In Silico, In Vitro, and In Vivo Evaluation of Caffeine-Coated Nanoparticles as a Promising Therapeutic Avenue for AML through NF-Kappa B and TRAIL Pathways Modulation
Abstract
1. Introduction
2. Results
2.1. Characterization of MSNPs and Drug Loading
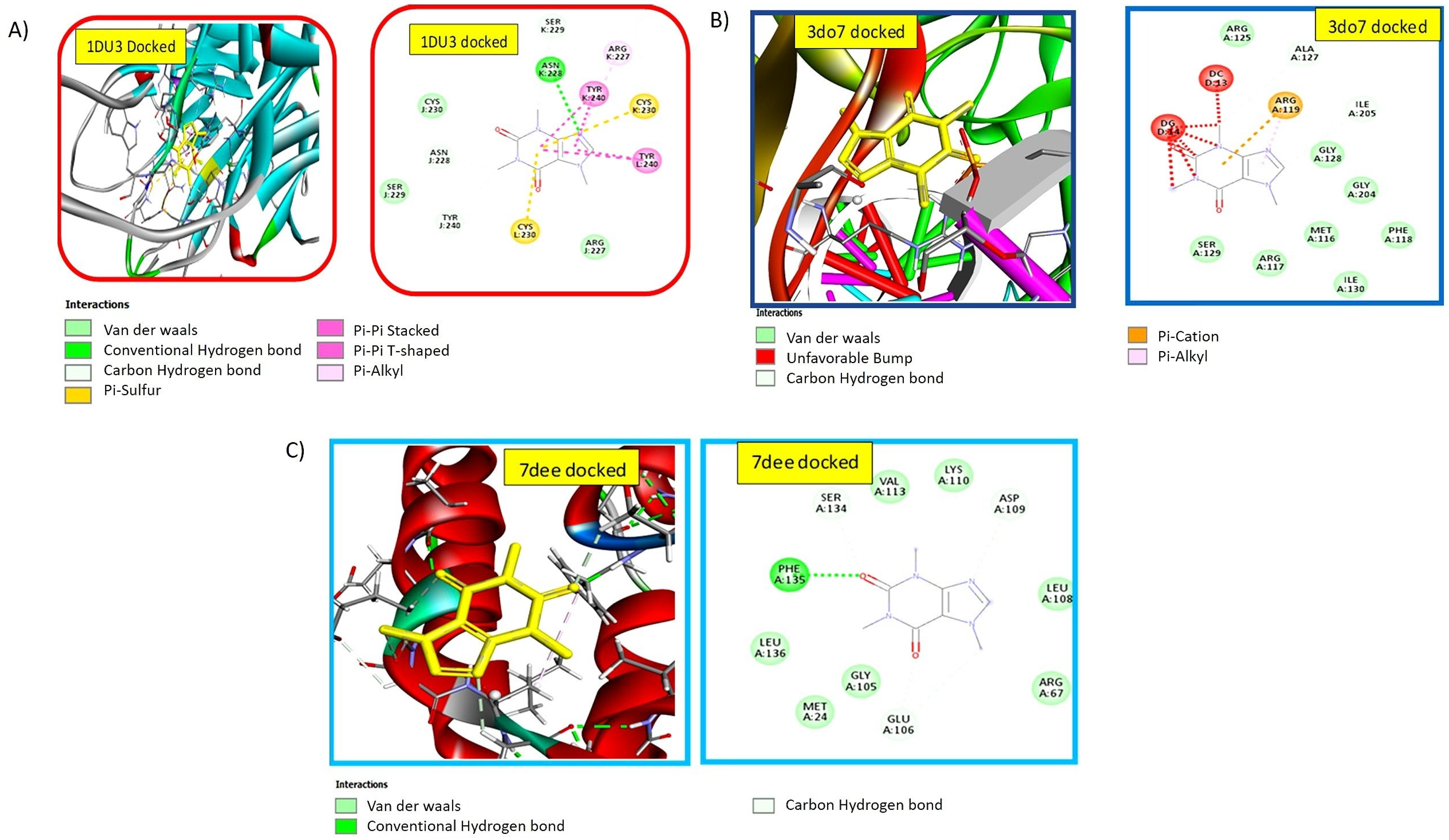
2.2. Nanomedicine Exhibiting Strong Interaction with Pro-Apoptotic TRAIL-DR5 Complex
2.3. Docking Studies Confirmed the Strong Binding Efficiency of the Ligand with Anti-Proliferative NF-kB p52/RelB/DNA Complex
2.4. Nanomedicine Exhibiting Strong Interaction with c-FLIP
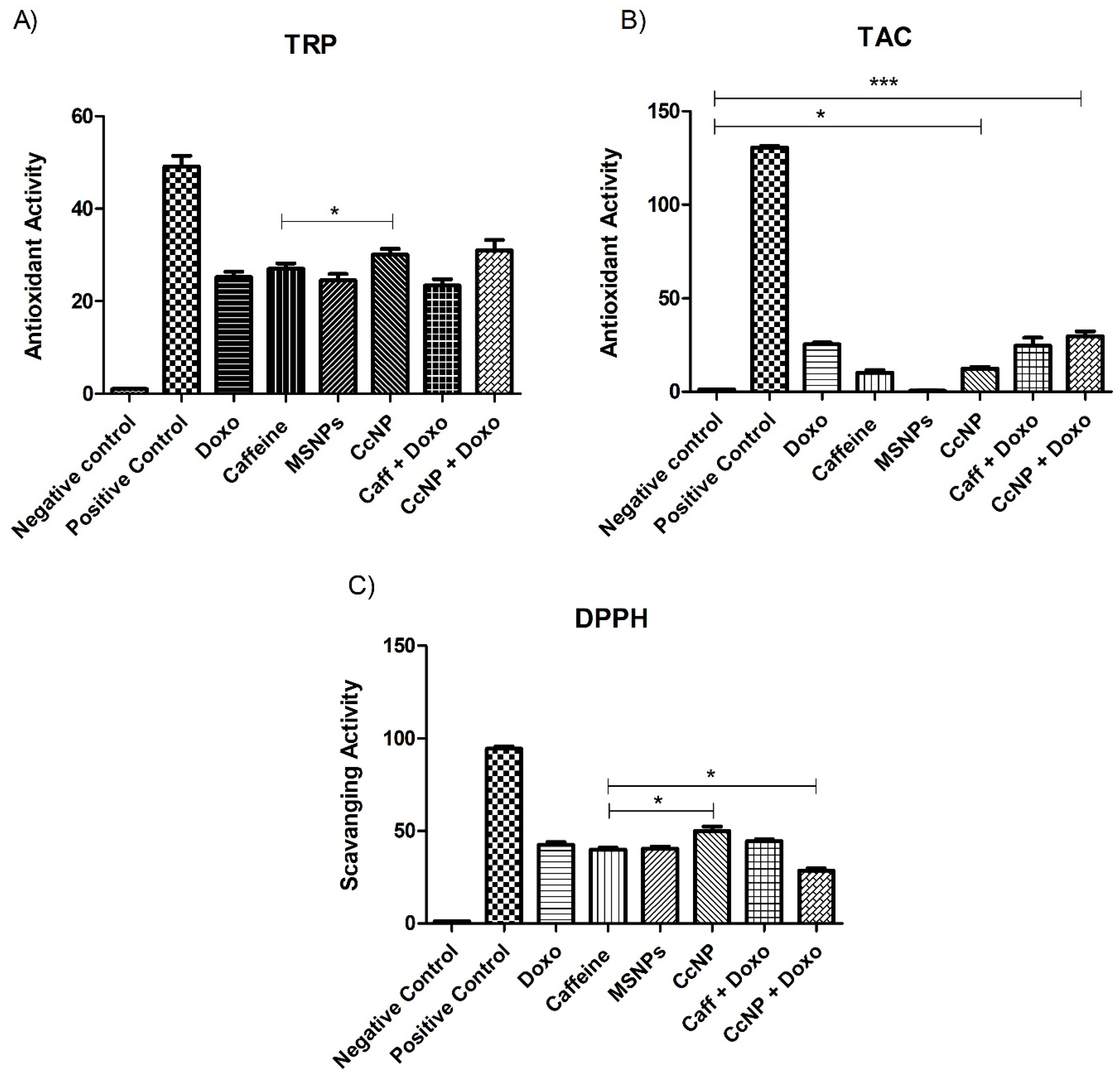
2.5. Increased Antioxidant Potential of Caffeine in the Form of Nanomedicine
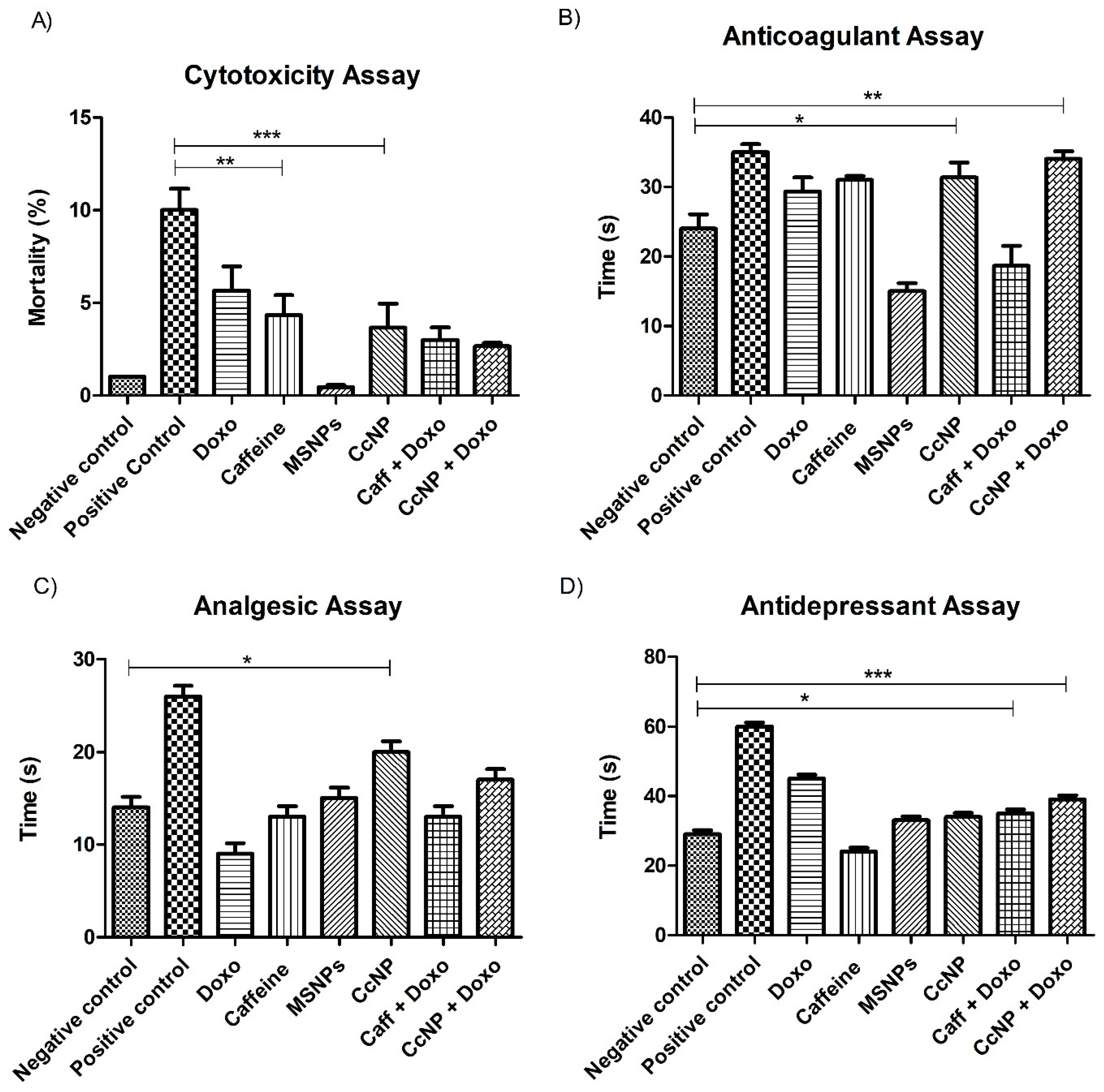
2.6. Enhanced Biological Potential of Nanomedicine as Anti-Depressant, Analgesic, and Anti-Coagulation Agent in Rat Models
2.7. Restoration of Liver, Heart, and Kidney Weights back to Normal after CcNP Treatment
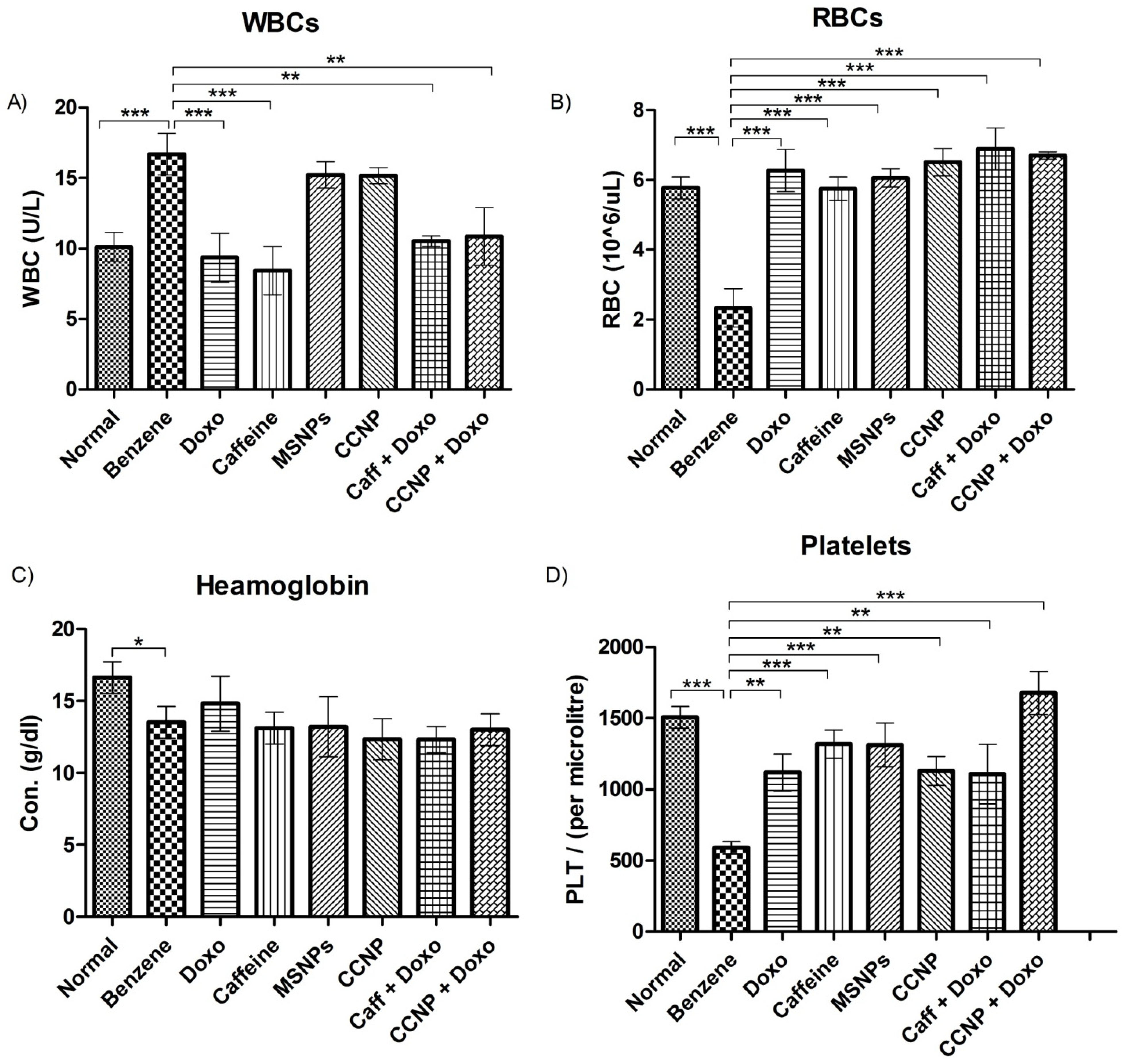
2.8. Restoration of Blood Parameters upon Nanomedicine Administration
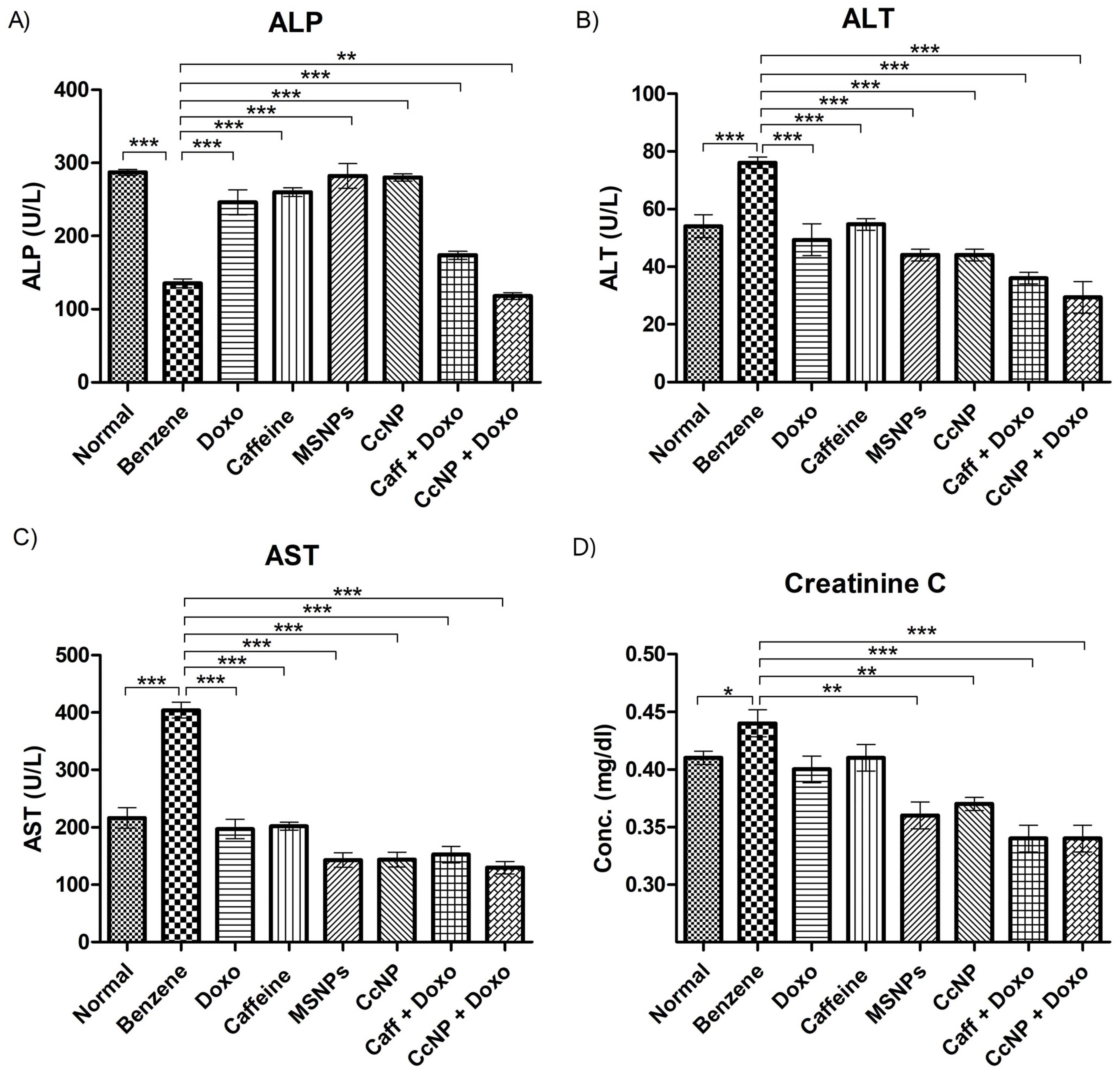
2.9. Enzymatic Activities Normalized after Drug Treatment
2.10. Suppression of STMN1 Expression by Nanomedicine
2.11. Upregulation of Tumor Suppressor Gene p53 by Nanomedicine
2.12. Regulation of Glycolysis by Nanomedicine in Leukemic Rats
2.13. Regulation of the mTOR Pathway by Nanomedicine
2.14. Nanomedicine Induces Anti-Proliferative Effects through NF-Kappa B Pathway Inhibition
2.15. Regulation of TRAIL Pathway by CcNP
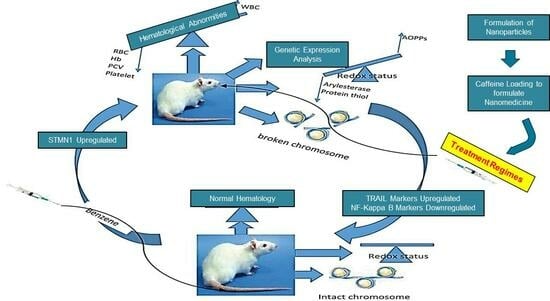
3. Discussion
4. Materials and Methods
4.1. Synthesis of Mesoporous Silica Nanoparticles (MCM-41 Generation)
4.2. Characterization of Nanoparticles
4.3. Drug Loading
4.4. Molecular Docking Studies
4.5. Cytotoxicity Assay
4.6. In Vitro Bioassays
4.7. In Vivo Bioassays
4.8. Experiment Design and Sprague-Dawley Model
4.9. Morphological Analysis
4.10. Blood Profiling
4.11. mRNA Extraction
4.12. cDNA Synthesis
4.13. RT PCR Expression Analysis
- STMN1 (F-TTGCCAGTGGATTGTGTAGAG, R-TTCTTTTGATCGAGGGCTGAG),
- P53 (F-TCCGACTATACCACTATCCACTAC, R-GCACAAACACGAACCTCAAAG),
- GAPDH (F-TCCAGTATGACTCTACCCACG, R-CACGACATACTCAGCACCAG),
