New Insights for Polyphenolic Compounds as Naturally Inspired Proteasome Inhibitors
Abstract
:1. Introduction
2. Results
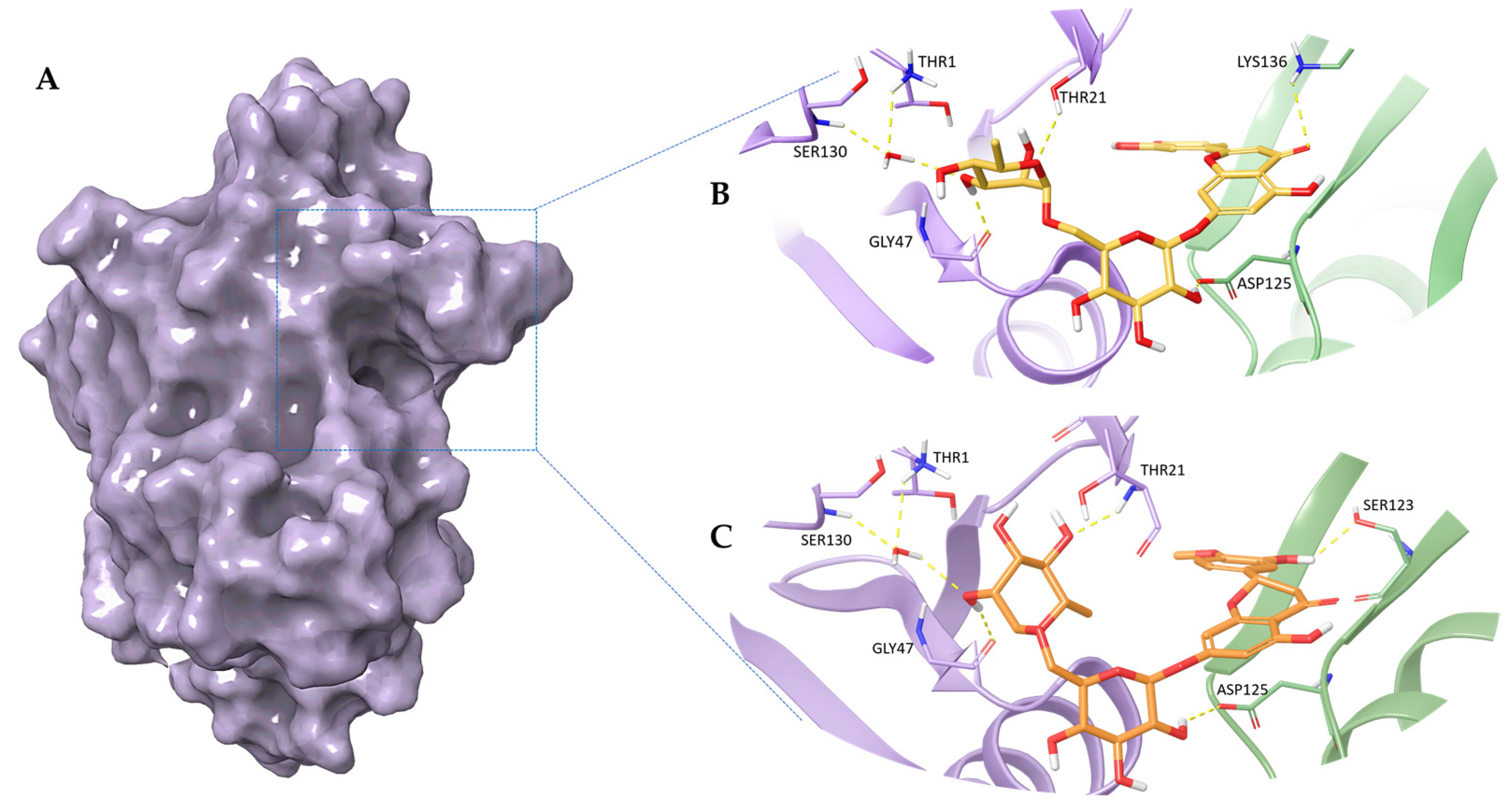
2.1. Molecular Docking Studies
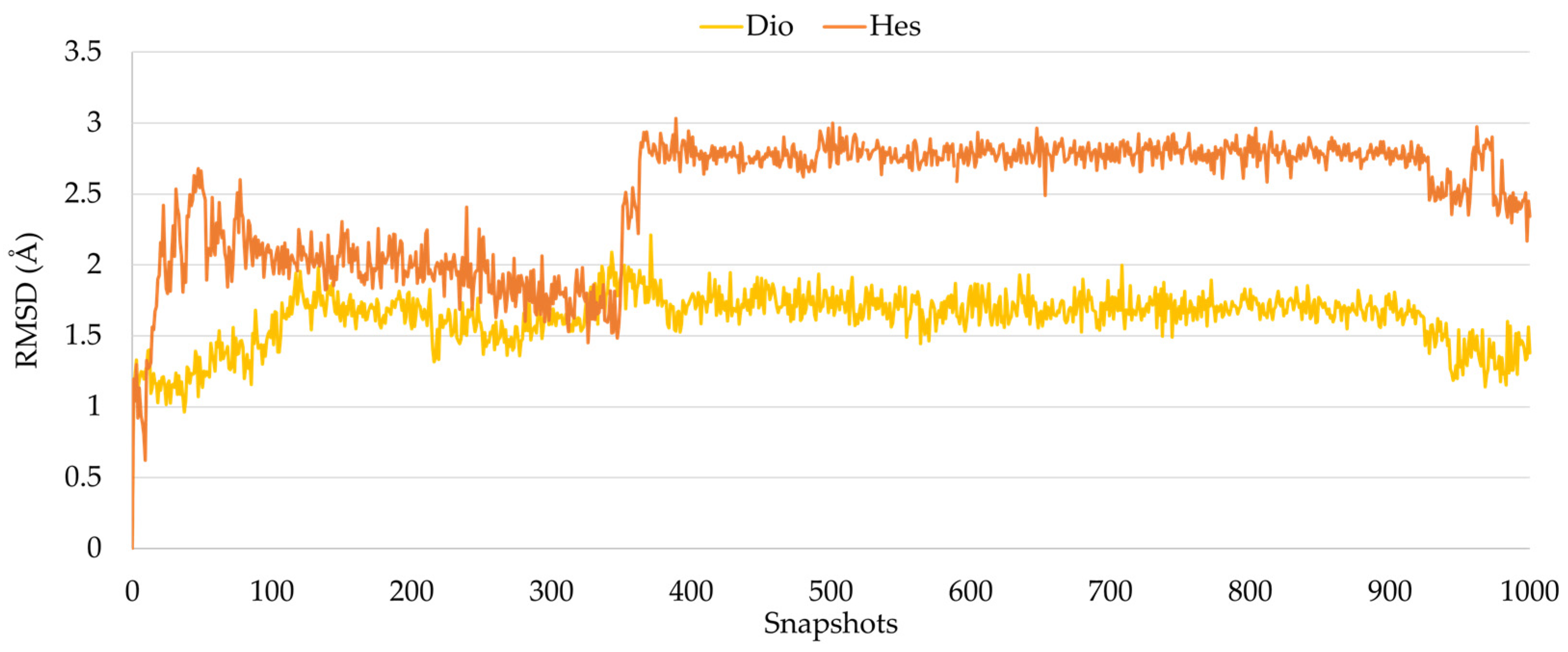
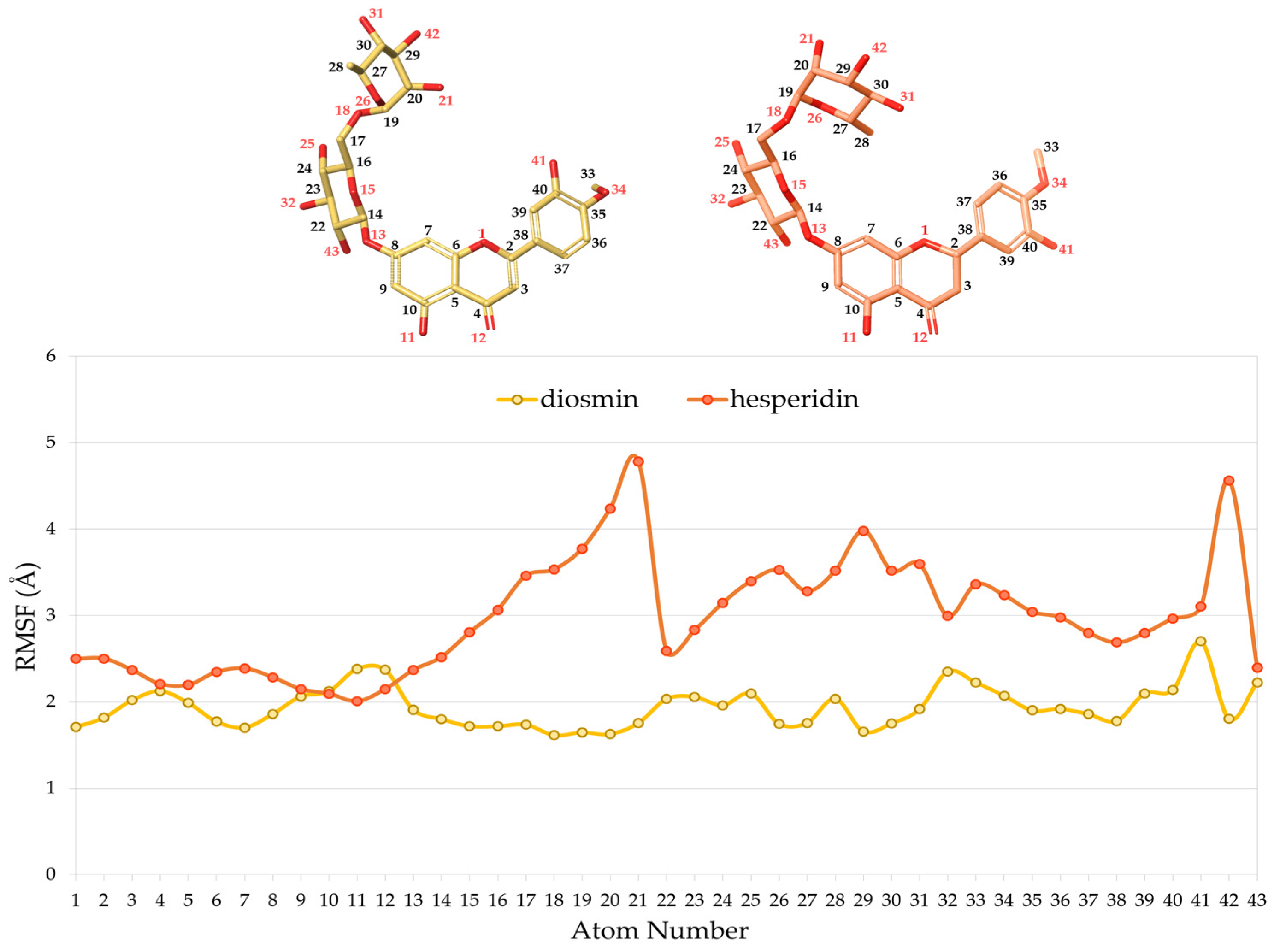
2.2. Molecular Dynamics Studies
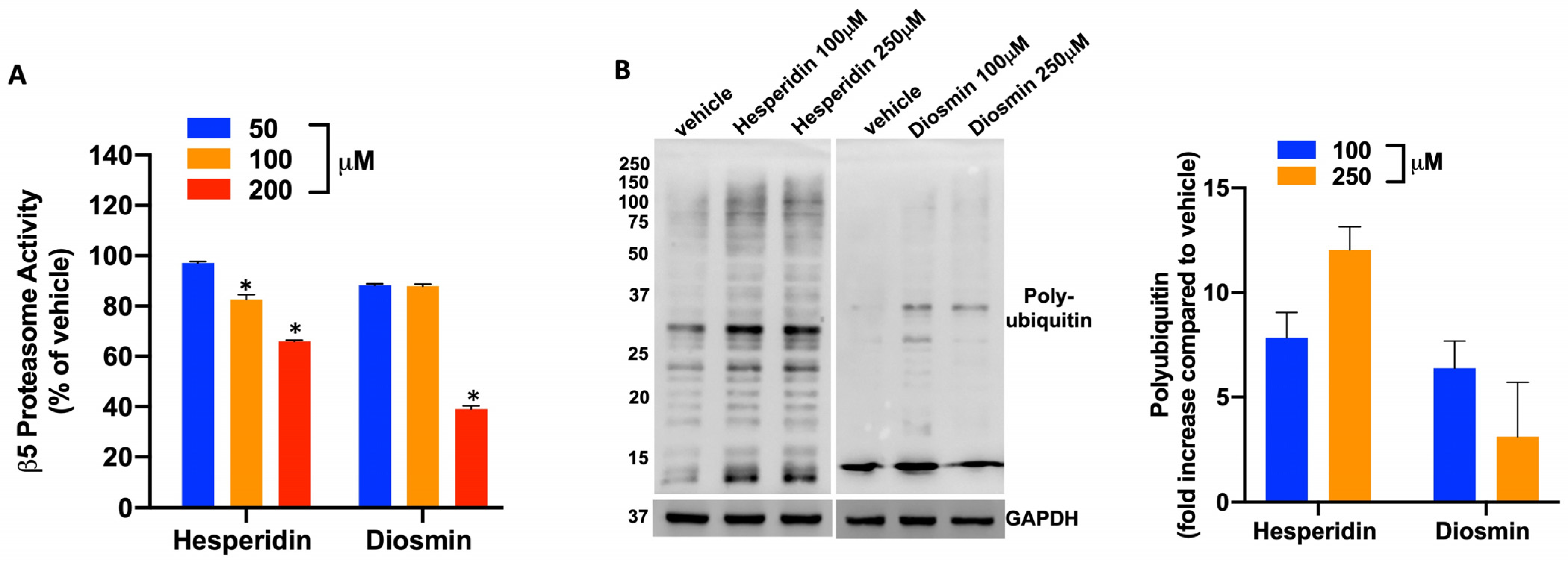
2.3. Hesperidin and Diosmin Inhibit β5 Proteasome Activity In Vitro and in Cell-Based Assays
2.4. Hesperidin and Diosmin Exert Anti-Tumor Activity against PI-Sensitive and PI-Resistant MM Cells
3. Discussion
4. Materials and Methods
4.1. Computational Studies
4.2. MM Cell Lines
4.3. Cell Viability Assay
4.4. Apoptosis Assay
4.5. Western Blotting (WB)
4.6. In Vitro Proteasome Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El Gharras, H. Polyphenols: Food sources, properties and applications—A review. Int. J. Food Sci. Technol. 2009, 44, 2512–2518. [Google Scholar] [CrossRef]
- Haminiuk, C.W.; Maciel, G.M.; Plata-Oviedo, M.S.; Peralta, R.M. Phenolic compounds in fruits—An overview. Int. J. Food Sci. Technol. 2012, 47, 2023–2044. [Google Scholar] [CrossRef]
- Pandey, K.B.; Rizvi, S.I. Plant polyphenols as dietary antioxidants in human health and disease. Oxidative Med. Cell. Longev. 2009, 2, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Lacueva, C.; Medina-Remon, A.; Llorach, R.; Urpi-Sarda, M.; Khan, N.; Chiva-Blanch, G.; Zamora-Ros, R.; Rotches-Ribalta, M.; Lamuela-Raventos, R.M. Phenolic compounds: Chemistry and occurrence in fruits and vegetables. In Fruit and Vegetable Phytochemicals: Chemistry, Nutritional Value and Stability; Wiley-Blackwell: Ames, IA, USA, 2010; pp. 53–56. [Google Scholar]
