N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives Exert Antimicrobial and Cytotoxic Effects via Aldehyde Dehydrogenase Pathway: Synthesis, In Silico and In Vitro Studies
Abstract
:1. Introduction
2. Results
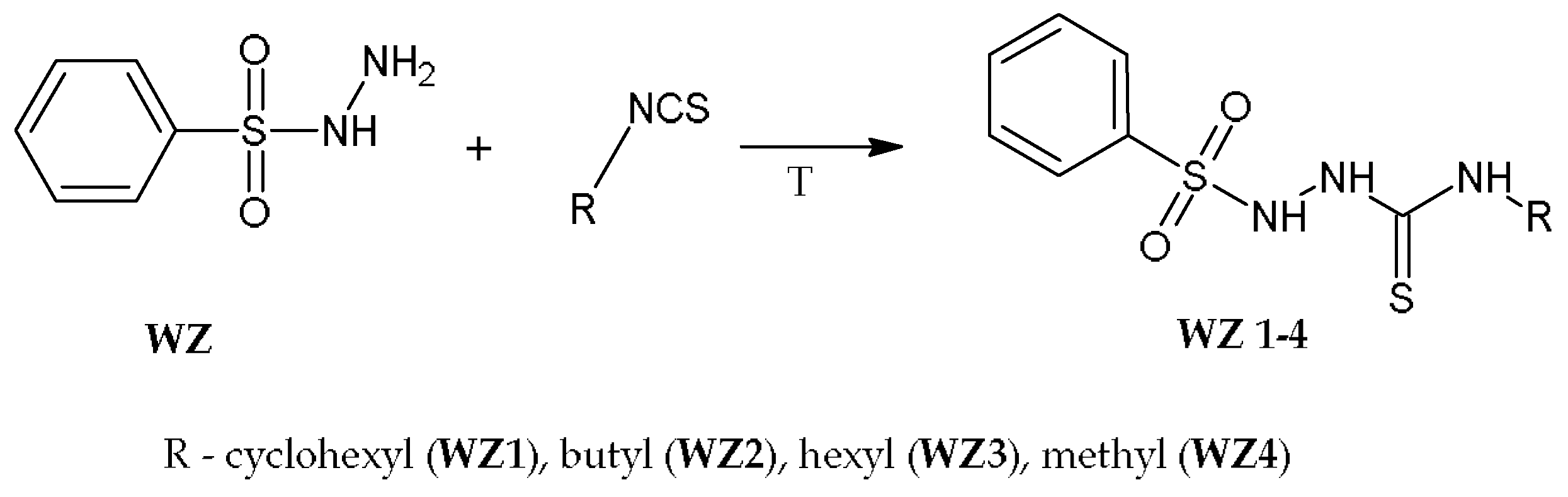
2.1. Chemistry
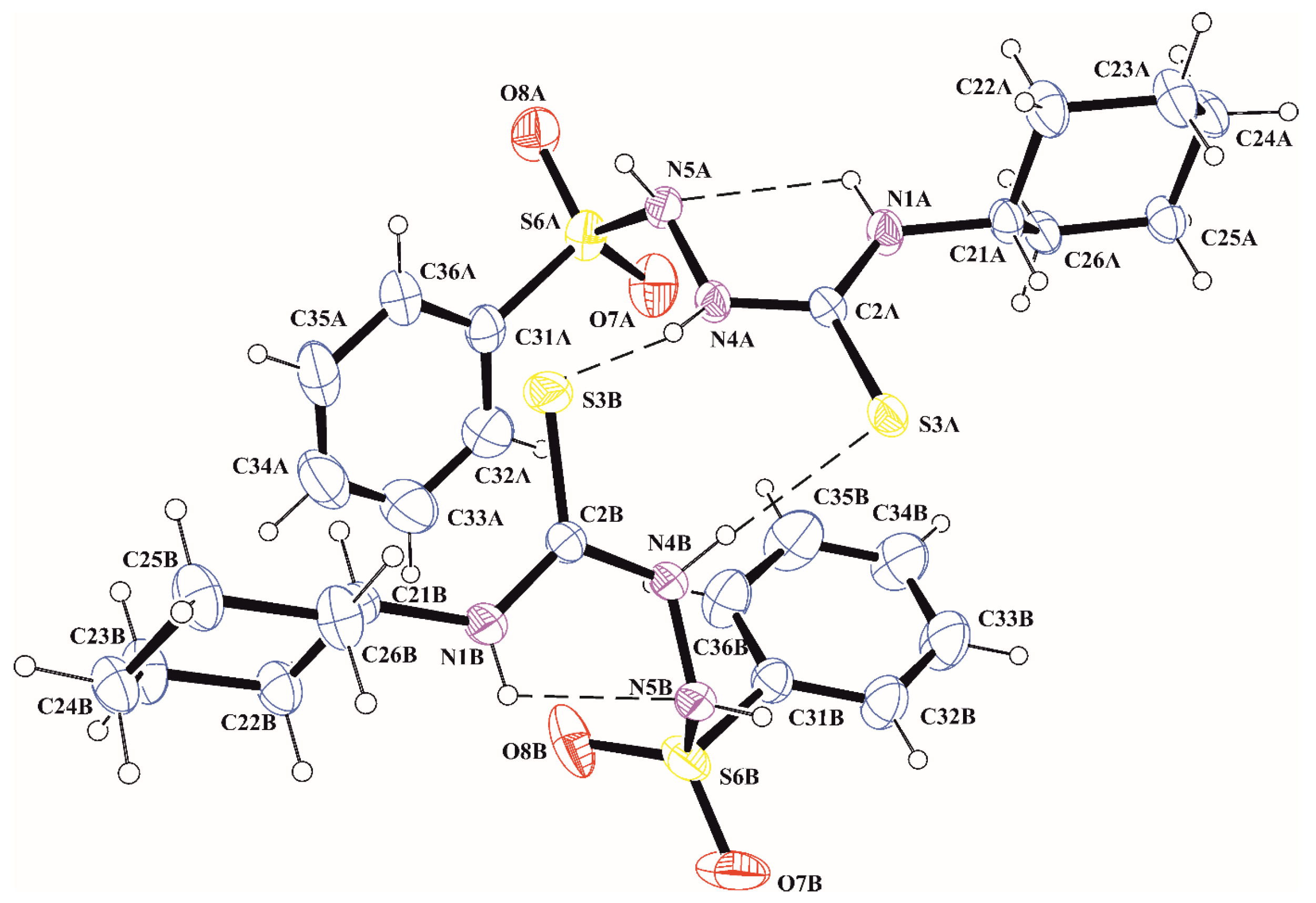
2.2. X-ray Investigation
2.3. In Silico Prediction of Cytotoxicity for Cancer
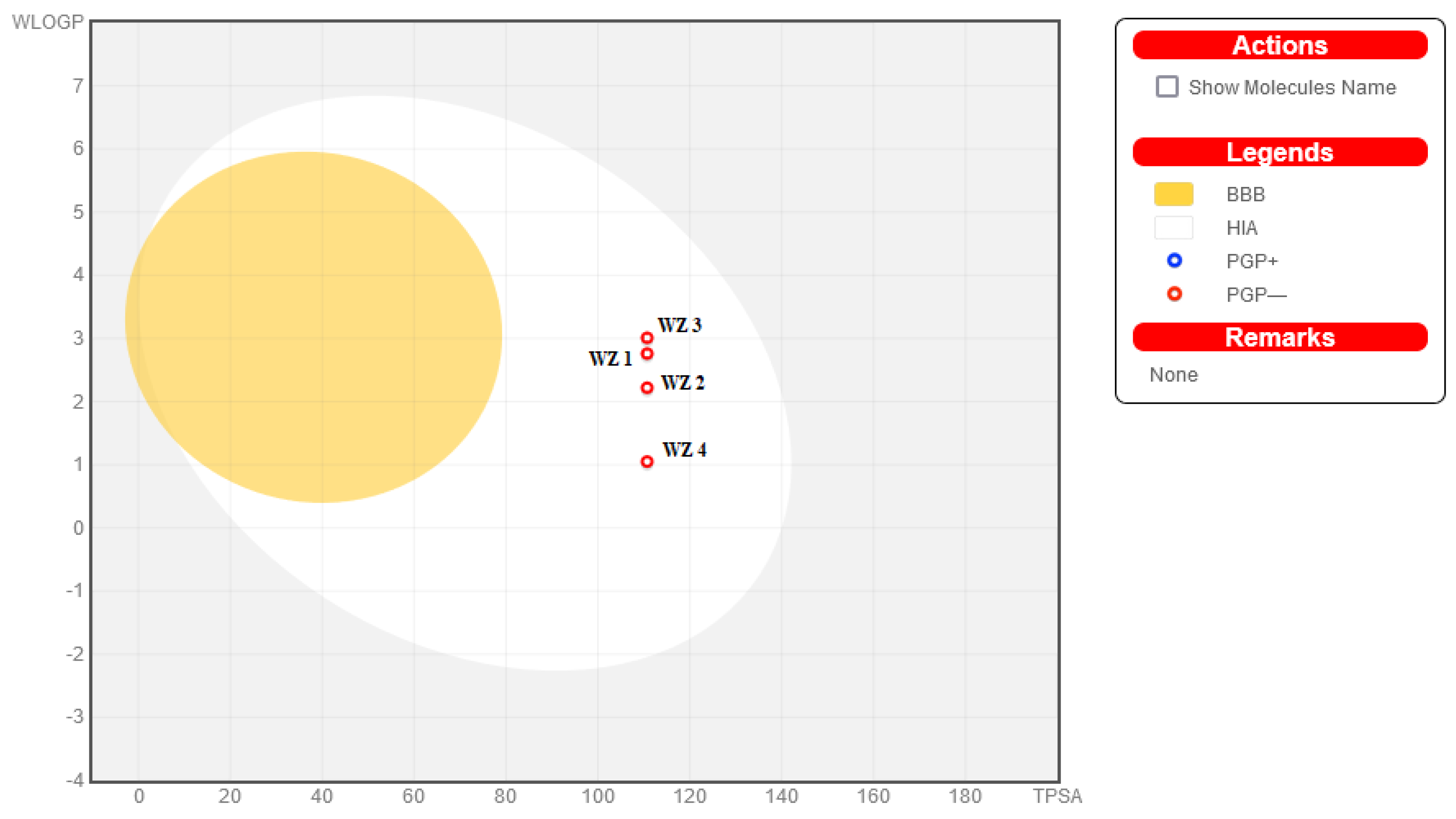
2.4. ADMET Analysis
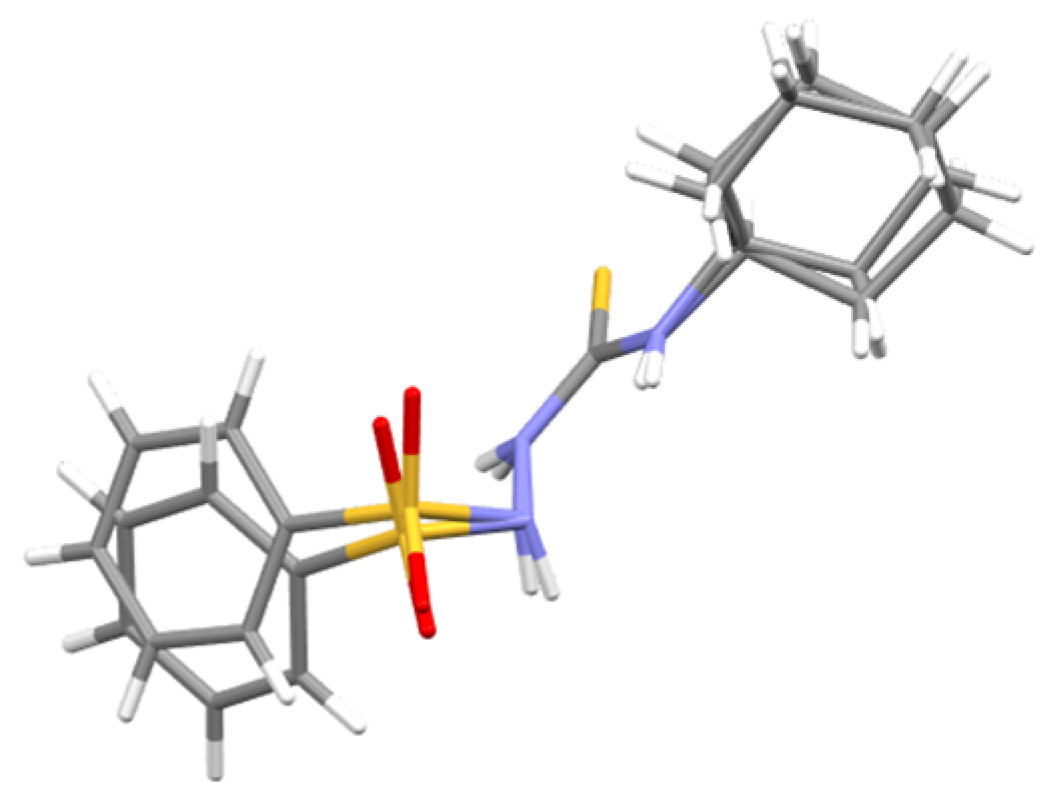
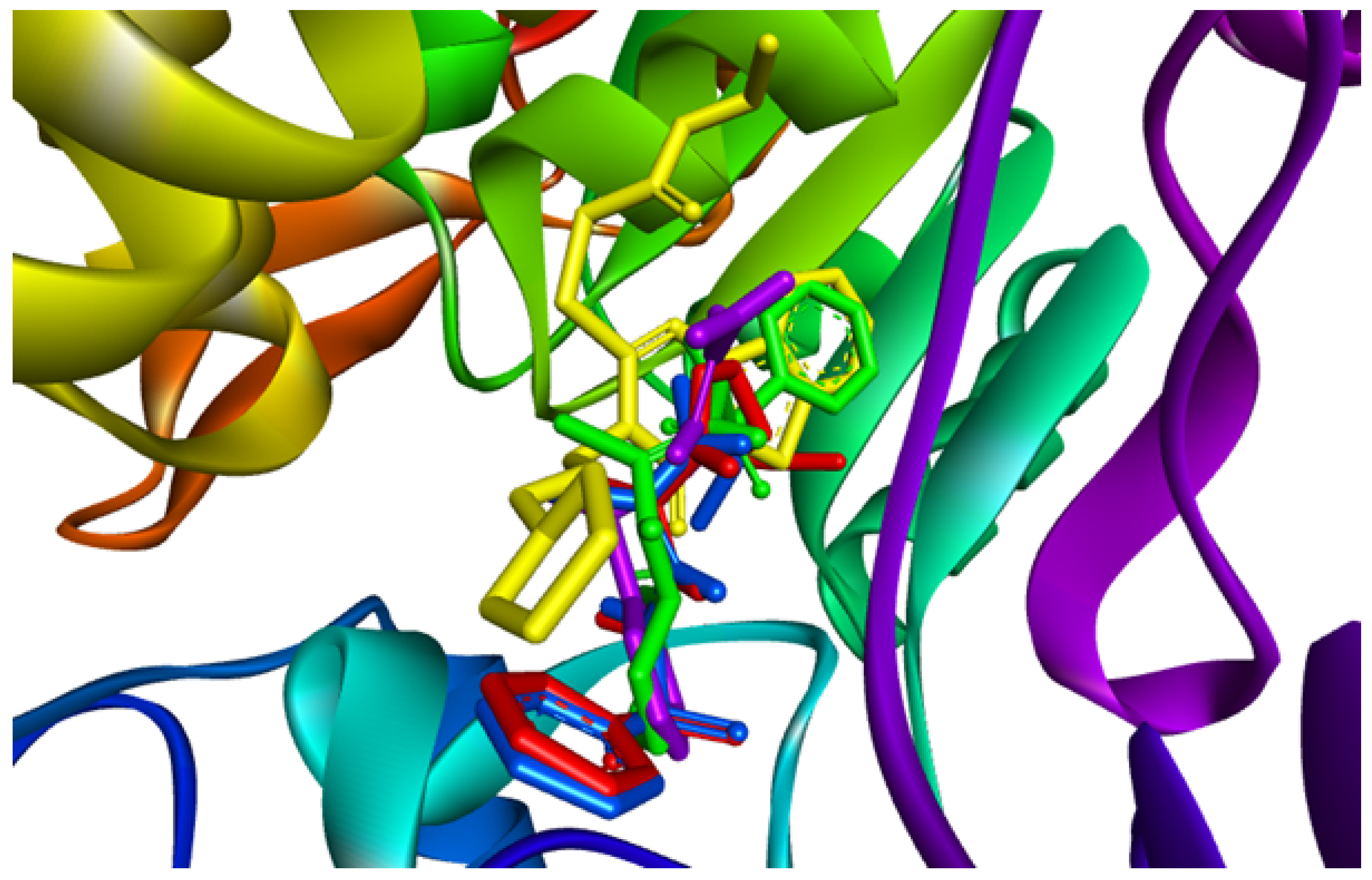
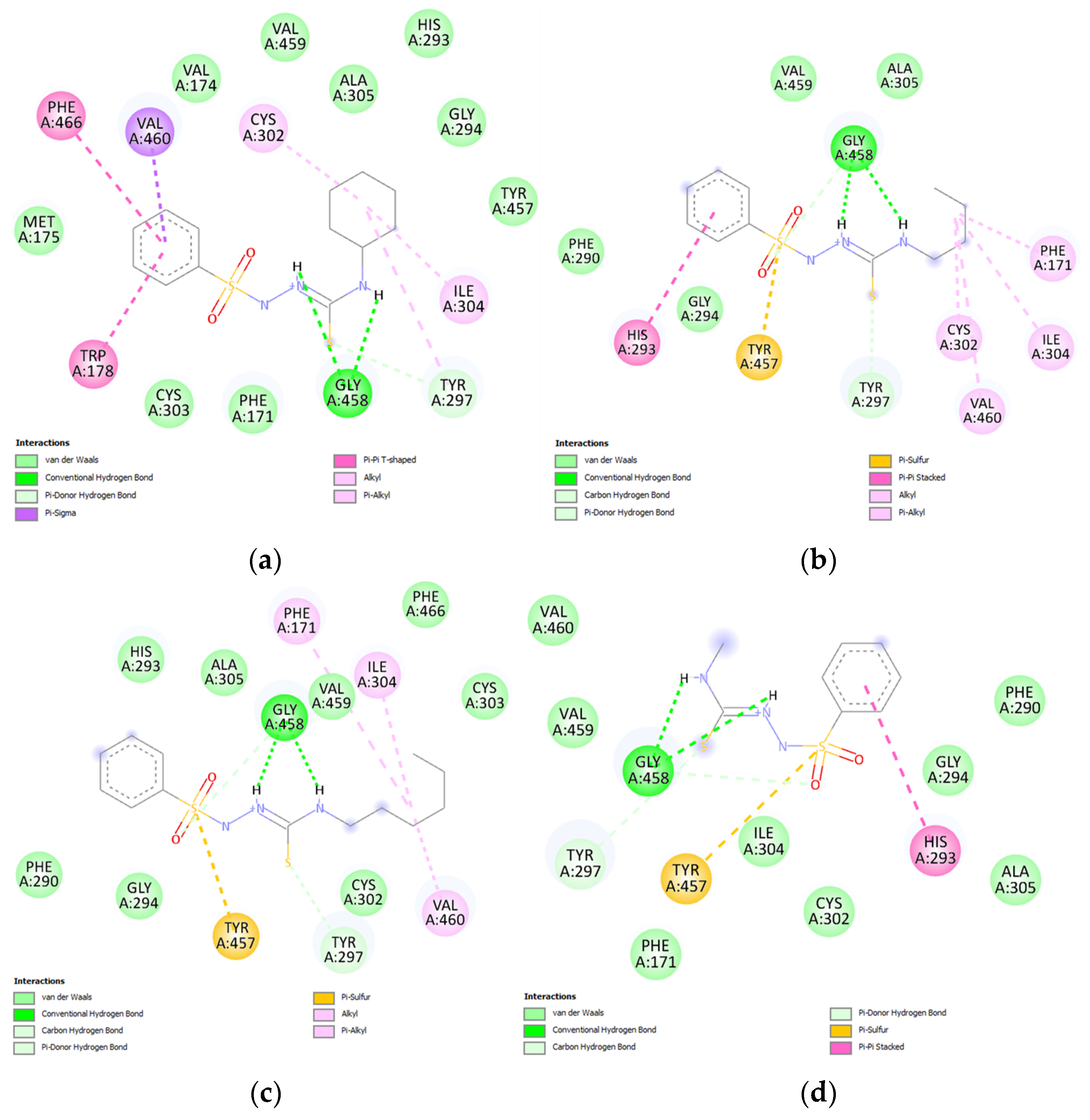
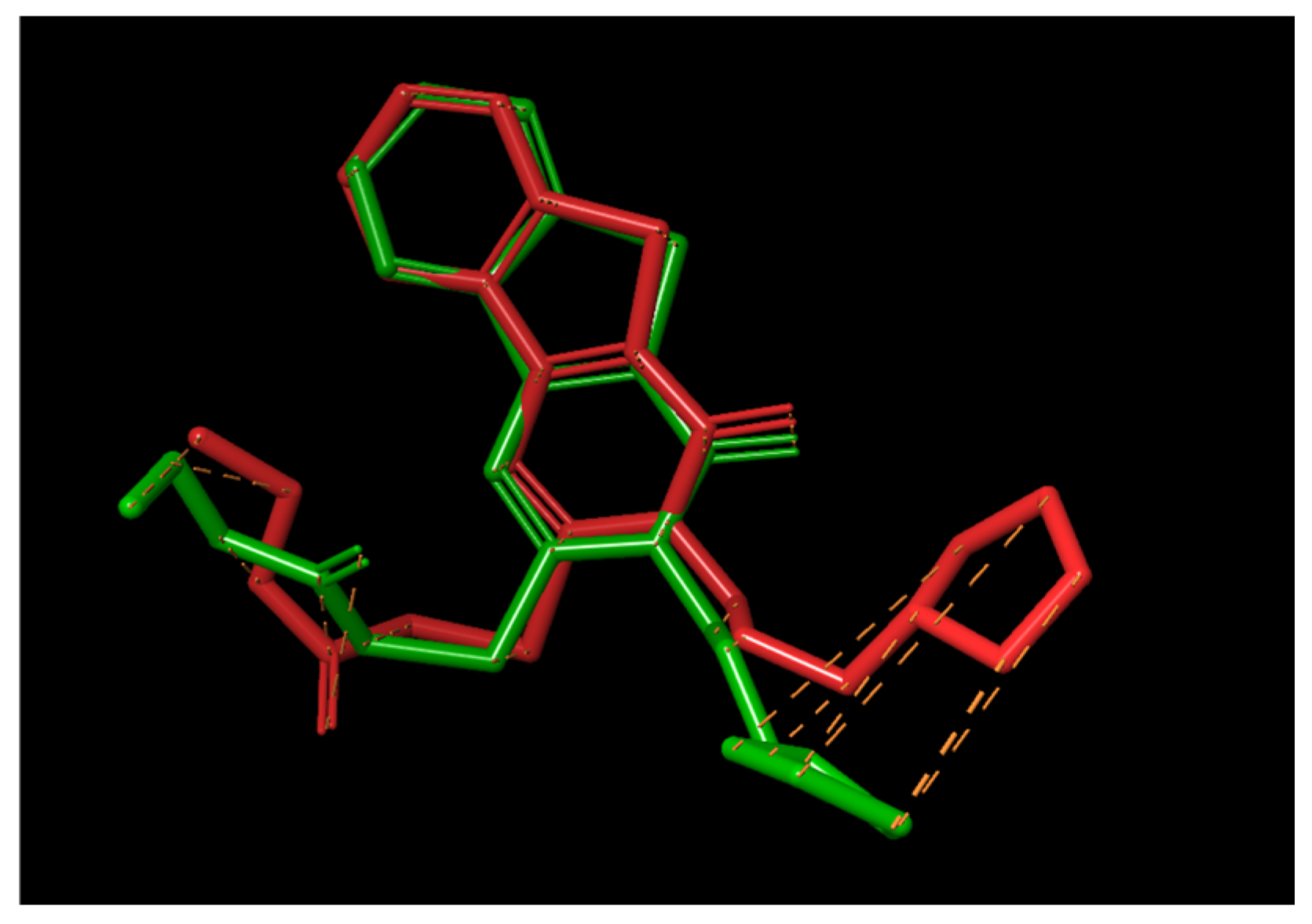
2.5. Molecular Docking Results
2.6. The Antimicrobial Activity Assessment
2.7. The Hemolytic Activity Assay
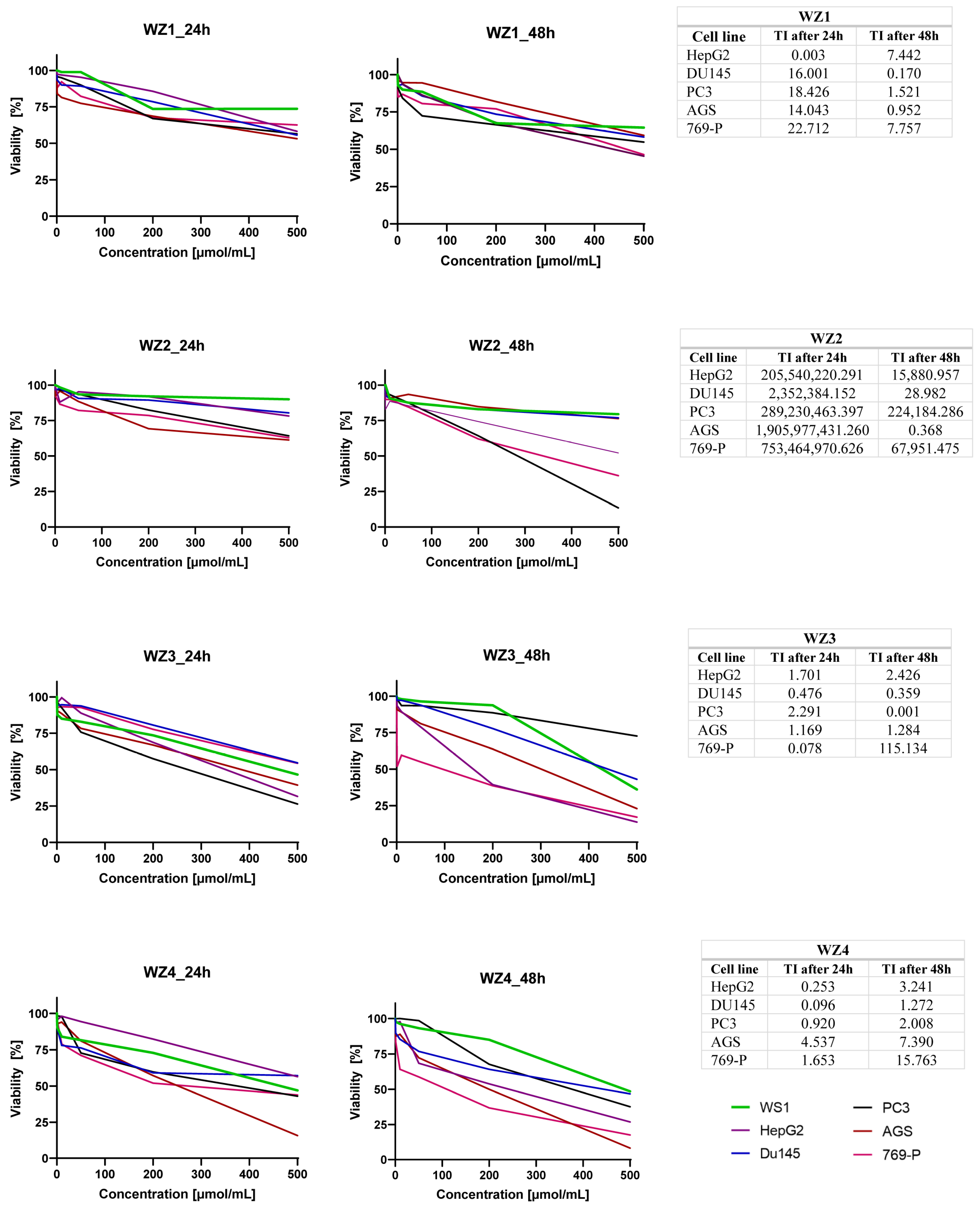
2.8. In Vitro Cell Assay
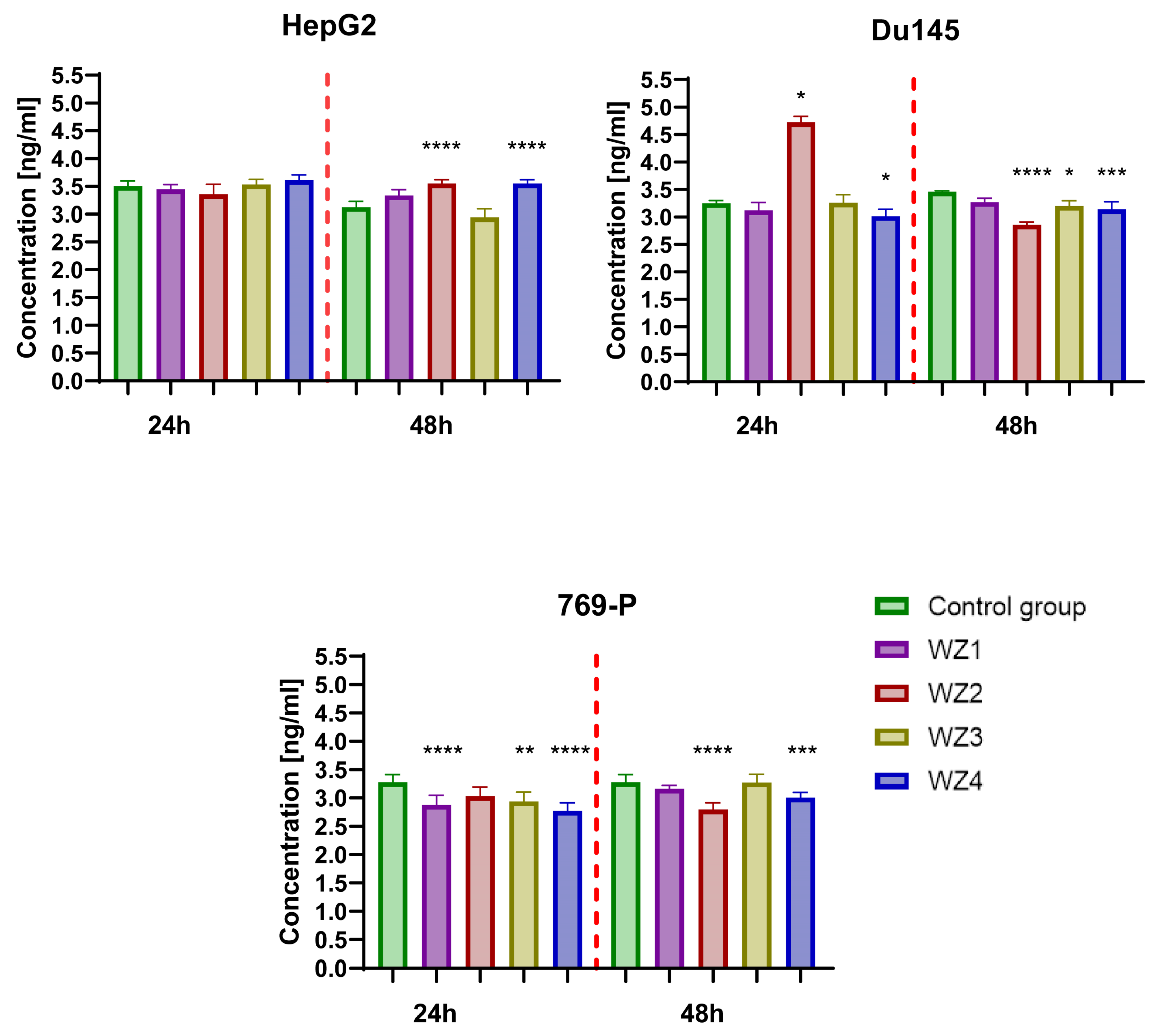
2.9. Determination of Concentration Aldehyde Dehydrogenase 1 Family, Member A1 (ALDH1A1)
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. General Synthesis Procedure for N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives WZ1–WZ4
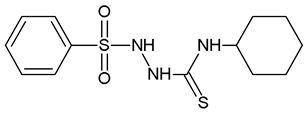
2-(Benzenesulfonyl)-N-Cyclohexylhydrazine-1-Carbothioamide (WZ1)
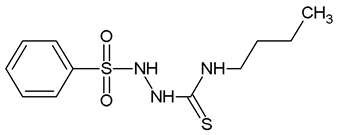
2-(Benzenesulfonyl)-N-Butylhydrazine-1-Carbothioamide (WZ2)
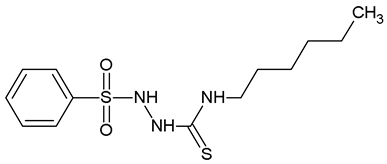
2-(Benzenesulfonyl)-N-Hexylhydrazine-1-Carbothioamide (WZ3)
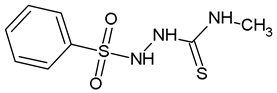
2-(Benzenesulfonyl)-N-Methylhydrazine-1-Carbothioamide (WZ4)
4.2. X-ray Structure Determination
4.3. In Silico Screening and ADMET Predictions
4.4. Molecular Docking
4.5. Antimicrobial Activity
4.5.1. Microorganisms
4.5.2. In Vitro Antimicrobial Activity Assay
4.5.3. Toxicity to Erythrocyte Assay
4.6. Anticancer Activity
4.6.1. Cell Culture
4.6.2. Antiproliferative Activity Assay
4.6.3. Determination of Concentration Aldehyde Dehydrogenase 1 Family, Member A1 (ALDH1A1)
4.6.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ahmadi, R.; Emami, S. Recent Applications of Vinyl Sulfone Motif in Drug Design and Discovery. Eur. J. Med. Chem. 2022, 234, 114255. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-M.; Hu, Y.-Y.; Fang, B.; Zhou, C.-H. Benzenesulfonyl Thiazoloimines as Unique Multitargeting Antibacterial Agents towards Enterococcus Faecalis. Eur. J. Med. Chem. 2023, 248, 115088. [Google Scholar] [CrossRef] [PubMed]
- Hearn, M.J.; Pugh, C.D.; Cynamon, M.H. Synthesis of Functionalized Sulfonamides as Antitubercular Agents. Phosphorus Sulfur Silicon Relat. Elem. 2023, 198, 733–751. [Google Scholar] [CrossRef]
- Bano, S.; Javed, K.; Ahmad, S.; Rathish, I.G.; Singh, S.; Alam, M.S. Synthesis and Biological Evaluation of Some New 2-Pyrazolines Bearing Benzene Sulfonamide Moiety as Potential Anti-Inflammatory and Anti-Cancer Agents. Eur. J. Med. Chem. 2011, 46, 5763–5768. [Google Scholar] [CrossRef] [PubMed]
- Kalariya, R.; Ojha, D.; Rana, S.; Rode, A.; Bhosale, R.; Yadav, J.S. Novel Fluorinated Amino Acid Derivatives as Potent Antitumor Agents against MCF-7 and HepG2 Cells: Synthesis, Characterization, in Vitro Assays and Molecular Docking Studies. Results Chem. 2023, 5, 100954. [Google Scholar] [CrossRef]
- Ghorab, M.M.; Ragab, F.A.; Heiba, H.I.; Soliman, A.M. Design and Synthesis of Some Novel 4-Chloro-N-(4-(1-(2-(2-Cyanoacetyl)Hydrazono)Ethyl)Phenyl) Benzenesulfonamide Derivatives as Anticancer and Radiosensitizing Agents. Eur. J. Med. Chem. 2016, 117, 8–18. [Google Scholar] [CrossRef]
