Chemotherapeutic Activity of Imidazolium-Supported Pd(II) o-Vanillylidene Diaminocyclohexane Complexes Immobilized in Nanolipid as Inhibitors for HER2/neu and FGFR2/FGF2 Axis Overexpression in Breast Cancer Cells
Abstract
1. Introduction
2. Results
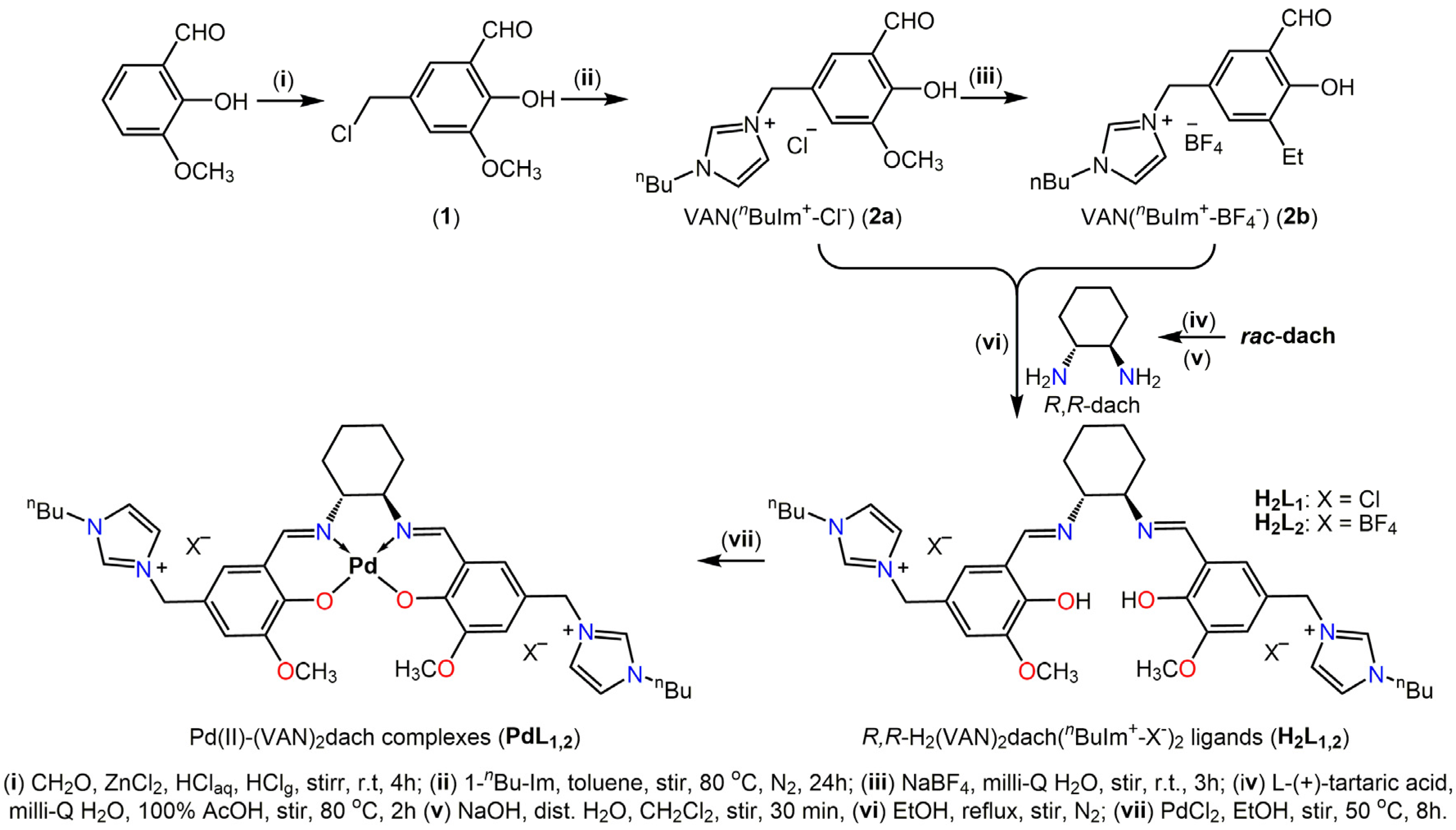
2.1. Chemistry of Synthesis
2.2. Characterization of New Materials
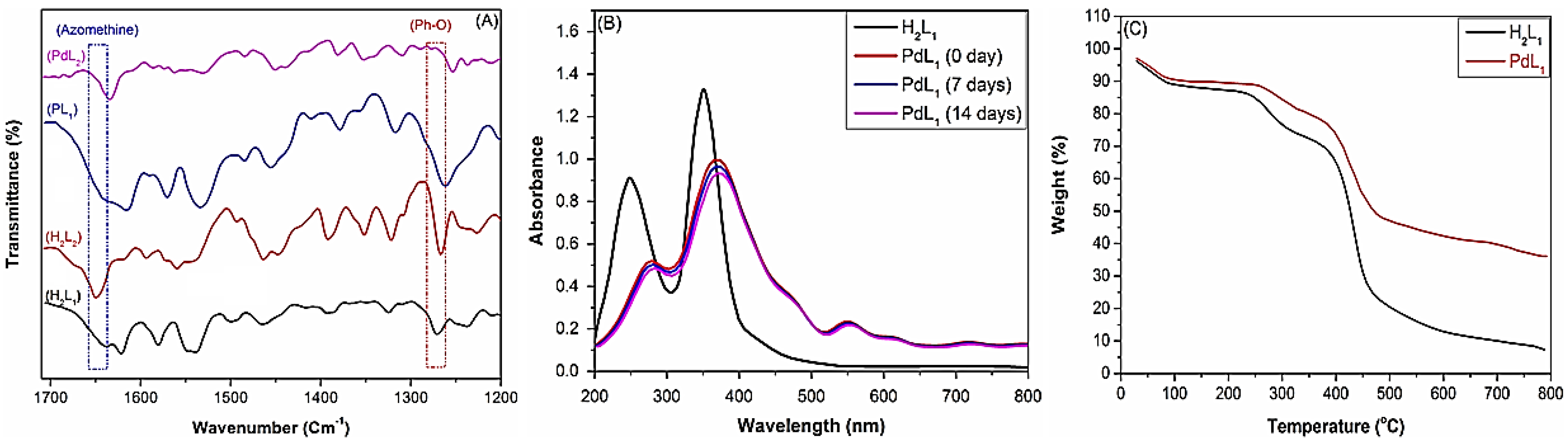
2.2.1. Physicochemical Characterization
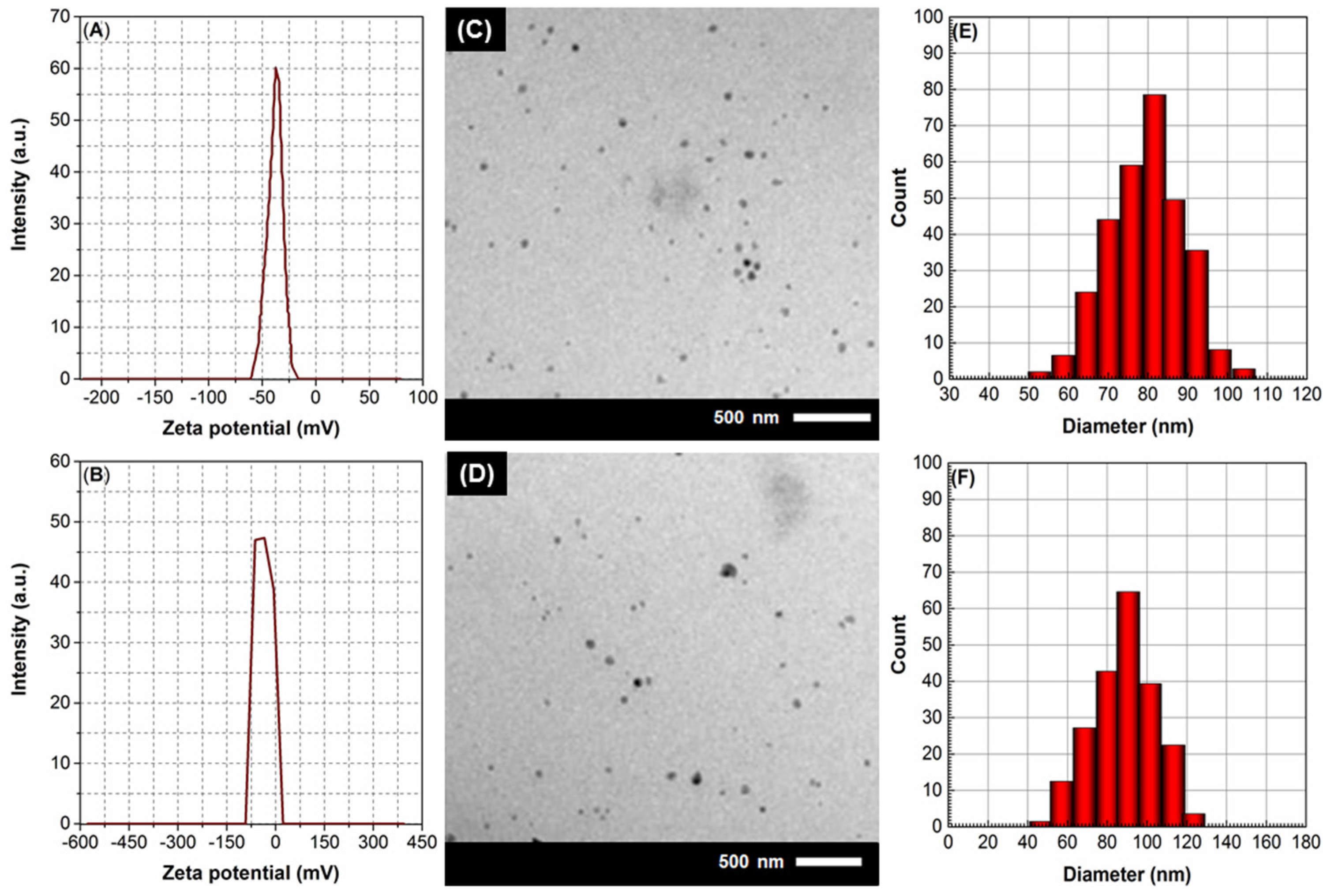
2.2.2. Morphological Characterization
2.2.3. Pharmacological Characterization
2.3. Pharmacology and Biological Activities
2.3.1. In Vitro Cytotoxicity
2.3.2. Comet Assay
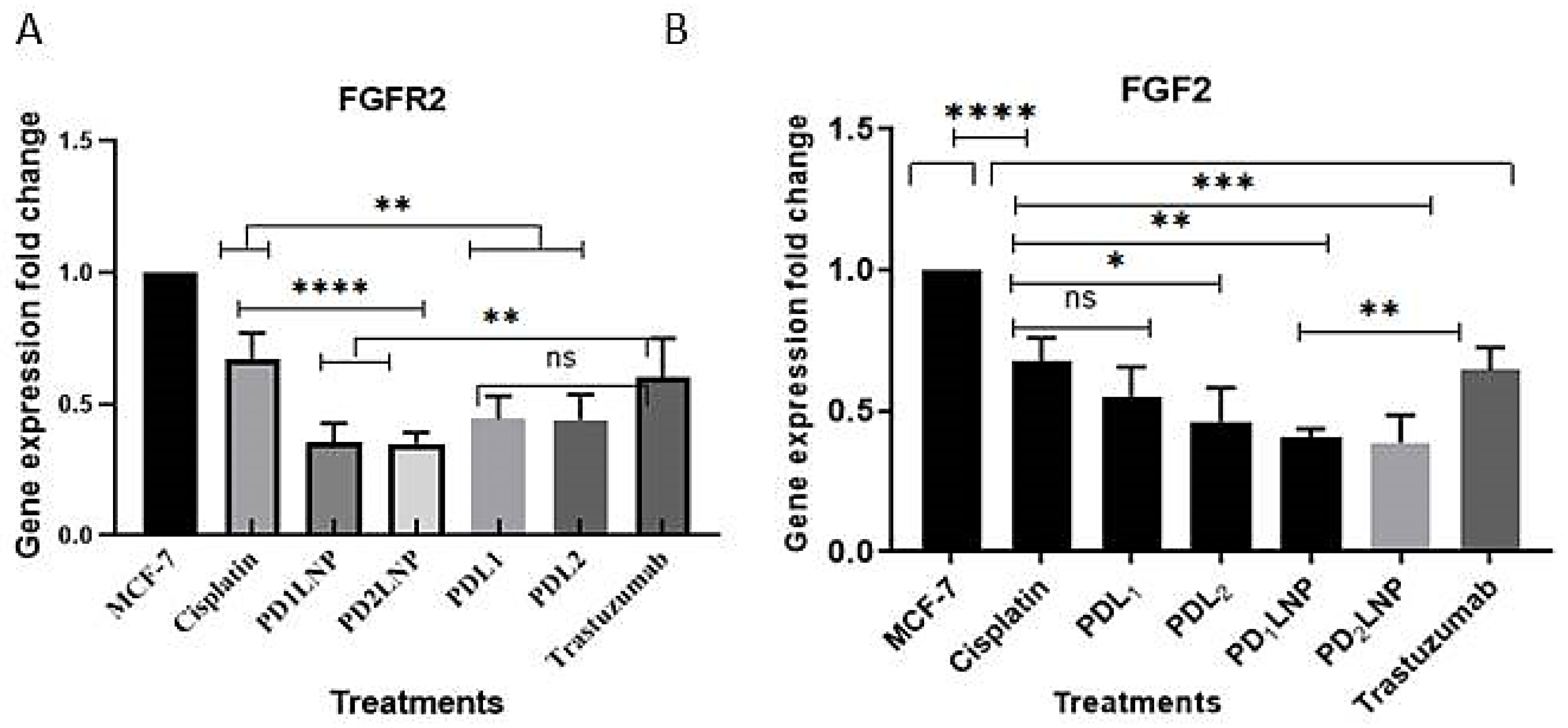
2.3.3. Quantitative Real-Time (qPCR) Estimation for FGFR2 and FGF2
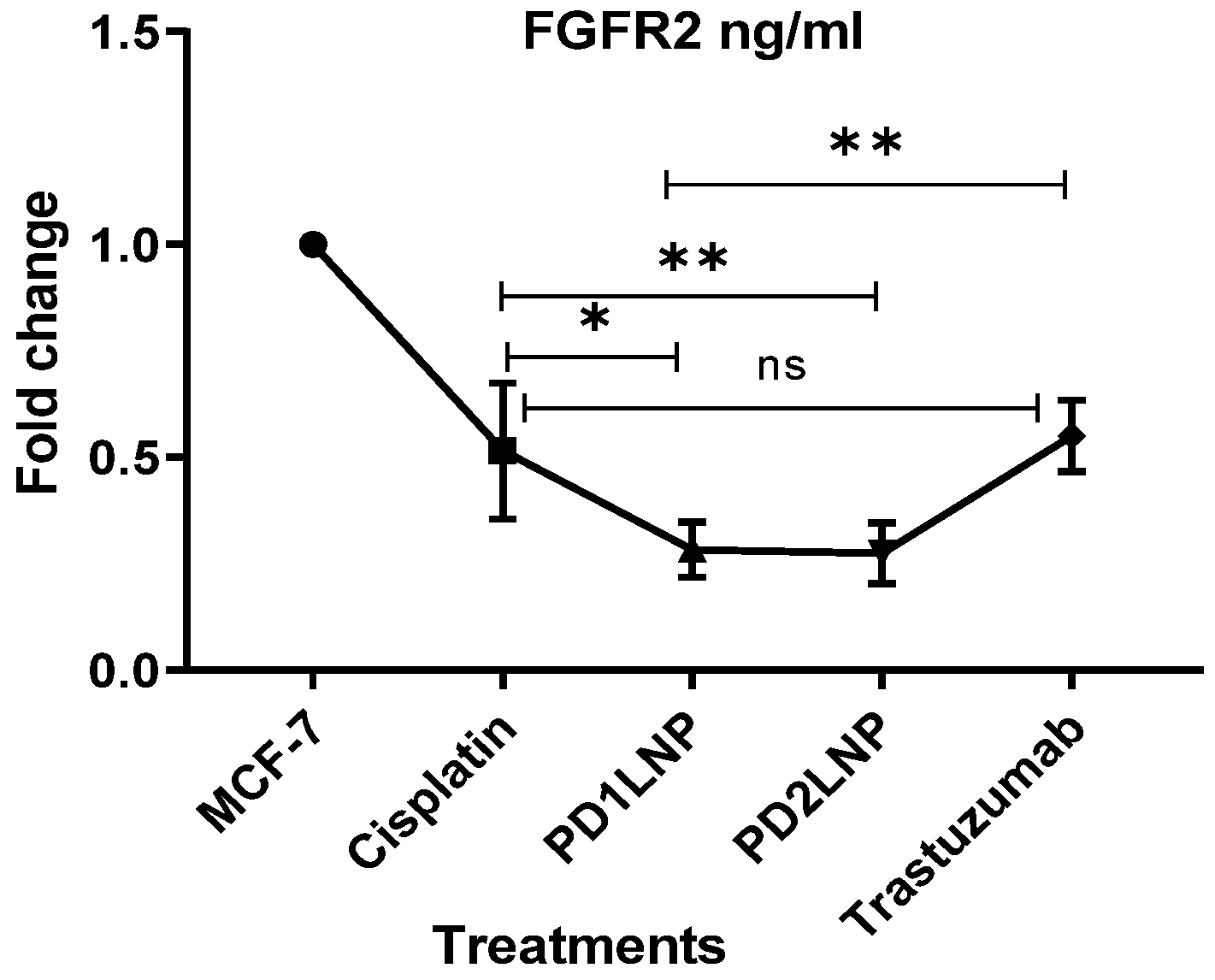
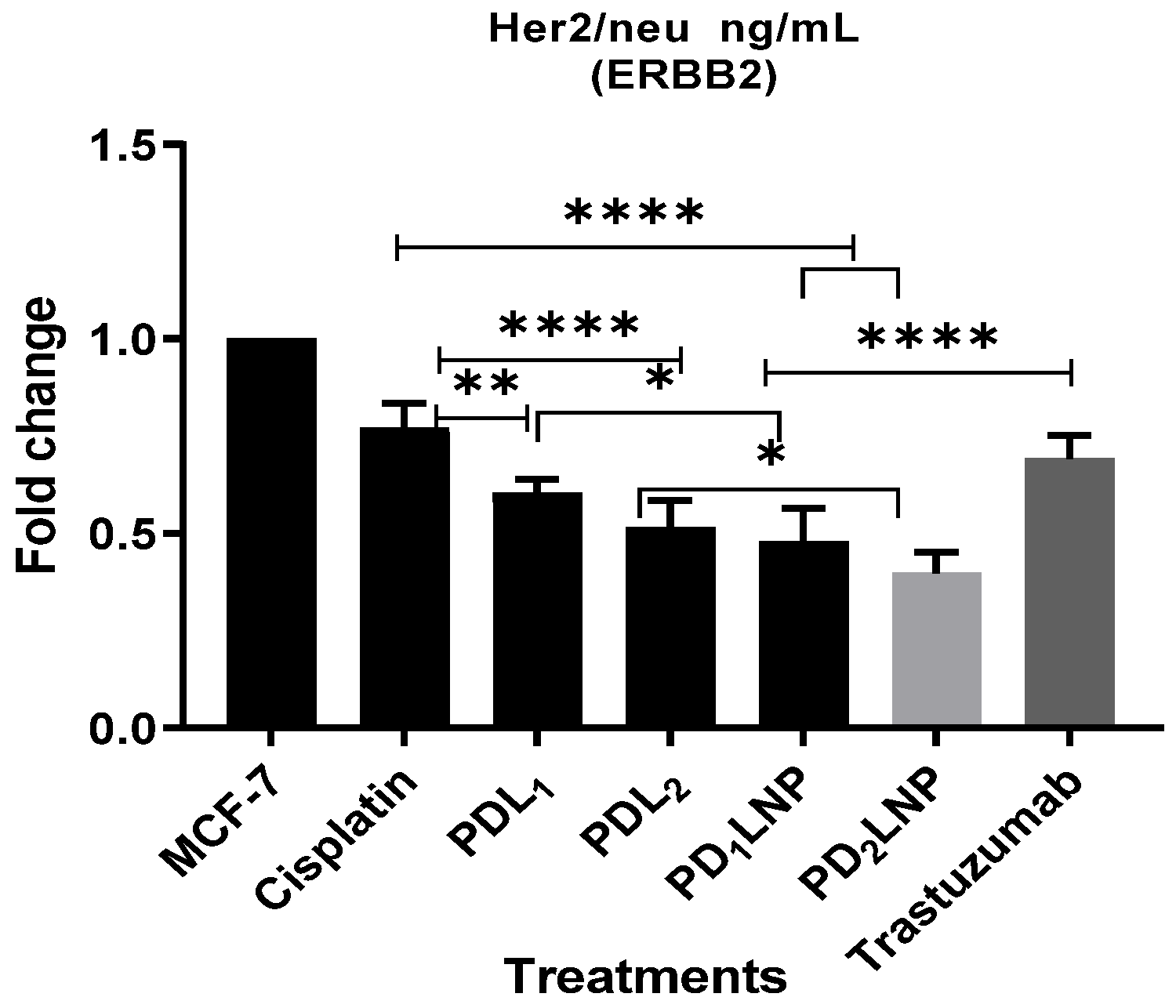
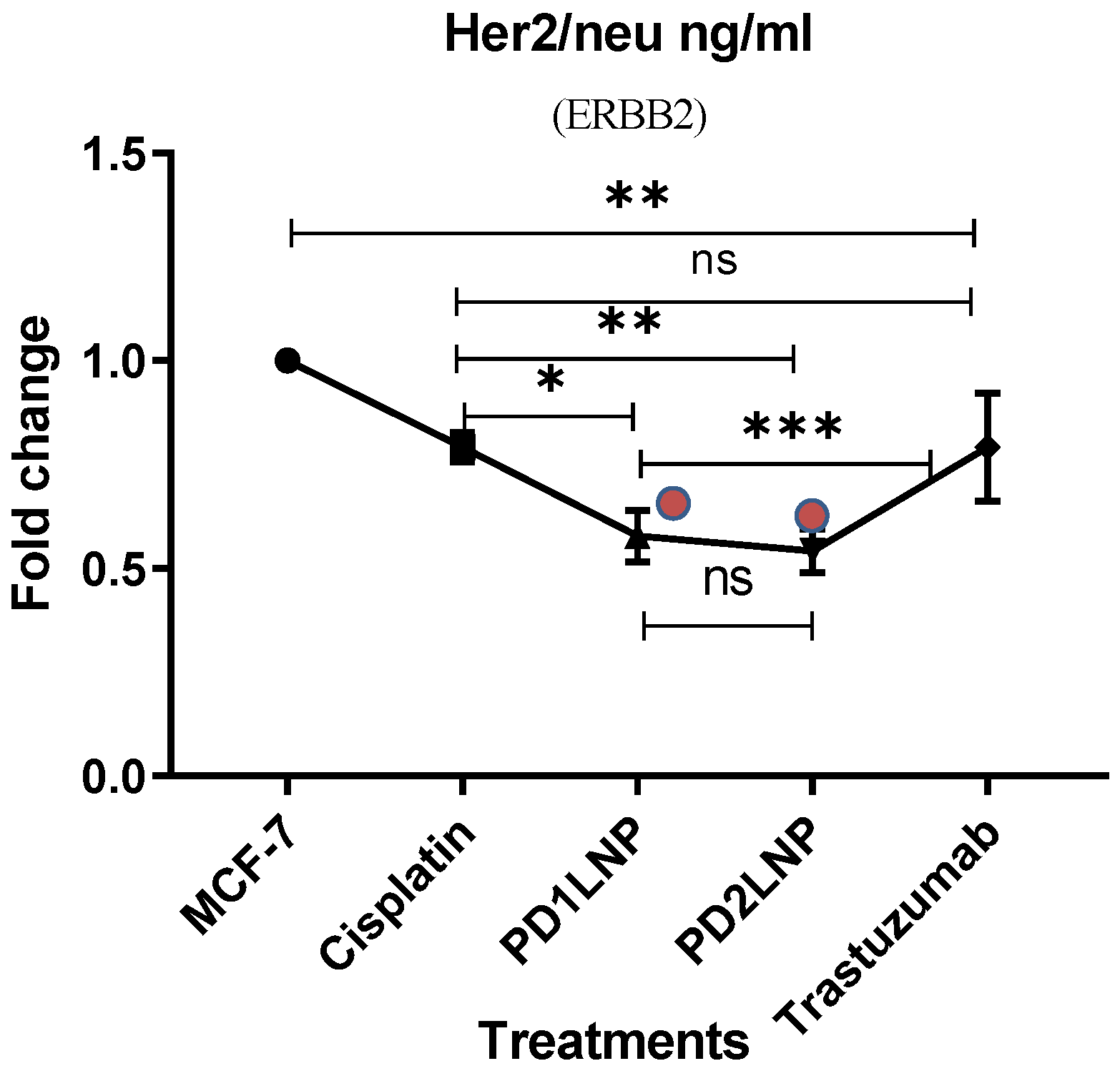
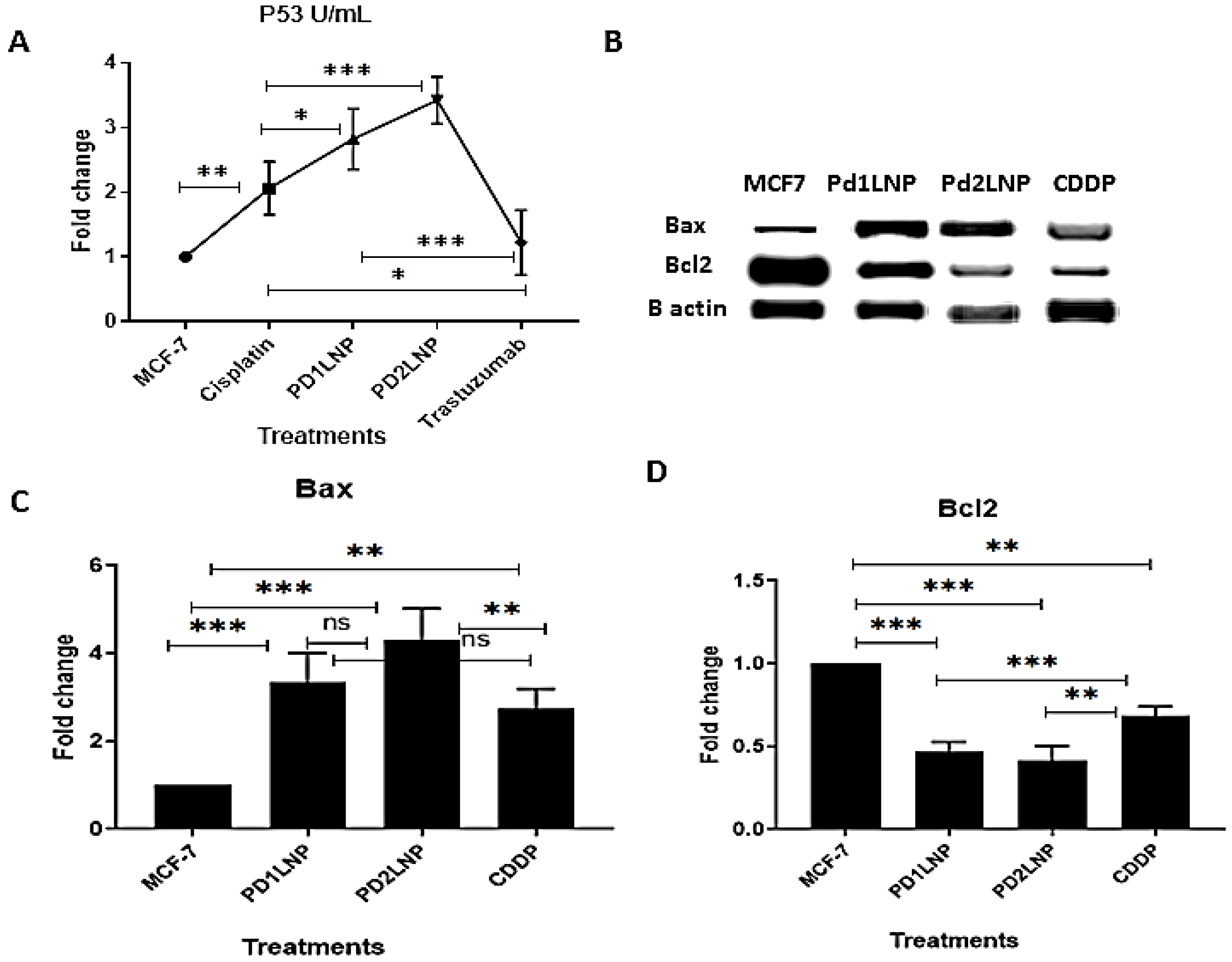
2.3.4. The Enzyme-Linked Immunosorbent Assay (ELISA) for FGFR2, Her2/neu, TGFβ1, and P53
3. Discussion
4. Materials and Methods
4.1. Synthesis of R,R-H2(VAN)2dach(nBuIm+-X–)2 Ligands (H2L1,2)
4.2. Synthesis of Pd(II)-(VAN)2dach Complexes (PdL1,2)
4.3. Preparation of PdL1,2 Complexes-Loaded LNPs (PdL1,2LNPs)
4.4. Characterization of PdL1,2LNPs
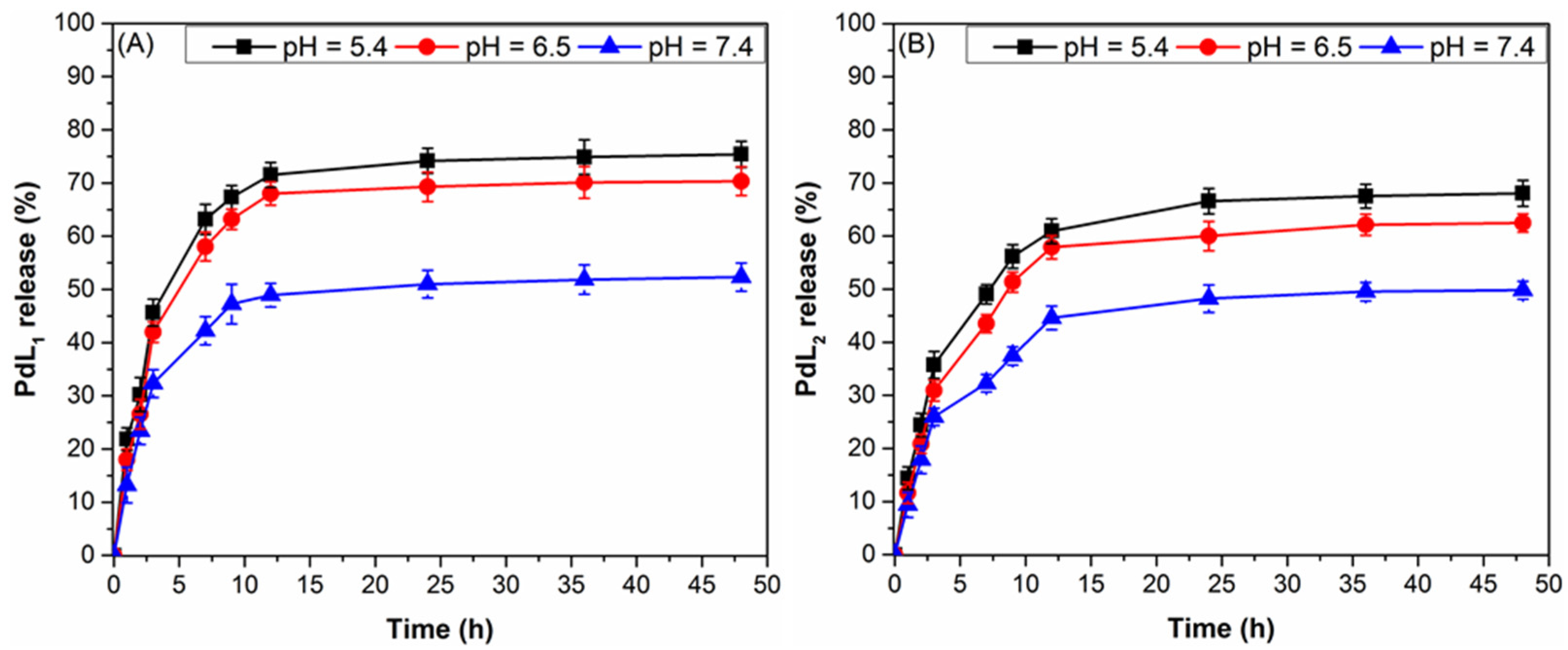
4.5. In Vitro PdL1,2 Release from PdL1,2LNPs
4.6. In Vitro Cytotoxicity Studies
4.6.1. Cell Cultures
4.6.2. Cytotoxic Effect Assay and Cell Proliferation using MTT
4.6.3. Cell Viability Assay
4.7. Comet Assay for DNA Damage Detection
4.8. The Enzyme-Linked Immunosorbent Assay (ELISA) for Her2/neu, FGFR2, TGFβ1, and P53
4.9. Western Blotting Analysis of Bax and Bcle Proteins in MCF-7 Cells
4.10. Quantitative Real-Time (qPCR) Estimation FGFR2 and FGF2
4.11. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lower, E.E.; Glass, E.; Blau, R.; Harman, S. HER-2/neu expression in primary and metastatic breast cancer. Breast Cancer Res. Treat. 2009, 113, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Zeng, L.; Lv, Y. BRCA1 overexpression attenuates breast cancer cell growth and migration by regulating the pyruvate kinase M2-mediated Warburg effect via the PI3K/AKT signaling pathway. PeerJ 2022, 10, e14052. [Google Scholar] [CrossRef]
- Hanker, A.B.; Pfefferle, A.D.; Balko, J.M.; Kuba, M.G.; Young, C.D.; Sánchez, V.; Sutton, C.R.; Cheng, H.; Perou, C.M.; Zhao, J.J. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc. Natl. Acad. Sci. USA 2013, 110, 14372–14377. [Google Scholar] [CrossRef] [PubMed]