- mTOR (F-AGTGAAAGTGAAGCCGAGAG), (R-CGACAAGGAGATAGAACGGAAG),
- Rel A (F-CTACGAGACCTTCAAGAGCATC, R-GATGTTGAAAAGGCATAGGGC),
- Rel B (F-CTTTTCTCAAGCTGACGTGC, R-AGATCTCCAGGTCCTCGTATG),
- DR5 (F-TCAACCCTGTGCCAATCC, R-ATGAACTCCTTCCAGCGTG),
- cFLIP (F-AGAAGCCCTCACCTTGTTTC, R-CTCTTGTCCTTGGCTACCTTG),
- TRAIL ligand (F-CACATTACCGGGATCACTCG, R-AGCTCTCCGTTTCTCAAGTG)
- and Cyt-c (F-CCCTAAGAGTCTGATCCTTTGTG, R-TCCAGTCTTATGCTTGCCTC).
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.; Laakso, M.; Ovaska, K.; Mirtti, T.; Lundin, J.; Rannikko, A.; Sankila, A.; Turunen, J.P.; Lundin, M.; Konsti, J.; et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO Rep. 2011, 30, 3962–3976. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposals for the classification of the acute leukaemias French-American-British (FAB) co-operative group. Br. J. Haematol. 1976, 33, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.T.; Jindal, A. Nano-natural Products as Anticancer Agents. In Anticancer Plants: Clinical Trials and Nanotechnology; Akhtar, M.S., Swamy, M.K., Eds.; Springer: Singapore, 2017; Volume 3, pp. 27–50. [Google Scholar]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef]
- Partridge, A.H.; Burstein, H.J.; Winer, E.P. Side effects of chemotherapy and combined chemohormonal therapy in women with early-stage breast cancer. JNCI Monogr. 2001, 2001, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Heckman, M.A.; Weil, J.; De Mejia, E.G. Caffeine (1,3,7-trimethylxanthine) in foods: A comprehensive review on consumption, functionality, safety, and regulatory matters. J. Food Sci. 2010, 75, R77–R87. [Google Scholar] [CrossRef] [PubMed]
- Calvo, I.; Gabrielli, N.; Iglesias-Baena, I.; Garcıa-Santamarina, S.; Hoe, K. Genome-Wide Screen of Genes Required for Caffeine Tolerance in fission yeast. PLoS ONE 2009, 4, e6619. [Google Scholar] [CrossRef]
- Monteiro, J.; Alves, M.G.; Oliveira, P.F.; Silva, B.M. Pharmacological potential of methylxanthines: Retrospective analysis and future expectations. Crit. Rev. Food Sci. Nutr. 2019, 59, 2597–2625. [Google Scholar] [CrossRef]
- Sinha, R.; Kim, G.J.; Nie, S.; Shin, D.M. Nanotechnology in cancer therapeutics: Bioconjugated nanoparticles for drug delivery. Mol. Cancer Ther. 2006, 5, 1909–1917. [Google Scholar] [CrossRef]