- Mokoena, T.P.; Maluleka, M.M.; Mampa, R.M.; Mphahlele, M.J.; Monchusi, B.A. Synthesis, Crystal Structures, Spectroscopic Characterization and in Vitro Evaluation of the 4-Sulfono-3-Methoxycinnamaldehydes as Potential α-Glucosidase and/or α-Amylase Inhibitors. J. Mol. Struct. 2023, 1271, 134119. [Google Scholar] [CrossRef]
- Taha, M.; Salahuddin, M.; Almandil, N.B.; Farooq, R.K.; Rahim, F.; Uddin, N.; Nawaz, M.; Alhibshi, A.H.; Anouar, E.H.; Khan, K.M. In Vitro and in Vivo Antidiabetics Study of New Oxadiazole Derivatives Along with Molecular Docking Study. Polycycl. Aromat. Compd. 2023, 43, 6911–6926. [Google Scholar] [CrossRef]
- Roy, S.; Sen, S.; Saha, S.; Deb, S.K.; Singh, B.; Biswas, G. Design, Synthesis and Molecular Docking Studies of 5-Fluoro 1-Aryl/Alkyl Sulfonyl Benzimidazole Derivatives for Treatment of Parkinson’s Disease. Phosphorus Sulfur Silicon Relat. Elem. 2023, 198, 336–344. [Google Scholar] [CrossRef]
- Scholar, E. Sulfonamides. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA, 2007; pp. 1–4. ISBN 978-0-08-055232-3. [Google Scholar]
- van Bambeke, F.; Mingeot-Leclercq, M.-P.; Glupczynski, Y.; Tulkens, P.M. 137—Mechanisms of Action. In Infectious Diseases, 4th ed.; Cohen, J., Powderly, W.G., Opal, S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1162–1180.e1. ISBN 978-0-7020-6285-8. [Google Scholar]
- Ovung, A.; Bhattacharyya, J. Sulfonamide Drugs: Structure, Antibacterial Property, Toxicity, and Biophysical Interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef]
- Nevagi, R.J.; Dhake, A.S.; Narkhede, H.I.; Kaur, P. Design, Synthesis and Biological Evaluation of Novel Thiosemicarbazide Analogues as Potent Anticonvulsant Agents. Bioorganic Chem. 2014, 54, 68–72. [Google Scholar] [CrossRef]
- Rane, R.A.; Naphade, S.S.; Bangalore, P.K.; Palkar, M.B.; Shaikh, M.S.; Karpoormath, R. Synthesis of Novel 4-Nitropyrrole-Based Semicarbazide and Thiosemicarbazide Hybrids with Antimicrobial and Anti-Tubercular Activity. Bioorg. Med. Chem. Lett. 2014, 24, 3079–3083. [Google Scholar] [CrossRef]
- Cihan-Üstündağ, G.; Gürsoy, E.; Naesens, L.; Ulusoy-Güzeldemirci, N.; Çapan, G. Synthesis and Antiviral Properties of Novel Indole-Based Thiosemicarbazides and 4-Thiazolidinones. Bioorg Med. Chem 2016, 24, 240–246. [Google Scholar] [CrossRef]
- Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 27 October 2023).
- CDC What Exactly is Antibiotic Resistance? Available online: https://www.cdc.gov/drugresistance/about.html (accessed on 27 October 2023).
- Habboush, Y.; Guzman, N. Antibiotic Resistance. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Wild, C.P.; Weiderpass, E.; Stewart, B.W. World Cancer Report: Cancer Research for Cancer Prevention; International Agency for Research on Cancer: Lyon, France, 2020; ISBN 978-92-832-0447-3. [Google Scholar]
- Amjad, M.T.; Chidharla, A.; Kasi, A. Cancer Chemotherapy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Debela, D.T.; Muzazu, S.G.; Heraro, K.D.; Ndalama, M.T.; Mesele, B.W.; Haile, D.C.; Kitui, S.K.; Manyazewal, T. New Approaches and Procedures for Cancer Treatment: Current Perspectives. SAGE Open Med 2021, 9, 20503121211034366. [Google Scholar] [CrossRef]
- Chacko, S.; Samanta, S. Novel Thiosemicarbazide Hybrids with Amino Acids and Peptides Against Hepatocellular Carcinoma: A Molecular Designing Approach Towards Multikinase Inhibitor. Curr. Comput. Aided Drug Des. 2015, 11, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Kozyra, P.; Korga-Plewko, A.; Karczmarzyk, Z.; Hawrył, A.; Wysocki, W.; Człapski, M.; Iwan, M.; Ostrowska-Leśko, M.; Fornal, E.; Pitucha, M. Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative. Biomolecules 2022, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Icharam Narkhede, H.; Shridhar Dhake, A.; Rikhabchand Surana, A. Synthesis and Screening of Thiosemicarbazide-Dithiocarbamate Conjugates for Antioxidant and Anticancer Activities. Bioorganic Chem. 2022, 124, 105832. [Google Scholar] [CrossRef] [PubMed]