- Meert, A.-P.; Martin, B.; Paesmans, M.; Berghmans, T.; Mascaux, C.; Verdebout, J.-M.; Delmotte, P.; Lafitte, J.-J.; Sculier, J.-P. The role of HER-2/neu expression on the survival of patients with lung cancer: A systematic review of the literature. Br. J. Cancer 2003, 89, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.e.; Esteva, F.J. Trastuzumab: Triumphs and tribulations. Oncogene 2007, 26, 3637–3643. [Google Scholar] [CrossRef] [PubMed]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Caires, A.C.F. Recent advances involving palladium (II) complexes for the cancer therapy. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2007, 7, 484–491. [Google Scholar] [CrossRef]
- Yan, W.; Zhou, Y.; Zhou, Z.; Ji, Z.; Li, H. Padeliporfin vascular-targeted photodynamic therapy versus active surveillance in men with low-risk prostate cancer. Lancet Oncol. 2017, 18, e187. [Google Scholar] [CrossRef] [PubMed]
- Kapdi, A.R.; Fairlamb, I.J.S. Anti-cancer palladium complexes: A focus on PdX2L2, palladacycles and related complexes. Chem. Soc. Rev. 2014, 43, 4751–4777. [Google Scholar] [CrossRef]
- Alotaibi, B.S.; Buabeid, M.; Ibrahim, N.A.; Kharaba, Z.J.; Ijaz, M.; Noreen, S.; Murtaza, G. Potential of nanocarrier-based drug delivery systems for brain targeting: A current review of literature. Int. J. Nanomed. 2021, 16, 7517–7533. [Google Scholar] [CrossRef]
- Elbehairi, S.E.I.; Alfaifi, M.Y.; Shati, A.A.; Alshehri, M.A.; Elshaarawy, R.F.M.; Hafez, H.S. Role of Pd (II)–chitooligosaccharides–Gboxin analog in oxidative phosphorylation inhibition and energy depletion: Targeting mitochondrial dynamics. Chem. Biol. Drug Des. 2020, 96, 1148–1161. [Google Scholar] [CrossRef]
- Kamal, I.; Khedr, A.I.M.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Elshaarawy, R.F.M.; Saad, A.S. Chemotherapeutic and chemopreventive potentials of ρ-coumaric acid–Squid chitosan nanogel loaded with Syzygium aromaticum essential oil. Int. J. Biol. Macromol. 2021, 188, 523–533. [Google Scholar] [CrossRef]
- Kumari, P.; Ghosh, B.; Biswas, S. Nanocarriers for cancer-targeted drug delivery. J. Drug Target. 2016, 24, 179–191. [Google Scholar] [CrossRef]
- Fei, W.; Li, C.; Tao, J.; Cai, X.; Yao, W.; Ye, Y.; Zhang, Y.; Yao, Y.; Song, Q.; Li, F. Construction of arsenic-metal complexes loaded nanodrugs for solid tumor therapy: A mini review. Int. J. Pharm. 2020, 583, 119385. [Google Scholar] [CrossRef]
- Hassan, Y.A.; Alfaifi, M.Y.; Shati, A.A.; Elbehairi, S.E.I.; Elshaarawy, R.F.M.; Kamal, I. Co-delivery of anticancer drugs via poly (ionic crosslinked chitosan-palladium) nanocapsules: Targeting more effective and sustainable cancer therapy. J. Drug Deliv. Sci. Technol. 2022, 69, 103151. [Google Scholar] [CrossRef]