- Abbas, M.; Saeed, F.; Anjum, F.M.; Afzaal, M.; Tufail, T.; Bashir, M.S.; Ishtiaq, A.; Hussain, S.; Suleria, H.A.R. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. [Google Scholar] [CrossRef]
- Cutrim, C.S.; Cortez, M.A.S. A review on polyphenols: Classification, beneficial effects and their application in dairy products. Int. J. Dairy Technol. 2018, 71, 564–578. [Google Scholar] [CrossRef]
- Leopoldini, M.; Russo, N.; Toscano, M. The Molecular Basis of Working Mechanism of Natural Polyphenolic Antioxidants. Food Chem. 2011, 125, 288–306. [Google Scholar] [CrossRef]
- Losada-Barreiro, S.; Bravo-Díaz, C. Free Radicals and Polyphenols: The Redox Chemistry of Neurodegenerative Diseases. Eur. J. Med. Chem. 2017, 133, 379–402. [Google Scholar] [CrossRef]
- Cassidy, L.; Fernandez, F.; Johnson, J.B.; Naiker, M.; Owoola, A.G.; Broszczak, D.A. Oxidative stress in alzheimer’s disease: A review on emergent natural polyphenolic therapeutics. Complement. Ther. Med. 2020, 49, 102294. [Google Scholar] [CrossRef]
- Perron, N.R.; Brumaghim, J.L. A Review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell Biochem. Biophys. 2009, 53, 75–100. [Google Scholar] [CrossRef]
- Costa, L.G.; Garrick, J.M.; Roquè, P.J.; Pellacani, C. Mechanisms of Neuroprotection by Quercetin: Counteracting Oxidative Stress and More. Oxidative Med. Cell. Longev. 2016, 2016, 2986796. [Google Scholar] [CrossRef]
- Marchese, E.; Orlandi, V.; Turrini, F.; Romeo, I.; Boggia, R.; Alcaro, S.; Costa, G. In Silico and In Vitro Study of Antioxidant Potential of Urolithins. Antioxidants 2023, 12, 697. [Google Scholar] [CrossRef]