- Slowing, I.I.; Trewyn, B.G.; Giri, S.; Lin, V.Y. Mesoporous silica nanoparticles for drug delivery and biosensing applications. Adv. Funct. Mater. 2007, 17, 1225–1236. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhi, Z.; Jiang, T.; Zhang, J.; Wang, Z.; Wang, S. Spherical mesoporous silica nanoparticles for loading and release of the poorly water-soluble drug telmisartan. J. Control Release 2010, 145, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, G.; Hernandez-Martinez, A.R.; Pool, H.; Molina, G.; Cruz-Soto, M.; Luna-Barcenas, G.; Estevez, M. Synthesis and functionalization of silica-based nanoparticles with fluorescent biocompounds extracted from Eysenhardtia polystachya for biological applications. Mater. Sci. Eng. 2015, 57, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Guo, J.; Shen, S.; Chang, B.; Zhang, Y.; Jiang, X.; Yang, W. Synthesis of discrete and dispersible hollow mesoporous silica nanoparticles with tailored shell thickness for controlled drug release. J. Mater. Chem. 2012, 22, 17636–17643. [Google Scholar] [CrossRef]
- Nandiyanto, A.B.D.; Kim, S.-G.; Iskandar, F.; Okuyama, K. Synthesis of spherical mesoporous silica nanoparticles with nanometer-size controllable pores and outer diameters. Microporous Mesoporous Mater. 2009, 120, 447–453. [Google Scholar] [CrossRef]
- Das, S.; Tripathi, N.; Preet, R.; Siddharth, S.; Nayak, A.; Bharatam, P.V.; Kundu, C.N. Quinacrine induces apoptosis in cancer cells by forming a functional bridge between TRAIL-DR5 complex and modulating the mitochondrial intrinsic cascade. Oncotarget 2017, 8, 248–267. [Google Scholar] [CrossRef] [PubMed]
- Fusco, A.J.; Huang, D.B.; Miller, D.; Wang, V.Y.; Vu, D.; Ghosh, G. NF-kappaB p52:RelB heterodimer recognizes two classes of kappaB sites with two distinct modes. EMBO Rep. 2009, 10, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Safa, A.R.; Day, T.W.; Wu, C.H. Cellular FLICE-like inhibitory protein (C-FLIP): A novel target for cancer therapy. Curr. Cancer Drug Targets 2008, 8, 37–46. [Google Scholar] [CrossRef]