- Pitucha, M.; Korga-Plewko, A.; Kozyra, P.; Iwan, M.; Kaczor, A.A. 2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA. Biomolecules 2020, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Pitucha, M.; Woś, M.; Miazga-Karska, M.; Klimek, K.; Mirosław, B.; Pachuta-Stec, A.; Gładysz, A.; Ginalska, G. Synthesis, Antibacterial and Antiproliferative Potential of Some New 1-Pyridinecarbonyl-4-Substituted Thiosemicarbazide Derivatives. Med. Chem. Res. 2016, 25, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Wos, M.; Miazga-Karska, M.; Kaczor, A.A.; Klimek, K.; Karczmarzyk, Z.; Kowalczuk, D.; Wysocki, W.; Ginalska, G.; Urbanczyk-Lipkowska, Z.; Morawiak, M.; et al. Novel Thiosemicarbazide Derivatives with 4-Nitrophenyl Group as Multi-Target Drugs: α-Glucosidase Inhibitors with Antibacterial and Antiproliferative Activity. Biomed. Pharmacother. 2017, 93, 1269–1276. [Google Scholar] [CrossRef]
- Geng, P.-F.; Liu, X.-Q.; Zhao, T.-Q.; Wang, C.-C.; Li, Z.-H.; Zhang, J.; Wei, H.-M.; Hu, B.; Ma, L.-Y.; Liu, H.-M. Design, Synthesis and in Vitro Biological Evaluation of Novel [1,2,3]Triazolo[4,5-d]Pyrimidine Derivatives Containing a Thiosemicarbazide Moiety. Eur. J. Med. Chem. 2018, 146, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Huang, J.; Cao, Y.; Zhang, Z.; He, F.; Lin, X.; Wu, Q.; Zhao, S. Synthesis and Biological Evaluation of 1-(2-(6-Methoxynaphthalen-2-Yl)-6-Methylnicotinoyl)-4-Substituted Semicarbazides/Thiosemicarbazides as Anti-Tumor Nur77 Modulators. Molecules 2022, 27, 1698. [Google Scholar] [CrossRef] [PubMed]
- Szopa, A.; Herbet, M.; Pachuta-Stec, A.; Lachowicz, J.; Pawłowski, K.; Iwan, M.; Jarecka-Florek, D.; Krasińska, O.; Serefko, A.; Poleszak, E.; et al. Evaluation of Developmental Toxicity in Zebrafish Embryos and Antiproliferative Potential against Human Tumor Cell Lines of New Derivatives Containing 4-Nitrophenyl Group. Toxicol. Appl. Pharmacol. 2023, 458, 116325. [Google Scholar] [CrossRef]
- Popovici, C.; Pavel, C.-M.; Sunel, V.; Cheptea, C.; Dimitriu, D.G.; Dorohoi, D.O.; David, D.; Closca, V.; Popa, M. Optimized Synthesis of New Thiosemicarbazide Derivatives with Tuberculostatic Activity. Int. J. Mol. Sci. 2021, 22, 12139. [Google Scholar] [CrossRef]
- Sudha Rani, M.; Krishnadevi, K.; Rajeswari, M.; Somaiah, N. Design, Synthesis and Anticancer Activity of Sulfonamide Derivatives of 1,2,3-Triazole-Indoles. Chem. Data Collect. 2023, 43, 100975. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of Bond Lengths Determined by X-Ray and Neutron Diffraction. Part 1. Bond Lengths in Organic Compounds. J. Chem. Soc. Perkin Trans. 1987, 2, S1–S19. [Google Scholar] [CrossRef]
- Cremer, D.; Pople, J.A. General Definition of Ring Puckering Coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- Lagunin, A.A.; Rudik, A.V.; Pogodin, P.V.; Savosina, P.I.; Tarasova, O.A.; Dmitriev, A.V.; Ivanov, S.M.; Biziukova, N.Y.; Druzhilovskiy, D.S.; Filimonov, D.A.; et al. CLC-Pred 2.0: A Freely Available Web Application for In Silico Prediction of Human Cell Line Cytotoxicity and Molecular Mechanisms of Action for Druglike Compounds. Int. J. Mol. Sci. 2023, 24, 1689. [Google Scholar] [CrossRef]