- Demina, P.A.; Khaydukov, K.V.; Babayeva, G.; Varaksa, P.O.; Atanova, A.V.; Stepanov, M.E.; Nikolaeva, M.E.; Krylov, I.V.; Evstratova, I.I.; Pokrovsky, V.S. Upconversion Nanoparticles Intercalated in Large Polymer Micelles for Tumor Imaging and Chemo/Photothermal Therapy. Int. J. Mol. Sci. 2023, 24, 10574. [Google Scholar] [CrossRef]
- Zhang, M.; Saint-Germain, C.; He, G.; Sun, R.W.-Y. Drug delivery systems for anti-cancer active complexes of some coinage metals. Curr. Med. Chem. 2018, 25, 493–505. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Lin, G.; He, Z.; Wang, Y. Metal complex-based liposomes: Applications and prospects in cancer diagnostics and therapeutics. J. Control. Release 2022, 348, 1066–1088. [Google Scholar] [CrossRef]
- Zhou, S.; Li, J.; Yu, J.; Wang, Y.; Liu, H.; Lin, G.; He, Z.; Wang, Y. Unique flower-like cur-metal complexes loaded liposomes for primary and metastatic breast cancer therapy. Mater. Sci. Eng. C 2021, 121, 111835. [Google Scholar] [CrossRef]
- Sheoran, S.; Arora, S.; Samsonraj, R.; Govindaiah, P. Lipid-based nanoparticles for treatment of cancer. Heliyon 2022, 8, e09403. [Google Scholar] [CrossRef]
- Pagano, K.; Listro, R.; Linciano, P.; Rossi, D.; Longhi, E.; Taraboletti, G.; Molinari, H.; Collina, S.; Ragona, L. Identification of a novel extracellular inhibitor of FGF2/FGFR signaling axis by combined virtual screening and NMR spectroscopy approach. Bioorganic Chem. 2023, 136, 106529. [Google Scholar] [CrossRef]
- Murillo, M.I.; Gaiddon, C.; Le Lagadec, R. Targeting of the intracellular redox balance by metal complexes towards anticancer therapy. Front. Chem. 2022, 10, 967337. [Google Scholar] [CrossRef]
- Kraft, J.C.; Freeling, J.P.; Wang, Z.; Ho, R.J.Y. Emerging research and clinical development trends of liposome and lipid nanoparticle drug delivery systems. J. Pharm. Sci. 2014, 103, 29–52. [Google Scholar] [CrossRef]
- Alfaifi, M.Y.; Zein, M.A.E.; Shati, A.A.; Alshehri, M.A.; Elbehairi, S.E.I.; Hafez, H.S.; Elshaarawy, R.F.M. Synthesis, photophysical behavior and biomolecular reactivity of new triphenylphosphonium-based Pd (II) salphens as new anticancer candidates. J. Photochem. Photobiol. A Chem. 2019, 385, 112083. [Google Scholar] [CrossRef]
- Wani, W.A.; Baig, U.; Shreaz, S.; Shiekh, R.A.; Iqbal, P.F.; Jameel, E.; Ahmad, A.; Mohd-Setapar, S.H.; Mushtaque, M.; Hun, L.T. Recent advances in iron complexes as potential anticancer agents. New J. Chem. 2016, 40, 1063–1090. [Google Scholar] [CrossRef]