- Jang, J.H.; Lee, T.J.; Sung, E.G.; Song, I.H.; Kim, J.Y. Dapagliflozin induces apoptosis by downregulating cFILP(L) and increasing cFILP(S) instability in Caki-1 cells. Oncol. Lett. 2022, 24, 401. [Google Scholar] [CrossRef]
- Rana, S.; Verma, Y. Biochemical toxicity of benzene. J. Environ. Biol. 2005, 26, 157–168. [Google Scholar]
- Hallenbeck, W.H.; Cunningham-Burns, K.M. Carbamates. In Pesticides and Human Health; Springer: New York, NY, USA, 1985; pp. 30–32. [Google Scholar]
- Greenwood, M.J.; Seftel, M.D.; Richardson, C.; Barbaric, D.; Barnett, M.J.; Bruyere, H.; Forrest, D.L.; Horsman, D.E.; Smith, C.; Song, K.; et al. Leukocyte count as a predictor of death during remission induction in acute myeloid leukemia. J. Leuk. 2006, 47, 1245–1252. [Google Scholar] [CrossRef]
- Ito, K.; Nakazato, T.; Miyakawa, Y.; Yamato, K.; Ikeda, Y.; Kizaki, M. Caffeine induces G2/M arrest and apoptosis via a novel p53-dependent pathway in NB4 promyelocytic leukemia cells. J. Cell Physiol. 2003, 196, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.; Bruhns, P.; Frazier, W.A.; Ravetch, J.V.; Oldenborg, P.-A. Platelet homeostasis is regulated by platelet expression of CD47 under normal conditions and in passive immune thrombocytopenia. Blood 2005, 105, 3577–3582. [Google Scholar] [CrossRef] [PubMed]
- Farrow, A.C.; Buchanan, G.R.; Zwiener, R.J.; Bowman, W.P.; Winick, N.J. Serum aminotransferase elevation during and following treatment of childhood acute lymphoblastic leukemia. J. Clin. Oncol. 1997, 15, 1560–1566. [Google Scholar] [CrossRef] [PubMed]
- Lahoti, A.; Kantarjian, H.; Salahudeen, A.K.; Ravandi, F.; Cortes, J.E.; Faderl, S.; O’Brien, S.; Wierda, W.; Mattiuzzi, G.N. Predictors and outcome of acute kidney injury in patients with acute myelogenous leukemia or high-risk myelodysplastic syndrome. Cancer 2010, 116, 4063–4068. [Google Scholar] [CrossRef] [PubMed]
- Handschuh, L.; Kaźmierczak, M.; Milewski, M.C.; Góralski, M.; Łuczak, M.; Wojtaszewska, M.; Uszczyńska-Ratajczak, B.; Lewandowski, K.; Komarnicki, M.; Figlerowicz, M. Gene expression profiling of acute myeloid leukemia samples from adult patients with, A.M.L.-M.1.; -M2 through boutique microarrays real-time, P.C.R.; droplet digital, P.C.R. Int. J. Oncol. 2018, 52, 656–678. [Google Scholar] [PubMed]