- Kozyra, P.; Adamczuk, G.; Karczmarzyk, Z.; Matysiak, J.; Podkościelna, B.; Humeniuk, E.; Wysocki, W.; Korga-Plewko, A.; Senczyna, B.; Pitucha, M. Novel Phenoxyacetylthiosemicarbazide Derivatives as Novel Ligands in Cancer Diseases. Toxicol. Appl. Pharmacol. 2023, 475, 116634. [Google Scholar] [CrossRef]
- Guan, L.; Yang, H.; Cai, Y.; Sun, L.; Di, P.; Li, W.; Liu, G.; Tang, Y. ADMET-Score—A Comprehensive Scoring Function for Evaluation of Chemical Drug-Likeness. Medchemcomm 2018, 10, 148–157. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A Webserver for the Prediction of Toxicity of Chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef]
- Czylkowska, A.; Lanka, S.; Szczesio, M.; Czarnecka, K.; Szymański, P.; Pitucha, M.; Drabińska, A.; Camargo, B.C.; Szczytko, J. New Derivatives of 5-((1-Methyl-Pyrrol-2-Yl) Methyl)-4-(Naphthalen-1-Yl)-1,2,4-Triazoline-3-Thione and Its Coordination Compounds with Anticancer Activity. Int. J. Mol. Sci. 2022, 23, 9162. [Google Scholar] [CrossRef] [PubMed]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Luna-Vázquez-Gómez, R.; Arellano-García, M.E.; García-Ramos, J.C.; Radilla-Chávez, P.; Salas-Vargas, D.S.; Casillas-Figueroa, F.; Ruiz-Ruiz, B.; Bogdanchikova, N.; Pestryakov, A. Hemolysis of Human Erythrocytes by ArgovitTM AgNPs from Healthy and Diabetic Donors: An In Vitro Study. Materials 2021, 14, 2792. [Google Scholar] [CrossRef]
- Ameryckx, A.; Pochet, L.; Wang, G.; Yildiz, E.; Saadi, B.E.; Wouters, J.; Van Bambeke, F.; Frédérick, R. Pharmacomodulations of the Benzoyl-Thiosemicarbazide Scaffold Reveal Antimicrobial Agents Targeting d-Alanyl-d-Alanine Ligase in Bacterio. Eur. J. Med. Chem. 2020, 200, 112444. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.A.; Abdallah, E.M.; Ahmed, S.A.; Rabee, M.M.; Bräse, S. Transition Metal Complexes of Thiosemicarbazides, Thiocarbohydrazides, and Their Corresponding Carbazones with Cu(I), Cu(II), Co(II), Ni(II), Pd(II), and Ag(I)—A Review. Molecules 2023, 28, 1808. [Google Scholar] [CrossRef] [PubMed]
- Namiecińska, E.; Sobiesiak, M.; Małecka, M.; Guga, P.; Rozalska, B.; Budzisz, E. Antimicrobial and Structural Properties of Metal Ions Complexes with Thiosemicarbazide Motif and Related Heterocyclic Compounds. Curr. Med. Chem. 2019, 26, 664–693. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, B.C.; Queiroz, P.A.; Baldin, V.P.; do Amaral, P.H.; Rodrigues, L.L.; Vandresen, F.; R Caleffi-Ferracioli, K.; de L Scodro, R.B.; Cardoso, R.F.; Siqueira, V.L. (-)-Camphene-Based Derivatives as Potential Antibacterial Agents against Staphylococcus aureus and Enterococcus spp. Future Microbiol. 2020, 15, 1527–1534. [Google Scholar] [CrossRef]
- Ünver, Y.; Sancak, K.; Çelik, F.; Birinci, E.; Küçük, M.; Soylu, S.; Burnaz, N.A. New Thiophene-1,2,4-Triazole-5(3)-Ones: Highly Bioactive Thiosemicarbazides, Structures of Schiff Bases and Triazole-Thiols. Eur. J. Med. Chem. 2014, 84, 639–650. [Google Scholar] [CrossRef]
- Özbek, N.; Katırcıoğlu, H.; Karacan, N.; Baykal, T. Synthesis, Characterization and Antimicrobial Activity of New Aliphatic Sulfonamide. Bioorganic Med. Chem. 2007, 15, 5105–5109. [Google Scholar] [CrossRef]
- Jessberger, N.; Dietrich, R.; Granum, P.E.; Märtlbauer, E. The Bacillus Cereus Food Infection as Multifactorial Process. Toxins 2020, 12, 701. [Google Scholar] [CrossRef] [PubMed]
- de Araújo Neto, L.N.; do Carmo Alves de Lima, M.; de Oliveira, J.F.; de Souza, E.R.; Buonafina, M.D.S.; Vitor Anjos, M.N.; Brayner, F.A.; Alves, L.C.; Neves, R.P.; Mendonça-Junior, F.J.B. Synthesis, Cytotoxicity and Antifungal Activity of 5-Nitro-Thiophene-Thiosemicarbazones Derivatives. Chem. Biol. Interact. 2017, 272, 172–181. [Google Scholar] [CrossRef]