- Giglio, R.V.; Patti, A.M.; Cicero, A.; Lippi, G.; Rizzo, M.; Toth, P.P.; Banach, M. Polyphenols: Potential Use in the Prevention and Treatment of Cardiovascular Diseases. Curr. Pharm. Des. 2018, 24, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, I.; Oliveira, J.; Pinho, A.; Carvalho, E. The Role of Nutraceutical Containing Polyphenols in Diabetes Prevention. Metabolites 2022, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, L.; Zhang, B.; Deng, Z.Y.; Luo, T. The Structure Basis of Phytochemicals as Metabolic Signals for Combating Obesity. Front. Nutr. 2022, 9, 913883. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Landívar, M.F.; Tapia-Quirós, P.; Vecino, X.; Reig, M.; Valderrama, C.; Granados, M.; Cortina, J.L.; Saurina, J. Polyphenols and their potential role to fight viral diseases: An overview. Sci. Total Environ. 2021, 801, 149719. [Google Scholar] [CrossRef] [PubMed]
- Shakoor, H.; Feehan, J.; Apostolopoulos, V.; Platat, C.; Al Dhaheri, A.S.; Ali, H.I.; Ismail, L.C.; Bosevski, M.; Stojanovska, L. Immunomodulatory Effects of Dietary Polyphenols. Nutrients 2021, 13, 728. [Google Scholar] [CrossRef] [PubMed]
- Bhullar, K.S.; Rupasinghe, H.P. Polyphenols: Multipotent therapeutic agents in neurodegenerative diseases. Oxidative Med. Cell. Longev. 2013, 2013, 891748. [Google Scholar] [CrossRef]
- Gupta, N.; Singh, S.; Chauhan, D.; Srivastava, R.; Singh, V.K. Exploring the Anticancer Potentials of Polyphenols: A Comprehensive Review of Patents in the Last Five Years. Recent Pat. Anticancer Drug Discov. 2023, 18, 3–10. [Google Scholar] [CrossRef]