- Wu, N.; Gao, N.; Fan, D.; Wei, J.; Zhang, J.; An, J. miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microbes Infect. 2014, 16, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Devasagayam, T.; Kamat, J.; Mohan, H.; Kesavan, P. Caffeine as an antioxidant: Inhibition of lipid peroxidation induced by reactive oxygen species. BBA-Biomembranes 1996, 1282, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Jiang, G.J.; Wang, K.; Miao, D.Q.; Guo, L.; Hou, Y.; Schatten, H.; Sun, Q.Y. Protein profile changes during porcine oocyte aging and effects of caffeine on protein expression patterns. PLoS ONE 2011, 6, e28996. [Google Scholar] [CrossRef]
- Fruchon, S.; Kheirallah, S.; Al Saati, T.; Ysebaert, L.; Laurent, C.; Leseux, L.; Fournie, J.J.; Laurent, G.; Bezombes, C. Involvement of the Syk–mTOR pathway in follicular lymphoma cell invasion and angiogenesis. Leukemia 2012, 26, 795–805. [Google Scholar] [CrossRef][Green Version]
- Martelli, A.M.; Evangelisti, C.; Chappell, W.; Abrams, S.L.; Bäsecke, J.; Stivala, F.; Donia, M.; Fagone, P.; Nicoletti, F.; Libra, M.; et al. Targeting the translational apparatus to improve leukemia therapy: Roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia 2011, 25, 1064–1079. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhou, P.; Wang, W.; Sun, A.; Guo, F. RelB, together with RelA, sustains cell survival and confers proteasome inhibitor sensitivity of chronic lymphocytic leukemia cells from bone marrow. Int. J. Mol. Med. 2014, 92, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Tapia, M.A.; González-Navarrete, I.; Dalmases, A.; Bosch, M.; Rodriguez-Fanjul, V.; Rolfe, M.; Ross, J.S.; Mezquita, J.; Mezquita, C.; Bachs, O.; et al. Inhibition of the canonical IKK/NFκB pathway sensitizes human cancer cells to doxorubicin. Cell Cycle 2007, 6, 2284–2292. [Google Scholar] [CrossRef] [PubMed]
- Szatkowska, P.; Koba, M.; Koslinski, P.; Szablewski, M. Molecularly imprinted polymers’ applications: A short review. Mini-Rev. Org. Chem. 2013, 10, 400–408. [Google Scholar] [CrossRef]