- Ashraf, T.; Ali, B.; Qayyum, H.; Haroone, M.S.; Shabbir, G. Pharmacological aspects of schiff base metal complexes: A critical review. Inorg. Chem. Commun. 2023, 150, 110449. [Google Scholar] [CrossRef]
- Tadele, K.T.; Tsega, T.W. Schiff Bases and their metal complexes as potential anticancer candidates: A review of recent works. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2019, 19, 1786–1795. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological activity of ionic liquids and their application in pharmaceutics and medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. MedChemComm 2018, 9, 409–436. [Google Scholar] [CrossRef]
- Alfaifi, M.Y.; Elbehairi, S.E.I.; Hafez, H.S.; Elshaarawy, R.F.M. Spectroscopic exploration of binding of new imidazolium-based palladium (II) saldach complexes with CT-DNA as anticancer agents against HER2/neu overexpression. J. Mol. Struct. 2019, 1191, 118–128. [Google Scholar] [CrossRef]
- Alkabli, J.; Rizk, M.A.; Elshaarawy, R.F.M.; El-Sayed, W.N. Ionic chitosan Schiff bases supported Pd (II) and Ru (II) complexes; production, characterization, and catalytic performance in Suzuki cross-coupling reactions. Int. J. Biol. Macromol. 2021, 184, 454–462. [Google Scholar] [CrossRef]
- Alfaifi, M.Y.; Elbehairi, S.E.I.; Elshaarawy, R.F.M.; Zein, M.A.E. Novel thiazolium ionic liquids-tagged bicyclo-palladium (II) Schiff base complexes; Synthesis, characterization and in vitro cytotoxicity toward ovarian cancer. J. Mol. Struct. 2022, 1249, 131594. [Google Scholar] [CrossRef]
- Ray, K.; Weyhermüller, T.; Neese, F.; Wieghardt, K. Electronic structure of square planar bis (benzene-1, 2-dithiolato) metal complexes [M (L) 2] z (z = 2−, 1−, 0; M = Ni, Pd, Pt, Cu, Au): An experimental, density functional, and correlated ab initio study. Inorg. Chem. 2005, 44, 5345–5360. [Google Scholar] [CrossRef]
- Refaee, A.A.; El-Naggar, M.E.; Mostafa, T.B.; Elshaarawy, R.F.M.; Nasr, A.M. Nano-bio finishing of cotton fabric with quaternized chitosan Schiff base-TiO2-ZnO nanocomposites for antimicrobial and UV protection applications. Eur. Polym. J. 2022, 166, 111040. [Google Scholar] [CrossRef]
- Carvalho, P.M.; Felício, M.R.; Santos, N.C.; Gonçalves, S.; Domingues, M.M. Application of light scattering techniques to nanoparticle characterization and development. Front. Chem. 2018, 6, 237. [Google Scholar] [CrossRef]
- Marques, M.P.M.; Batista de Carvalho, A.L.M.; Mamede, A.P.; Dopplapudi, A.; Rudić, S.; Tyagi, M.; Garcia Sakai, V.; Batista de Carvalho, L.A.E. A new look into the mode of action of metal-based anticancer drugs. Molecules 2020, 25, 246. [Google Scholar] [CrossRef]