- Pishawikar, S.A.; More, H.N. Synthesis, Docking and in-Vitro Screening of Mannich Bases of Thiosemicarbazide for Anti-Fungal Activity. Arab. J. Chem. 2017, 10, S2714–S2722. [Google Scholar] [CrossRef]
- Yamaguchi, M.U.; Barbosa da Silva, A.P.; Ueda-Nakamura, T.; Dias Filho, B.P.; Conceição da Silva, C.; Nakamura, C.V. Effects of a Thiosemicarbazide Camphene Derivative on Trichophyton Mentagrophytes. Molecules 2009, 14, 1796–1807. [Google Scholar] [CrossRef]
- Zhang, X.; Lei, P.; Sun, T.; Jin, X.; Yang, X.; Ling, Y. Design, Synthesis, and Fungicidal Activity of Novel Thiosemicarbazide Derivatives Containing Piperidine Fragments. Molecules 2017, 22, 2085. [Google Scholar] [CrossRef]
- Siwek, A.; Stefańska, J.; Dzitko, K.; Ruszczak, A. Antifungal Effect of 4-Arylthiosemicarbazides against Candida Species. Search for Molecular Basis of Antifungal Activity of Thiosemicarbazide Derivatives. J. Mol. Model. 2012, 18, 4159–4170. [Google Scholar] [CrossRef] [PubMed]
- Henley-Smith, C.J.; Steffens, F.E.; Botha, F.S.; Lall, N. Predicting the Influence of Multiple Components on Microbial Inhibition Using a Logistic Response Model—A Novel Approach. BMC Complement. Altern. Med. 2014, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Zohra, M.; Fawzia, A. Hemolytic Activity of Different Herbal Extracts Used in Algeria. Int. J. Pharm. Sci. Res. 2014, 5, 495–500. [Google Scholar]
- Wei, F.; Wang, S.; Gou, X. A Review for Cell-Based Screening Methods in Drug Discovery. Biophys. Rep. 2021, 7, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef] [PubMed]
- Parks, R.E.; Kidder, G.W.; Dewey, V.C. Thiosemicarbazide Toxicity in Mice. Proc. Soc. Exp. Biol. Med. 1952, 79, 287–289. [Google Scholar] [CrossRef]
- Tennekoon, G.E. Pulmonary Oedema Due to Thiosemicarbazide. J. Pathol. Bacteriol. 1954, 67, 341–347. [Google Scholar] [CrossRef]
- Li, X.; Xu, Q.; Fu, X.; Luo, W. ALDH1A1 Overexpression Is Associated with the Progression and Prognosis in Gastric Cancer. BMC Cancer 2014, 14, 705. [Google Scholar] [CrossRef]
- Muralikrishnan, V.; Fang, F.; Given, T.C.; Podicheti, R.; Chtcherbinine, M.; Metcalfe, T.X.; Sriramkumar, S.; O’Hagan, H.M.; Hurley, T.D.; Nephew, K.P. A Novel ALDH1A1 Inhibitor Blocks Platinum-Induced Senescence and Stemness in Ovarian Cancer. Cancers 2022, 14, 3437. [Google Scholar] [CrossRef]
- Yue, H.; Hu, Z.; Hu, R.; Guo, Z.; Zheng, Y.; Wang, Y.; Zhou, Y. ALDH1A1 in Cancers: Bidirectional Function, Drug Resistance, and Regulatory Mechanism. Front. Oncol. 2022, 12, 918778. [Google Scholar] [CrossRef]
- Püschel, J.; Dubrovska, A.; Gorodetska, I. The Multifaceted Role of Aldehyde Dehydrogenases in Prostate Cancer Stem Cells. Cancers 2021, 13, 4703. [Google Scholar] [CrossRef]
- Januchowski, R.; Wojtowicz, K.; Sterzyſska, K.; Sosiſska, P.; Andrzejewska, M.; Zawierucha, P.; Nowicki, M.; Zabel, M. Inhibition of ALDH1A1 Activity Decreases Expression of Drug Transporters and Reduces Chemotherapy Resistance in Ovarian Cancer Cell Lines. Int. J. Biochem. Cell Biol. 2016, 78, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, X.; Zhan, Y.; Jiang, W.; Liu, X.; Wang, X.; Wu, B. Increased Expression of ALDH1A1 Protein Is Associated with Poor Prognosis in Clear Cell Renal Cell Carcinoma. Med. Oncol. 2013, 30, 574. [Google Scholar] [CrossRef] [PubMed]