- Ahn, Y.; Lee, S.; Park, C.S.; Jang, H.J.; Lee, J.H.; Park, B.G.; Park, Y.S.; Shin, W.S.; Kwon, D. Diethylamino-curcumin mimic with trizolyl benzene enhances TRAIL-mediated cell death on human glioblastoma cells. Mol. Cell Toxicol. 2018, 14, 241–245. [Google Scholar] [CrossRef]
- Sayers, T.J.; Brooks, A.D.; Koh, C.Y.; Ma, W.; Seki, N.; Raziuddin, A.; Blazar, B.R.; Zhang, X.; Elliott, P.J.; Murphy, W.J. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL-mediated apoptosis by reducing levels of c-FLIP. Blood 2003, 102, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Mérino, D.; Lalaoui, N.; Morizot, A.; Schneider, P.; Solary, E.; Micheau, O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol. Cell Biol. 2006, 26, 7046–7055. [Google Scholar] [CrossRef] [PubMed]
- El-Gazzar, A.; Perco, P.; Eckelhart, E.; Anees, M.; Sexl, V.; Mayer, B.; Liu, Y.; Mikulits, W.; Horvat, R.; Pangerl, T.; et al. Natural immunity enhances the activity of a DR5 agonistic antibody and carboplatin in the treatment of ovarian cancer. Mol. Cancer Ther. 2010, 9, 1007–1018. [Google Scholar] [CrossRef]
- Wu, S.-H.; Mou, C.-Y.; Lin, H.-P. Synthesis of mesoporous silica nanoparticles. Chem. Soc. Rev. 2013, 42, 3862–3875. [Google Scholar] [CrossRef] [PubMed]
- Basak, M.; Rahman, M.L.; Ahmed, M.F.; Biswas, B.; Sharmin, N. The use of X-ray diffraction peak profile analysis to determine the structural parameters of cobalt ferrite nanoparticles using Debye-Scherrer, Williamson-Hall, Halder-Wagner and Size-strain plot: Different precipitating agent approach. J. Alloys Compd. 2022, 895, 162694. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Zhu, R.; Liu, Q.; Fei, J.; Wang, S. Anti-inflammatory activity of curcumin-loaded solid lipid nanoparticles in IL-1β transgenic mice subjected to the lipopolysaccharide-induced sepsis. Biomaterials 2015, 53, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Yamashita, K.; Itoh, Y.; Yoshino, K.; Nozawa, S.; Kasukawa, H. Comparative studies of polyethylene glycol-modified liposomes prepared using different PEG-modification methods. BBA-Biomembranes 2012, 1818, 2801–2807. [Google Scholar] [CrossRef] [PubMed]