- Sato, M.R.; Oshiro Junior, J.A.; Machado, R.T.A.; de Souza, P.C.; Campos, D.L.; Pavan, F.R.; da Silva, P.B.; Chorilli, M. Nanostructured lipid carriers for incorporation of copper (II) complexes to be used against Mycobacterium tuberculosis. Drug Des. Dev. Ther. 2017, 11, 909–921. [Google Scholar] [CrossRef]
- Marquele-Oliveira, F.; de Almeida Santana, D.C.; Taveira, S.F.; Vermeulen, D.M.; Moraes de Oliveira, A.R.; da Silva, R.S.; Lopez, R.F.V. Development of nitrosyl ruthenium complex-loaded lipid carriers for topical administration: Improvement in skin stability and in nitric oxide release by visible light irradiation. J. Pharm. Biomed. Anal. 2010, 53, 843–851. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, Z.-R.; Sun, X.; Zuo, J.; Zhao, D.; Gong, T. Mechanisms of Phospholipid Complex Loaded Nanoparticles Enhancing the Oral Bioavailability. Mol. Pharm. 2010, 7, 565–575. [Google Scholar] [CrossRef]
- Dianat, S.; Bordbar, A.-K.; Tangestaninejad, S.; Yadollahi, B.; Amiri, R.; Zarkesh-Esfahani, S.-H.; Habibi, P. In vitro antitumor activity of free and nano-encapsulated Na5[PMo10V2O40]·nH2O and its binding properties with ctDNA by using combined spectroscopic methods. J. Inorg. Biochem. 2015, 152, 74–81. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Piasecka, D.; Braun, M.; Kitowska, K.; Mieczkowski, K.; Kordek, R.; Sadej, R.; Romanska, H. FGFs/FGFRs-dependent signalling in regulation of steroid hormone receptors–implications for therapy of luminal breast cancer. J. Exp. Clin. Cancer Res. 2019, 38, 230. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Palapetta, S.M.; Sandhya, V.K.; Sahu, A.; Alipoor, A.; Balakrishnan, L.; Advani, J.; George, B.; Kini, K.R.; Geetha, N.P. A network map of FGF-1/FGFR signaling system. J. Signal Transduct. 2014, 2014, 962962. [Google Scholar] [CrossRef] [PubMed]
- Wesche, J.; Haglund, K.; Haugsten, E.M. Fibroblast growth factors and their receptors in cancer. Biochem. J. 2011, 437, 199–213. [Google Scholar] [CrossRef]
- Katoh, Y.; Katoh, M. FGFR2-related pathogenesis and FGFR2-targeted therapeutics. Int. J. Mol. Med. 2009, 23, 307–311. [Google Scholar] [CrossRef]
- Ahmad, I.; Iwata, T.; Leung, H.Y. Mechanisms of FGFR-mediated carcinogenesis. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 850–860. [Google Scholar] [CrossRef]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef]