- Juli, G.; Oliverio, M.; Bellizzi, D.; Gallo Cantafio, M.E.; Grillone, K.; Passarino, G.; Colica, C.; Nardi, M.; Rossi, M.; Procopio, A.; et al. Anti-tumor Activity and Epigenetic Impact of the Polyphenol Oleacein in Multiple Myeloma. Cancers 2019, 11, 990. [Google Scholar] [CrossRef]
- Todoerti, K.; Gallo Cantafio, M.E.; Oliverio, M.; Juli, G.; Rocca, C.; Citraro, R.; Tassone, P.; Procopio, A.; De Sarro, G.; Neri, A.; et al. Oleil Hydroxytyrosol (HTOL) Exerts Anti-Myeloma Activity by Antagonizing Key Survival Pathways in Malignant Plasma Cells. Int. J. Mol. Sci. 2021, 22, 11639. [Google Scholar] [CrossRef]
- Niedzwiecki, A.; Roomi, M.W.; Kalinovsky, T.; Rath, M. Anticancer Efficacy of Polyphenols and Their Combinations. Nutrients 2016, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Attri, D.C.; Sati, P.; Dhyani, P.; Szopa, A.; Sharifi-Rad, J.; Hano, C.; Calina, D.; Cho, W.C. Recent updates on anticancer mechanisms of polyphenols. Front. Cell Dev. Biol. 2022, 10, 1005910. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Wehmer, M.; Rudack, T.; Beck, F.; Aufderheide, A.; Pfeifer, G.; Plitzko, J.M.; Förster, F.; Schulten, K.; Baumeister, W.; Sakata, E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2017, 114, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Ito, S. Proteasome Inhibitors for the Treatment of Multiple Myeloma. Cancers 2020, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- van de Donk, N.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.P.; Dou, Q. Bortezomib as the First Proteasome Inhibitor Anticancer Drug: Current Status and Future Perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Herndon, T.M.; Deisseroth, A.; Kaminskas, E.; Kane, R.C.; Koti, K.M.; Rothmann, M.D.; Habtemariam, B.; Bullock, J.; Bray, J.D.; Hawes, J.; et al. U.S. Food and Drug Administration approval: Carfilzomib for the treatment of multiple myeloma. Clin. Cancer Res. 2013, 19, 4559–4563. [Google Scholar] [CrossRef]