- Abourbih, S.; Sircar, K.; Tanguay, S.; Kassouf, W.; Aprikian, A.; Mansure, J.; Brimo, F. Aldehyde Dehydrogenase 1 Expression in Primary and Metastatic Renal Cell Carcinoma: An Immunohistochemistry Study. World J. Surg. Oncol. 2013, 11, 298. [Google Scholar] [CrossRef]
- Morrison, H. Aldehyde Dehydrogenase. In Enzyme Active Sites and Their Reaction Mechanisms; Elsevier: Amsterdam, The Netherlands, 2021; pp. 21–26. ISBN 978-0-12-821067-3. [Google Scholar]
- Tanaka, K.; Tomita, H.; Hisamatsu, K.; Nakashima, T.; Hatano, Y.; Sasaki, Y.; Osada, S.; Tanaka, T.; Miyazaki, T.; Yoshida, K.; et al. ALDH1A1-Overexpressing Cells Are Differentiated Cells but Not Cancer Stem or Progenitor Cells in Human Hepatocellular Carcinoma. Oncotarget 2015, 6, 24722–24732. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, X.; Liao, X.; Han, C.; Yu, T.; Qin, W.; Zhu, G.; Su, H.; Yu, L.; Liu, X.; et al. Aldehyde Dehydrogenase 1 (ALDH1) Isoform Expression and Potential Clinical Implications in Hepatocellular Carcinoma. PLoS ONE 2017, 12, e0182208. [Google Scholar] [CrossRef]
- CrysAlisPro 1.171.39.16b; Rigaku Oxford Diffraction: Oxford, UK, 2015.
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An Update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Morgan, C.A.; Hurley, T.D. Characterization of Two Distinct Structural Classes of Selective Aldehyde Dehydrogenase 1A1 Inhibitors. J. Med. Chem. 2015, 58, 1964–1975. [Google Scholar] [CrossRef] [PubMed]
- Avogadro: An Open-Source Molecular Builder and Visualization Tool. Version 1.2.0. Available online: http://avogadro.cc/ (accessed on 6 October 2023).
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.M.D.; Shakil, S.; Haneef, M. A Simple Click by Click Protocol to Perform Docking: AutoDock 4.2 Made Easy for Non-Bioinformaticians. EXCLI J. 2013, 12, 831–857. [Google Scholar]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Schrödinger Release 2022-4: Maestro; Schrödinger, LLC: New York, NY, USA, 2022.
- BIOVIA, Dassault Systèmes, Discovery Studio Visualizer, V21. 1.0.20298; Dassault Systèmes: San Diego, CA, USA, 2021.
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Determination of Minimum Inhibitory Concentrations (MICs) of Antibacterial Agents by Broth Dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [CrossRef]
- Wayne, P.A. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; M27-S4; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2012. [Google Scholar]
- O’Donnell, F.; Smyth, T.J.P.; Ramachandran, V.N.; Smyth, W.F. A Study of the Antimicrobial Activity of Selected Synthetic and Naturally Occurring Quinolines. Int. J. Antimicrob. Agents 2010, 35, 30–38. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Wald-Dickler, N.; Holtom, P.; Spellberg, B. Busting the Myth of “Static vs Cidal”: A Systemic Literature Review. Clin. Infect. Dis. 2018, 66, 1470–1474. [Google Scholar] [CrossRef]
- Biernasiuk, A.; Berecka-Rycerz, A.; Gumieniczek, A.; Malm, M.; Łączkowski, K.Z.; Szymańska, J.; Malm, A. The Newly Synthesized Thiazole Derivatives as Potential Antifungal Compounds against Candida Albicans. Appl. Microbiol. Biotechnol. 2021, 105, 6355–6367. [Google Scholar] [CrossRef]
- Turecka, K.; Chylewska, A.; Kawiak, A.; Waleron, K.F. Antifungal Activity and Mechanism of Action of the Co(III) Coordination Complexes With Diamine Chelate Ligands Against Reference and Clinical Strains of Candida Spp. Front. Microbiol. 2018, 9, 1594. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Rodrigues, C.F.; Araújo, D.; Rodrigues, M.E.; Henriques, M. Candida Species Biofilms’ Antifungal Resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Deepa, P.R.; Vandhana, S.; Jayanthi, U.; Krishnakumar, S. Therapeutic and Toxicologic Evaluation of Anti-Lipogenic Agents in Cancer Cells Compared with Non-Neoplastic Cells. Basic Clin. Pharmacol. Toxicol. 2012, 110, 494–503. [Google Scholar] [CrossRef] [PubMed]