- Hanker, A.B.; Garrett, J.T.; Estrada, M.V.; Moore, P.D.; Ericsson, P.G.; Koch, J.P.; Langley, E.; Singh, S.; Kim, P.S.; Frampton, G.M. HER2-overexpressing breast cancers amplify FGFR signaling upon acquisition of resistance to dual therapeutic blockade of HER2. Clin. Cancer Res. 2017, 23, 4323–4334. [Google Scholar] [CrossRef]
- Ray, S.; Mohan, R.; Singh, J.K.; Samantaray, M.K.; Shaikh, M.M.; Panda, D.; Ghosh, P. Anticancer and antimicrobial metallopharmaceutical agents based on palladium, gold, and silver N-heterocyclic carbene complexes. J. Am. Chem. Soc. 2007, 129, 15042–15053. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, S.; Obrenčević, K.; Bugarčić, Ž.D.; Popović, I.; Žakula, J.; Petrović, B. New bimetallic palladium (II) and platinum (II) complexes: Studies of the nucleophilic substitution reactions, interactions with CT-DNA, bovine serum albumin and cytotoxic activity. Dalton Trans. 2016, 45, 12444–12457. [Google Scholar] [CrossRef]
- Wei, W.; Liu, W.; Serra, S.; Asa, S.L.; Ezzat, S. The breast cancer susceptibility FGFR2 provides an alternate mode of HER2 activation. Oncogene 2015, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Tsurutani, J.; Sakai, K.; Kaneda, H.; Fujisaka, Y.; Takeda, M.; Watatani, M.; Arao, T.; Satoh, T.; Okamoto, I. Switching addictions between HER2 and FGFR2 in HER2-positive breast tumor cells: FGFR2 as a potential target for salvage after lapatinib failure. Biochem. Biophys. Res. Commun. 2011, 407, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Parris, A.B.; Howard, E.W.; Zhao, M.; Ma, Z.; Guo, Z.; Xing, Y.; Yang, X. FGFR inhibitor, AZD4547, impedes the stemness of mammary epithelial cells in the premalignant tissues of MMTV-ErbB2 transgenic mice. Sci. Rep. 2017, 7, 11306. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Nogueira, P.; Mancino, M.; Fuster, G.; López-Plana, A.; Jauregui, P.; Almendro, V.; Enreig, E.; Menéndez, S.; Rojo, F.; Noguera-Castells, A. Tumor-associated fibroblasts promote HER2-targeted therapy resistance through FGFR2 activation. Clin. Cancer Res. 2020, 26, 1432–1448. [Google Scholar] [CrossRef] [PubMed]
- Siena, S.; Sartore-Bianchi, A.; Marsoni, S.; Hurwitz, H.I.; McCall, S.J.; Penault-Llorca, F.; Srock, S.; Bardelli, A.; Trusolino, L. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann. Oncol. 2018, 29, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Novakova, O.; Chen, H.; Vrana, O.; Rodger, A.; Sadler, P.J.; Brabec, V. DNA interactions of monofunctional organometallic ruthenium (II) antitumor complexes in cell-free media. Biochemistry 2003, 42, 11544–11554. [Google Scholar] [CrossRef] [PubMed]