- Shirley, M. Ixazomib: First Global Approval. Drugs 2016, 76, 405–411. [Google Scholar] [CrossRef]
- McConkey, D.J.; Zhu, K. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Res. Updates 2008, 11, 164–179. [Google Scholar] [CrossRef]
- Paradzik, T.; Bandini, C.; Mereu, E.; Labrador, M.; Taiana, E.; Amodio, N.; Neri, A.; Piva, R. The Landscape of Signaling Pathways and Proteasome Inhibitors Combinations in Multiple Myeloma. Cancers 2021, 13, 1235. [Google Scholar] [CrossRef]
- Taiana, E.; Gallo Cantafio, M.E.; Favasuli, V.K.; Bandini, C.; Viglietto, G.; Piva, R.; Neri, A.; Amodio, N. Genomic Instability in Multiple Myeloma: A “Non-Coding RNA” Perspective. Cancers 2021, 13, 2127. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Q.; Dou, Q.P.; Yang, H. Discovery of Natural Proteasome Inhibitors as Novel Anticancer Therapeutics: Current Status and Perspectives. Curr. Protein. Pept. Sci. 2018, 19, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, F.A.; Costa, G.; Gallo Cantafio, M.E.; Torcasio, R.; Trapasso, F.; Alcaro, S.; Viglietto, G.; Amodio, N. Natural Agents as Novel Potential Source of Proteasome Inhibitors with Anti-Tumor Activity: Focus on Multiple Myeloma. Molecules 2023, 28, 1438. [Google Scholar] [CrossRef] [PubMed]
