, 204, 107406. [Google Scholar] [CrossRef]
- Jia, S.-J.; Gao, K.-Q.; Zhao, M. Epigenetic Regulation in Monocyte/Macrophage: A Key Player during Atherosclerosis. Cardiovasc. Ther. 2017, 35, e12262. [Google Scholar] [CrossRef]
- Findeisen, H.M.; Gizard, F.; Zhao, Y.; Qing, H.; Heywood, E.B.; Jones, K.L.; Cohn, D.; Bruemmer, D. Epigenetic Regulation of Vascular Smooth Muscle Cell Proliferation and Neointima Formation by Histone Deacetylase Inhibition. Arter. Thromb. Vasc. Biol. 2011, 31, 851–860. [Google Scholar] [CrossRef]



| Epigenetic Modifications | Enzyme or Targets | Macrophage Type | Function | CVDs | References |
|---|---|---|---|---|---|
| DNA methylation | DNMT1, DNA hydroxymethylation, CpG hypermethylation | M1/M2 | Endothelial cell dysfunction, Macrophage differentiation | Atherosclerosis, Cardiac hypertrophy, Hcy-induced atherosclerosis | [51,52,53,54,55,56,57,58,59,60,61,62] |
| Histone modifications (Methylation/ Acetylation) | ANP, BNP, HDAC3, SIRT-6/1, EZH-2 | M1/M2 | Chromatin remodeling, Gene transcription | Myocardial remodeling, Carotid artery stenosis, Atherosclerosis | [63,64,65,66,67,68,69,70,71,72,73,74] |
| RNA regulation (m6A/m5C/m1A) | CPEB4, TTP, METTL3/14, YTHDF2/3, FTO, DNMT2, ICAM-1 | M1/M2 | Gene expression, Encoding pro-inflammatory and anti-inflammatory factors | Atherosclerosis, Acute coronary syndrome | [75,76,77,78,79,80,81,82,83,84,85,86,87] |
| Non-coding RNAs miRNAs | MAML1, NOTCH, SIRP α, KBTBD7, PDK1, NOX2, NF-κB | M1/M2 | Gene regulation by inhibiting miRNAs or miRNA/RBP sponge transcription | Atherosclerosis, Myocardial infraction, Cardiac hypertrophy, Myocarditis, Heart failure | [88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110] |
| lncRNAs | API5 | M1/M2 | Epigenetic and transcriptional modifications, Chromatin remodeling, RNA splicing | Atherosclerosis, carotid artery disease | [111,112,113,114,115,116,117,118,119,120,121] |
| circRNAs | SP1, PARP | M1/M2 | Gene regulation by inhibiting miRNAs or miRNA/RBP sponge transcription | Myocardial infarction, Ventricular dysfunction, Myocardial fibrosis | [122,123,124] |
| CVDs | miRNA | lncRNA | circRNA | Targets | Macrophage Regulatory Expression | References |
|---|---|---|---|---|---|---|
| Atherosclerosis | miR-133b-3p, miR-17-5p, miR-378a-3p, miR-let7, miR-205-5p, | lncRNA-MALAT1, lncRNA-MIAT, lncRNA- CERNA1, lncRNA-RAPIA | circCdyl | MAML1, NOTCH, ABCA1, SIRP α, NF-kB, API5 | Increased M1expression and M1/M2 imbalance | [95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110] |
| Myocardial infarction | miR-21-5p, miR-375-3p, miR-204-5p, miR-148b-3p, miR-106b-5p | lncRNA-TCONS-00019715, lncRNA- Mirt2 | - | KBTBD7, MAPK, p38/NF-kB, PDK1, NOX2, PPARγ | M1/M2 imbalance | [111,112,113,114,115,116,117,118,119,120,121] |
| Ischemia-reperfusion injury | miR-182 | - | - | TLR4/NF-κB/PI3K/Akt | M1/M2 imbalance | [122,123,124] |
| Viral myocarditis | miR-155-5p, miR-21-5p, miR-146b-5p, miR-223 | lncRNA-NRON, lncRNA-AK085865, lncRNA-MEG3 | circCdyl | TLR, Pknox, TRAF6/ NF-κB, IRF4/ C/EBP-δ | M1/M2 imbalance | [124] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komal, S.; Han, S.-N.; Cui, L.-G.; Zhai, M.-M.; Zhou, Y.-J.; Wang, P.; Shakeel, M.; Zhang, L.-R. Epigenetic Regulation of Macrophage Polarization in Cardiovascular Diseases. Pharmaceuticals 2023, 16, 141. https://doi.org/10.3390/ph16020141
Komal S, Han S-N, Cui L-G, Zhai M-M, Zhou Y-J, Wang P, Shakeel M, Zhang L-R. Epigenetic Regulation of Macrophage Polarization in Cardiovascular Diseases. Pharmaceuticals. 2023; 16(2):141. https://doi.org/10.3390/ph16020141
Chicago/Turabian StyleKomal, Sumra, Sheng-Na Han, Liu-Gen Cui, Miao-Miao Zhai, Yue-Jiao Zhou, Pei Wang, Muhammad Shakeel, and Li-Rong Zhang. 2023. "Epigenetic Regulation of Macrophage Polarization in Cardiovascular Diseases" Pharmaceuticals 16, no. 2: 141. https://doi.org/10.3390/ph16020141
APA StyleKomal, S., Han, S.-N., Cui, L.-G., Zhai, M.-M., Zhou, Y.-J., Wang, P., Shakeel, M., & Zhang, L.-R. (2023). Epigenetic Regulation of Macrophage Polarization in Cardiovascular Diseases. Pharmaceuticals, 16(2), 141. https://doi.org/10.3390/ph16020141