- Yasui, H.; Takeno, A.; Hara, H.; Imamura, H.; Akamatsu, H.; Fujitani, K.; Nakane, M.; Kondoh, C.N.; Yukisawa, S.; Nasu, J. Prospective analysis of the expression status of FGFR2 and HER2 in colorectal and gastric cancer populations: DS-Screen Study. Int. J. Color. Dis. 2022, 37, 1393–1402. [Google Scholar] [CrossRef]
- Cui, W.; Fowlis, D.J.; Bryson, S.; Duffie, E.; Ireland, H.; Balmain, A.; Akhurst, R.J. TGFβ1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell 1996, 86, 531–542. [Google Scholar] [CrossRef]
- Bierie, B.; Stover, D.G.; Abel, T.W.; Chytil, A.; Gorska, A.E.; Aakre, M.; Forrester, E.; Yang, L.; Wagner, K.-U.; Moses, H.L. Transforming growth factor–β regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 2008, 68, 1809–1819. [Google Scholar] [CrossRef]
- Mohan, N.; Shen, Y.; Endo, Y.; ElZarrad, M.K.; Wu, W.J. Trastuzumab, but not pertuzumab, dysregulates HER2 signaling to mediate inhibition of autophagy and increase in reactive oxygen species production in human cardiomyocytes. Mol. Cancer Ther. 2016, 15, 1321–1331. [Google Scholar] [CrossRef]
- Chakraborty, K.; Tripathi, A.; Mishra, S.; Mallick, A.M.; Roy, R.S. Emerging concepts in designing next-generation multifunctional nanomedicine for cancer treatment. Biosci. Rep. 2022, 42, BSR20212051. [Google Scholar] [CrossRef] [PubMed]
- Loayza-Puch, F.; Drost, J.; Rooijers, K.; Lopes, R.; Elkon, R.; Agami, R. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome Biol. 2013, 14, R32. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef] [PubMed]
- Reifenberger, G.; Reifenberger, J.; Ichimura, K.; Meltzer, P.S.; Collins, V.P. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: Preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res. 1994, 54, 4299–4303. [Google Scholar] [PubMed]
- Moldoveanu, T.; Czabotar, P.E. BAX, BAK, and BOK: A coming of age for the BCL-2 family effector proteins. Cold Spring Harb. Perspect. Biol. 2020, 12, a036319. [Google Scholar] [CrossRef]
- Gao, E.; Liu, C.; Zhu, M.; Lin, H.; Wu, Q.; Liu, L. Current development of Pd (II) complexes as potential antitumor agents. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2009, 9, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Keter, F.K.; Kanyanda, S.; Lyantagaye, S.S.L.; Darkwa, J.; Rees, D.J.G.; Meyer, M. In vitro evaluation of dichloro-bis (pyrazole) palladium (II) and dichloro-bis (pyrazole) platinum (II) complexes as anticancer agents. Cancer Chemother. Pharmacol. 2008, 63, 127–138. [Google Scholar] [CrossRef]