- Maia, E.H.B.; Assis, L.C.; de Oliveira, T.A.; da Silva, A.M.; Taranto, A.G. Structure-Based Virtual Screening: From Classical to Artificial Intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Bogucka-Kocka, A.; Woźniak, M.; Feldo, M.; Kocki, J.; Szewczyk, K. Diosmin—Isolation techniques, determination in plant material and pharmaceutical formulations, and clinical use. Nat. Prod. Commun. 2013, 8, 1934578X1300800435. [Google Scholar] [CrossRef]
- Mustafa, S.; Akbar, M.; Khan, M.A.; Sunita, K.; Parveen, S.; Pawar, J.S.; Massey, S.; Agarwal, N.R.; Husain, S.A. Plant metabolite diosmin as the therapeutic agent in human diseases. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100122. [Google Scholar] [CrossRef]
- Schrader, J.; Henneberg, F.; Mata, R.A.; Tittmann, K.; Schneider, T.R.; Stark, H.; Bourenkov, G.; Chari, A. The Inhibition Mechanism of Human 20S Proteasomes Enables Next-Generation Inhibitor Design. Science 2016, 353, 594–598. [Google Scholar] [CrossRef]
- Malacrida, A.; Cavalloro, V.; Martino, E.; Costa, G.; Ambrosio, F.A.; Alcaro, S.; Rigolio, R.; Cassetti, A.; Miloso, M.; Collina, S. Anti-multiple myeloma potential of secondary metabolites from Hibiscus sabdariffa—Part 2. Molecules 2021, 26, 6596. [Google Scholar] [CrossRef]
- Listro, R.; Malacrida, A.; Ambrosio, F.A.; Rossino, G.; Di Giacomo, M.; Cavalloro, V.; Garbagnoli, M.; Linciano, P.; Rossi, D.; Cavaletti, G.; et al. From Nature to Synthetic Compounds: Novel 1(N),2,3 Trisubstituted-5-oxopyrrolidines Targeting Multiple Myeloma Cells. Int. J. Mol. Sci. 2022, 23, 13061. [Google Scholar] [CrossRef] [PubMed]
- Guedes, R.A.; Grilo, J.H.; Carvalho, A.N.; Fernandes, P.M.P.; Ressurreição, A.S.; Brito, V.; Santos, A.O.; Silvestre, S.; Gallerani, E.; Gama, M.J.; et al. New Scaffolds of Proteasome Inhibitors: Boosting Anticancer Potential by Exploiting the Synergy of In Silico and In Vitro Methodologies. Pharmaceuticals 2023, 16, 1096. [Google Scholar] [CrossRef] [PubMed]