- Vojoudi, H.; Badiei, A.; Amiri, A.; Banaei, A.; Ziarani, G.; Schenk-Joß, K. Efficient device for the benign removal of organic pollutants from aqueous solutions using modified mesoporous magnetite nanostructures. J. Phys. Chem. Solids 2018, 113, 210–219. [Google Scholar] [CrossRef]
- Kondapuram, S.; Sarvagalla, S.; Coumar, M. Docking-Based Virtual Screening Using PyRx Tool: Autophagy Target Vps34 as a Case Study. In Molecular Docking for Computer-Aided Drug Design; Academic Press: Cambridge, MA, USA, 2021; pp. 463–477. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, H.; Zarbafian, S.; Villar, E.; Mottarella, S.; Beglov, D.; Vajda, S.; Paschalidis, I.C.; Vakili, P.; Kozakov, D. Energy Minimization on Manifolds for Docking Flexible Molecules. J. Chem. Theory Comput. 2015, 11, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systemes BIOVIA. Discovery Studio Modelling Environment, Release 4.5; Scientific Research Publishing (SCIRP): Los Angeles, CA, USA, 2015; pp. 98–104. [Google Scholar]
- Rath, S.; Sahu, M.C.; Dubey, D.; Debata, N.K.; Padhy, R.N. Which value should be used as the lethal concentration 50 (LC 50) with bacteria? Interdiscip. Sci. Comput. Life Sci. 2011, 3, 138–143. [Google Scholar] [CrossRef]
- Phatak, R.S.; Pratinidhi, A.K.; Hendre, A.S. Screening of some Indian household spices for comparative studies of antioxidant and antiradical activities by using in-vitro models. Screening 2015, 8, 431–438. [Google Scholar]
- Moein, S.; Moein, M.; Khoshnoud, M.J.; Kalanteri, T. In vitro antioxidant properties evaluation of 10 Iranian medicinal plants by different methods. Iran. Red. Crescent Med. J. 2012, 14, 771. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nozaki-Taguchi, N.; Chiba, T. Analgesic effect of intrathecally administered orexin-A in the rat formalin test and in the rat hot plate test. Br. J. Pharmacol. 2002, 137, 170–176. [Google Scholar] [CrossRef]
- Samuelson, B.T.; Cuker, A.; Siegal, D.M.; Crowther, M.; Garcia, D.A. Laboratory assessment of the anticoagulant activity of direct oral anticoagulants: A systematic review. Chest 2017, 151, 127–138. [Google Scholar] [CrossRef]
- Care IoLARCo; Animals UoL. Guide for the Care and Use of Laboratory Animals; US Department of Health and Human Services, Public Health Service, National: Washington, DC, USA, 1986.