- Miklášová, N.; Fischer-Fodor, E.; Lönnecke, P.; Schrepler, M.P.; Virag, P.; Tatomir, C.; Cernea, V.I.; Hey-Hawkins, E.; Silaghi-Dumitrescu, L. Antiproliferative effect and genotoxicity of novel synthesized palladium complexes with organoarsenic ligands. J. Inorg. Biochem. 2009, 103, 1739–1747. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Błasiak, J.; Jałoszynski, P.; Trzeciak, A.; Szyfter, K. In vitro studies on the genotoxicity of the organophosphorus insecticide malathion and its two analogues. Mutat. Res. /Genet. Toxicol. Environ. Mutagen. 1999, 445, 275–283. [Google Scholar] [CrossRef]

| Treatment | Cell Line | PdL1 | PdL2 | PdL1LNPs | PdL2LNPs | CDDP | TRZ |
|---|---|---|---|---|---|---|---|
| IC50 (μg/mL) | MCF-7 | 16.28 ± 0.56 | 23.72 ± 1.25 | 1.93 ± 0.29 | 2.03 ± 0.21 | 5.51 ± 0.31 | 2.41 ± 0.27 |
| Hela | 53.85 ± 0.78 | 72.34 ± 0.89 | 38.96 ± 1.73 | 41.28 ± 1.53 | 75.25 ± 3.75 | 19.72 ± 1.89 | |
| SI (%) * | 3.31 | 3.05 | 20.19 | 20.33 | 13.66 | 8.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awaji, A.A.; Rizk, M.A.; Alsaiari, R.A.; Alqahtani, N.F.; Al-Qadri, F.A.; Alkorbi, A.S.; Hafez, H.S.; Elshaarawy, R.F.M. Chemotherapeutic Activity of Imidazolium-Supported Pd(II) o-Vanillylidene Diaminocyclohexane Complexes Immobilized in Nanolipid as Inhibitors for HER2/neu and FGFR2/FGF2 Axis Overexpression in Breast Cancer Cells. Pharmaceuticals 2023, 16, 1711. https://doi.org/10.3390/ph16121711
Awaji AA, Rizk MA, Alsaiari RA, Alqahtani NF, Al-Qadri FA, Alkorbi AS, Hafez HS, Elshaarawy RFM. Chemotherapeutic Activity of Imidazolium-Supported Pd(II) o-Vanillylidene Diaminocyclohexane Complexes Immobilized in Nanolipid as Inhibitors for HER2/neu and FGFR2/FGF2 Axis Overexpression in Breast Cancer Cells. Pharmaceuticals. 2023; 16(12):1711. https://doi.org/10.3390/ph16121711
Chicago/Turabian StyleAwaji, Aeshah A., Moustafa A. Rizk, Raiedhah A. Alsaiari, Norah F. Alqahtani, Fatima A. Al-Qadri, Ali S. Alkorbi, Hani S. Hafez, and Reda F. M. Elshaarawy. 2023. "Chemotherapeutic Activity of Imidazolium-Supported Pd(II) o-Vanillylidene Diaminocyclohexane Complexes Immobilized in Nanolipid as Inhibitors for HER2/neu and FGFR2/FGF2 Axis Overexpression in Breast Cancer Cells" Pharmaceuticals 16, no. 12: 1711. https://doi.org/10.3390/ph16121711
APA StyleAwaji, A. A., Rizk, M. A., Alsaiari, R. A., Alqahtani, N. F., Al-Qadri, F. A., Alkorbi, A. S., Hafez, H. S., & Elshaarawy, R. F. M. (2023). Chemotherapeutic Activity of Imidazolium-Supported Pd(II) o-Vanillylidene Diaminocyclohexane Complexes Immobilized in Nanolipid as Inhibitors for HER2/neu and FGFR2/FGF2 Axis Overexpression in Breast Cancer Cells. Pharmaceuticals, 16(12), 1711. https://doi.org/10.3390/ph16121711