| D–H…A | D–H | H…A | D…A | D–H…A |
|---|---|---|---|---|
| N1A–H1A…N5A | 0.88 (3) | 2.33 (4) | 2.672 (3) | 104 (3) |
| N1B–H1B…N5B | 0.84 (3) | 2.24 (4) | 2.681 (3) | 113 (3) |
| N4A–H4A…S3B i | 0.86 (4) | 2.49 (4) | 3.333 (3) | 170 (3) |
| N4B–H4B…S3A i | 0.87 (4) | 2.42 (4) | 3.281 (3) | 177 (3) |
| N5A–H5A…S3A ii | 0.95 (4) | 2.35 (4) | 3.296 (3) | 174 (2) |
| N5B–H5B…S3B iii | 0.85 (3) | 2.45 (4) | 3.259 (3) | 158 (3) |
| Compound | Pa | Pi | Cell-Line | Tissue/Organ |
|---|---|---|---|---|
| WZ1 | 0.896 | 0.005 | HepG2 | Liver |
| 0.688 | 0.008 | DU-145 | Prostate | |
| WZ2 | 0.860 | 0.005 | HepG2 | Liver |
| 0.620 | 0.011 | DU-145 | Prostate | |
| WZ3 | 0.857 | 0.005 | HepG2 | Liver |
| 0.607 | 0.012 | DU-145 | Prostate | |
| WZ4 | 0.856 | 0.005 | HepG2 | Liver |
| 0.696 | 0.008 | DU-145 | Prostate |
| Comp. | Pa | Pi | Mechanism of Action |
|---|---|---|---|
| WZ1 | 0.898 | 0.005 | Aldehyde dehydrogenase 1A1 inhibitor |
| 0.775 | 0.034 | Lysosomal alpha-glucosidase inhibitor | |
| 0.686 | 0.003 | Threonine aspartase 1 inhibitor | |
| 0.644 | 0.027 | Tyrosyl-DNA phosphodiesterase 1 inhibitor | |
| WZ2 | 0.856 | 0.008 | Aldehyde dehydrogenase 1A1 inhibitor |
| 0.720 | 0.045 | Lysosomal alpha-glucosidase inhibitor | |
| 0.657 | 0.025 | Tyrosyl-DNA phosphodiesterase inhibitor | |
| WZ3 | 0.835 | 0.010 | Aldehyde dehydrogenase 1A1 inhibitor |
| 0.666 | 0.056 | Lysosomal alpha-glucosidase inhibitor | |
| 0.638 | 0.028 | Tyrosyl-DNA phosphodiesterase 1 inhibitor | |
| WZ4 | 0.944 | 0.005 | Aldehyde dehydrogenase 1A1 inhibitor |
| 0.777 | 0.033 | Lysosomal alpha-glucosidase inhibitor | |
| 0.732 | 0.017 | Tyrosyl-DNA phosphodiesterase 1 inhibitor | |
| 0.637 | 0.046 | Pyruvate kinase PKM inhibitor |
| Potential Inhibition | Compound | |||
|---|---|---|---|---|
| WZ1 | WZ2 | WZ3 | WZ4 | |
| CYP1A2 | No | No | No | No |
| CYP2C19 | Yes | No | Yes | No |
| CYP2C9 | No | No | Yes | No |
| CYP2D6 | No | No | No | No |
| CYP3A4 | No | No | No | No |
| Docked Ligands | ALDH1A1 (PDB 4X4L) | |
|---|---|---|
| Binding Energy (kcal/mol) | Ki (μM) | |
| Native ligand | −8.16 | 1.04 |
| WZ1 | −7.12 | 6.06 |
| WZ2 | −5.77 | 58.46 |
| WZ3 | −6.14 | 31.82 |
| WZ4 | −5.47 | 98.49 |
| Species | MIC (MBC or MFC) [µg/mL] and {MBC/MIC or MFC/MIC} Values of the Studied Compounds and Positive Controls | |||||
|---|---|---|---|---|---|---|
| WZ1 | WZ2 | WZ3 | WZ4 | CIP/VA* /NY** | ||
| Gram-positive bacteria | Staphylococcus aureus ATCC 43300 | 500 (>2000) {>4} | 2000 (>2000) {>1} | 250 (500) {2} | 500 (2000) {4} | 0.24 (0.24) {1} |
| Staphylococcus aureus ATCC 25923 | 500 (>2000) {>4} | 1000 (>2000) {>2} | 250 (1000) {4} | 500 (>2000) {>4} | 0.48 (0.48) {1} | |
| Staphylococcus aureus ATCC 29213 | 500 (>2000) {>4} | 2000 (>2000) {>1} | 500 (500) {1} | 1000 (>2000) {>2} | 0.48 (0.48) {1} | |
| Staphylococcus epidermidis ATCC 12228 | 250 (>2000) {>8} | 1000 (>2000) {>2} | 250 (250) {1} | 125 (500) {4} | 0.12 (0.12) {1} | |
| Enterococcus faecalis ATCC 29212 | 2000 (>2000) {>1} | >2000 (>2000) {>1} | 500 (>2000) {>4} | 1000 (>2000) {>2} | 0.98 * (1.95) {2} | |
| Micrococcus luteus ATCC 10240 | 500 (>2000) {>4} | 1000 (2000) {2} | 250 (500) {2} | 250 (500) {2} | 0.98 (1.95) {2} | |
| Bacillus subtilis ATCC 6633 | 500 (>2000) {>4} | 1000 (>2000) {>2} | 125 (500) {4} | 125 (2000) {16} | 0.03 (0.03) {1} | |