- Sathpathi, S.; Mohanty, A.K.; Satpathi, P.; Mishra, S.K.; Behera, P.K.; Patel, G.; Dondorp, A.M. Comparing Leishman and Giemsa staining for the assessment of peripheral blood smear preparations in a malaria-endemic region in India. Malar. J. 2014, 13, 512. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P. Reagents and Methods for Isolation of Purified RNA. U.S. Patent 7,794,932, 14 September 2010. [Google Scholar]
- Stellwagen, N.C. DNA gel electrophoresis. In Nucleic Acid Electrophoresis; Springer: Berlin/Heidelberg, Germany, 1998; pp. 1–53. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddique, M.H.; Bukhari, S.; Khan, I.U.; Essa, A.; Ali, Z.; Sabir, U.; Ayoub, O.; Saadia, H.; Yaseen, M.; Sultan, A.; et al. In Silico, In Vitro, and In Vivo Evaluation of Caffeine-Coated Nanoparticles as a Promising Therapeutic Avenue for AML through NF-Kappa B and TRAIL Pathways Modulation. Pharmaceuticals 2023, 16, 1742. https://doi.org/10.3390/ph16121742
Siddique MH, Bukhari S, Khan IU, Essa A, Ali Z, Sabir U, Ayoub O, Saadia H, Yaseen M, Sultan A, et al. In Silico, In Vitro, and In Vivo Evaluation of Caffeine-Coated Nanoparticles as a Promising Therapeutic Avenue for AML through NF-Kappa B and TRAIL Pathways Modulation. Pharmaceuticals. 2023; 16(12):1742. https://doi.org/10.3390/ph16121742
Chicago/Turabian StyleSiddique, Muhammad Hamid, Sidra Bukhari, Inam Ullah Khan, Asiya Essa, Zain Ali, Usama Sabir, Omiya Ayoub, Haleema Saadia, Muhammad Yaseen, Aneesa Sultan, and et al. 2023. "In Silico, In Vitro, and In Vivo Evaluation of Caffeine-Coated Nanoparticles as a Promising Therapeutic Avenue for AML through NF-Kappa B and TRAIL Pathways Modulation" Pharmaceuticals 16, no. 12: 1742. https://doi.org/10.3390/ph16121742
APA StyleSiddique, M. H., Bukhari, S., Khan, I. U., Essa, A., Ali, Z., Sabir, U., Ayoub, O., Saadia, H., Yaseen, M., Sultan, A., Murtaza, I., Kerr, P. G., Bhat, M. A., & Anees, M. (2023). In Silico, In Vitro, and In Vivo Evaluation of Caffeine-Coated Nanoparticles as a Promising Therapeutic Avenue for AML through NF-Kappa B and TRAIL Pathways Modulation. Pharmaceuticals, 16(12), 1742. https://doi.org/10.3390/ph16121742