- Sliva, J. Diosmin—Still an important modality in the treatment of venous insufficiency. Vnitr. Lek. 2019, 65, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Feldo, M.; Wójciak-Kosior, M.; Sowa, I.; Kocki, J.; Bogucki, J.; Zubilewicz, T.; Kęsik, J.; Bogucka-Kocka, A. Effect of Diosmin Administration in Patients with Chronic Venous Disorders on Selected Factors Affecting Angiogenesis. Molecules 2019, 24, 3316. [Google Scholar] [CrossRef] [PubMed]
- Struckmann, J.R.; Nicolaides, A.N. Flavonoids: A Review of the Pharmacology and Therapeutic Efficacy of Daflon 500 mg in Patients with Chronic Venous Insufficiency and Related Disorders. Angiology 1994, 45, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Lichota, A.; Gwozdzinski, L.; Gwozdzinski, K. Therapeutic potential of natural compounds in inflammation and chronic venous insufficiency. Eur. J. Med. Chem. 2019, 176, 68–91. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Landis-Piwowar, K.; Chan, T.H.; Dou, Q.P. Green tea polyphenols as proteasome inhibitors: Implication in chemoprevention. Curr. Cancer Drug Targets 2011, 11, 296–306. [Google Scholar] [CrossRef]
- Shen, M.; Chan, T.H.; Dou, Q.P. Targeting tumor ubiquitin-proteasome pathway with polyphenols for chemosensitization. Anti-Cancer Agents Med. Chem. 2012, 12, 891–901. [Google Scholar] [CrossRef]
- Mujtaba, T.; Dou, Q.P. Black Tea Polyphenols Inhibit Tumor Proteasome Activity. In Vivo 2012, 26, 197–202. [Google Scholar]
- Schrödinger Release 2018-1: Protein Preparation Wizard; Schrödinger LLC: New York, NY, USA, 2018.