| Bacillus cereus ATCC 10876 | 500 (>2000) {>4} | 1000 (2000) {2} | 250 (250) {1} | 62.5 (125) {2} | 0.06 (0.12) {2} | |
| Gram-negative bacteria | Bordetella bronchiseptica ATCC 4617 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 500 (>2000) {>4} | >2000 (>2000) {>1} | 0.98 (0.98) {1} |
| Klebsiella pneumoniae ATCC 13883 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 1000 (>2000) {>2} | >2000 (>2000) {>1} | 0.12 (0.24) {2} | |
| Proteus mirabilis ATCC 12453 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 1000 (>2000) {>2} | >2000 (>2000) {>1} | 0.03 (0.03) {1) | |
| Salmonella Typhimurium ATCC 14028 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 1000 (>2000) {>2} | >2000 (>2000) {>1} | 0.06 (0.06) {1} | |
| Escherichia coli ATCC 25922 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 500 (>2000) {>4} | >2000 (>2000) {>1} | 0.004 (0.008) {2} | |
| Pseudomonas aeruginosa ATCC 9027 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 1000 (1000) {1} | >2000 (>2000) {>1} | 0.48 (0.98) {2} | |
| Fungi | Candida albicans ATCC 2091 | >2000 (>2000) {>1} | 2000 (>2000) {>1} | 500 (1000) {2} | 250 (1000) {4} | 0.24 ** (0.24) {1} |
| Candida albicans ATCC 10231 | >2000 (>2000) {>1} | 2000 (>2000) {>1} | 500 (1000) {2} | 500 (1000) {2} | 0.48 ** (0.48) {1} | |
| Candida parapsilosis ATCC 22019 | >2000 (>2000) {>1} | 2000 (>2000) {>1} | 2000 (2000) {1} | 125 (1000) {8} | 0.24 ** (0.48) {2} | |
| Candida glabrata ATCC 90030 | >2000 (>2000) {>1} | >2000 (>2000) {>1} | 2000 (2000) {1} | 125 (500) {4} | 0.24 ** (0.48) {2} | |
| Candida krusei ATCC 14243 | >2000 (>2000) {>1} | 2000 (>2000) {>1} | 500 (1000) {2} | 500 (2000) {4} | 0.24 ** (0.24) {1} | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walczak-Nowicka, L.; Biernasiuk, A.; Ziemichód, W.; Karczmarzyk, Z.; Kwaśnik, M.; Kozyra, P.; Wysocki, W.; Stenzel-Bembenek, A.; Kowalczuk, D.; Herbet, M.; et al. N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives Exert Antimicrobial and Cytotoxic Effects via Aldehyde Dehydrogenase Pathway: Synthesis, In Silico and In Vitro Studies. Pharmaceuticals 2023, 16, 1706. https://doi.org/10.3390/ph16121706
Walczak-Nowicka L, Biernasiuk A, Ziemichód W, Karczmarzyk Z, Kwaśnik M, Kozyra P, Wysocki W, Stenzel-Bembenek A, Kowalczuk D, Herbet M, et al. N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives Exert Antimicrobial and Cytotoxic Effects via Aldehyde Dehydrogenase Pathway: Synthesis, In Silico and In Vitro Studies. Pharmaceuticals. 2023; 16(12):1706. https://doi.org/10.3390/ph16121706
Chicago/Turabian StyleWalczak-Nowicka, Lucja, Anna Biernasiuk, Wojciech Ziemichód, Zbigniew Karczmarzyk, Mateusz Kwaśnik, Paweł Kozyra, Waldemar Wysocki, Agnieszka Stenzel-Bembenek, Dorota Kowalczuk, Mariola Herbet, and et al. 2023. "N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives Exert Antimicrobial and Cytotoxic Effects via Aldehyde Dehydrogenase Pathway: Synthesis, In Silico and In Vitro Studies" Pharmaceuticals 16, no. 12: 1706. https://doi.org/10.3390/ph16121706
APA StyleWalczak-Nowicka, L., Biernasiuk, A., Ziemichód, W., Karczmarzyk, Z., Kwaśnik, M., Kozyra, P., Wysocki, W., Stenzel-Bembenek, A., Kowalczuk, D., Herbet, M., & Pitucha, M. (2023). N-Substituted 2-(Benzenosulfonyl)-1-Carbotioamide Derivatives Exert Antimicrobial and Cytotoxic Effects via Aldehyde Dehydrogenase Pathway: Synthesis, In Silico and In Vitro Studies. Pharmaceuticals, 16(12), 1706. https://doi.org/10.3390/ph16121706