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Schrödinger Release 2018-1: Prime; Schrödinger LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: Glide; Schrödinger LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: LigPrep; Schrödinger LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: MacroModel; Schrödinger LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: Desmond Molecular Dynamics System; D.E. Shaw Research: New York, NY, USA, 2018.
- Hou, T.J.; Wang, J.M.; Li, Y.Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Morelli, E.; Hunter, Z.R.; Fulciniti, M.; Gullà, A.; Perrotta, I.D.; Zuccalà, V.; Federico, C.; Juli, G.; Manzoni, M.; Ronchetti, D.; et al. Therapeutic activation of G protein-coupled estrogen receptor 1 in Waldenström Macroglobulinemia. Exp. Hematol. Oncol. 2022, 11, 54. [Google Scholar] [CrossRef]

| Name | DrugBank ID | 2D Structure | D-Score * |
|---|---|---|---|
| Diosmin | DB08995 | 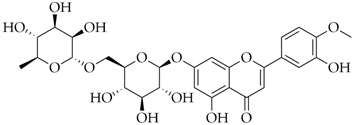 | −8.26 |
| Hesperidin | DB04703 | 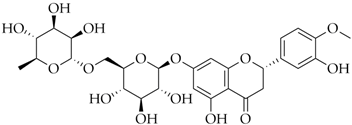 | −8.14 |
| Polydatin | DB11263 | 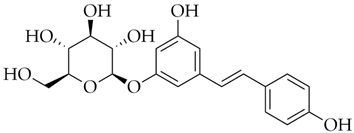 | −7.99 |
| Cromoglicic Acid | DB01003 | 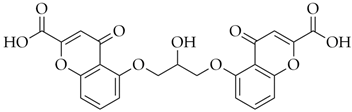 | −7.98 |
| Curcumin | DB11672 |  | −7.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchese, E.; Gallo Cantafio, M.E.; Ambrosio, F.A.; Torcasio, R.; Valentino, I.; Trapasso, F.; Viglietto, G.; Alcaro, S.; Costa, G.; Amodio, N. New Insights for Polyphenolic Compounds as Naturally Inspired Proteasome Inhibitors. Pharmaceuticals 2023, 16, 1712. https://doi.org/10.3390/ph16121712
Marchese E, Gallo Cantafio ME, Ambrosio FA, Torcasio R, Valentino I, Trapasso F, Viglietto G, Alcaro S, Costa G, Amodio N. New Insights for Polyphenolic Compounds as Naturally Inspired Proteasome Inhibitors. Pharmaceuticals. 2023; 16(12):1712. https://doi.org/10.3390/ph16121712
Chicago/Turabian StyleMarchese, Emanuela, Maria Eugenia Gallo Cantafio, Francesca Alessandra Ambrosio, Roberta Torcasio, Ilenia Valentino, Francesco Trapasso, Giuseppe Viglietto, Stefano Alcaro, Giosuè Costa, and Nicola Amodio. 2023. "New Insights for Polyphenolic Compounds as Naturally Inspired Proteasome Inhibitors" Pharmaceuticals 16, no. 12: 1712. https://doi.org/10.3390/ph16121712
APA StyleMarchese, E., Gallo Cantafio, M. E., Ambrosio, F. A., Torcasio, R., Valentino, I., Trapasso, F., Viglietto, G., Alcaro, S., Costa, G., & Amodio, N. (2023). New Insights for Polyphenolic Compounds as Naturally Inspired Proteasome Inhibitors. Pharmaceuticals, 16(12), 1712. https://doi.org/10.3390/ph16121712










