Oncogenic Potential of Replication Factor C Subunit 4: Correlations with Tumor Progression and Assessment of Potential Inhibitors
Abstract
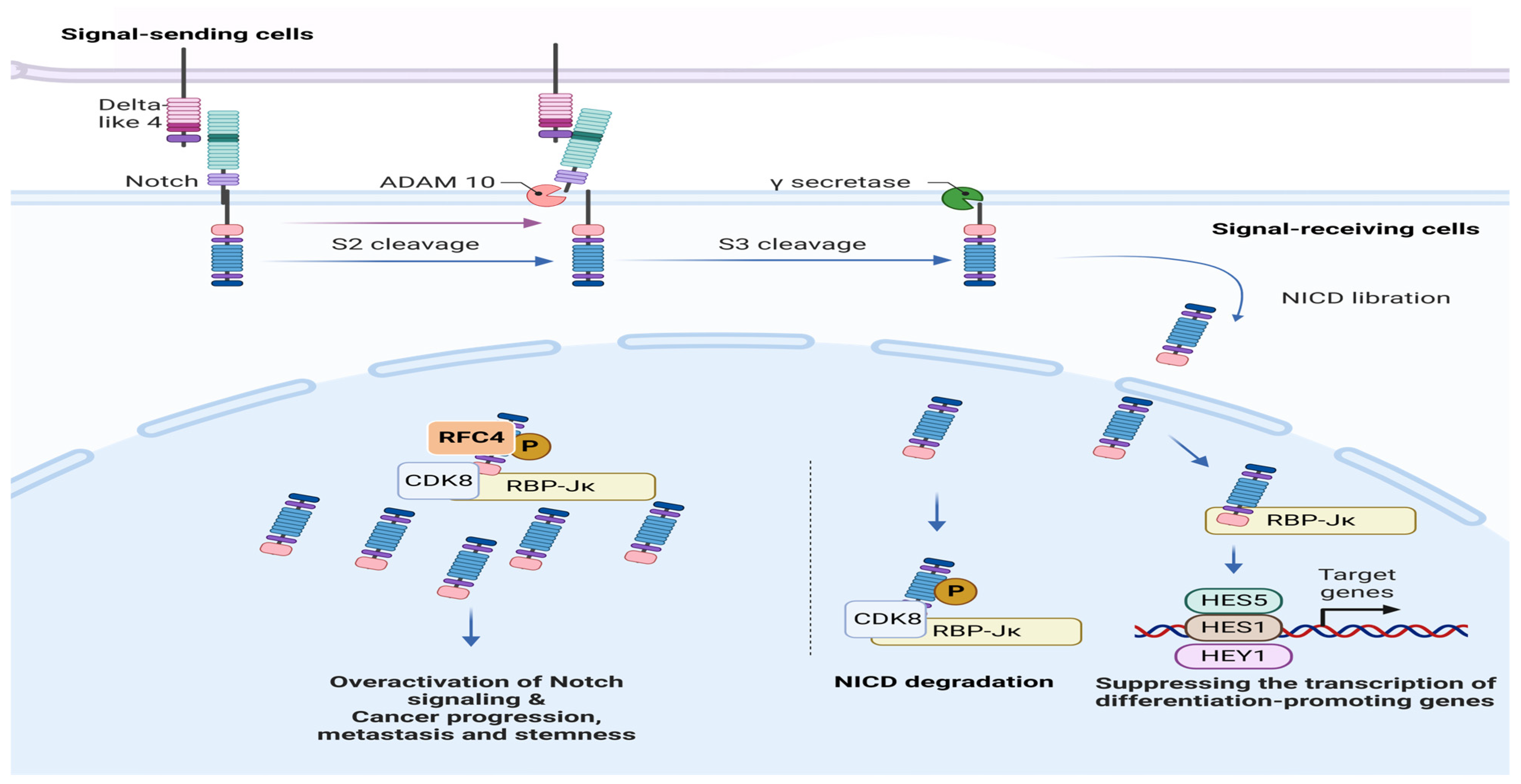
1. Introduction
2. Results
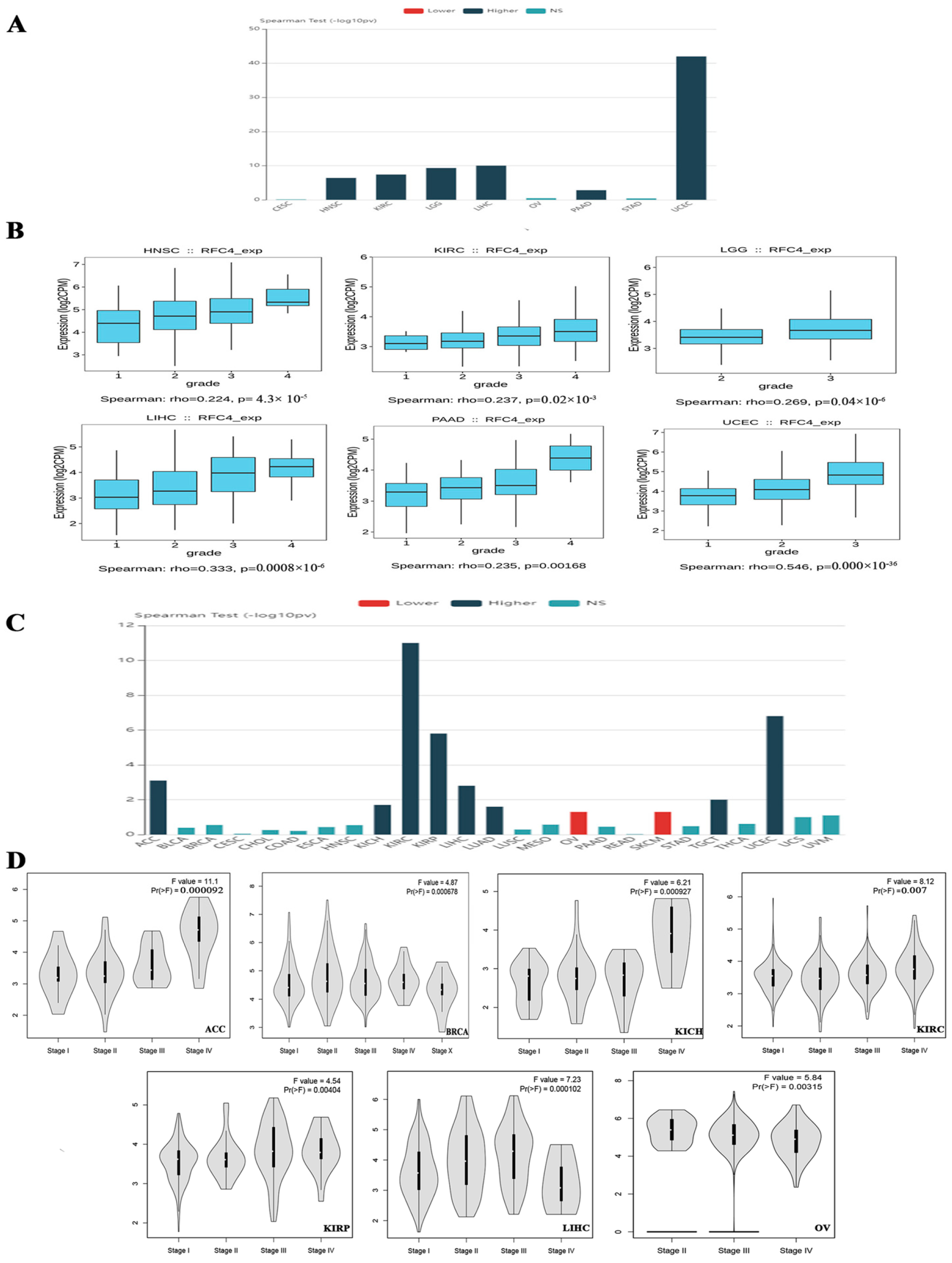
2.1. RFC4 Is Overexpressed in Several Human Tumors Compared to Normal Tissue
2.2. The Association between RFC4 Expression and Tumor Stage and Grade in Multiple Malignancies in Humans
2.3. Assessment of Differential RFC4 Protein Levels
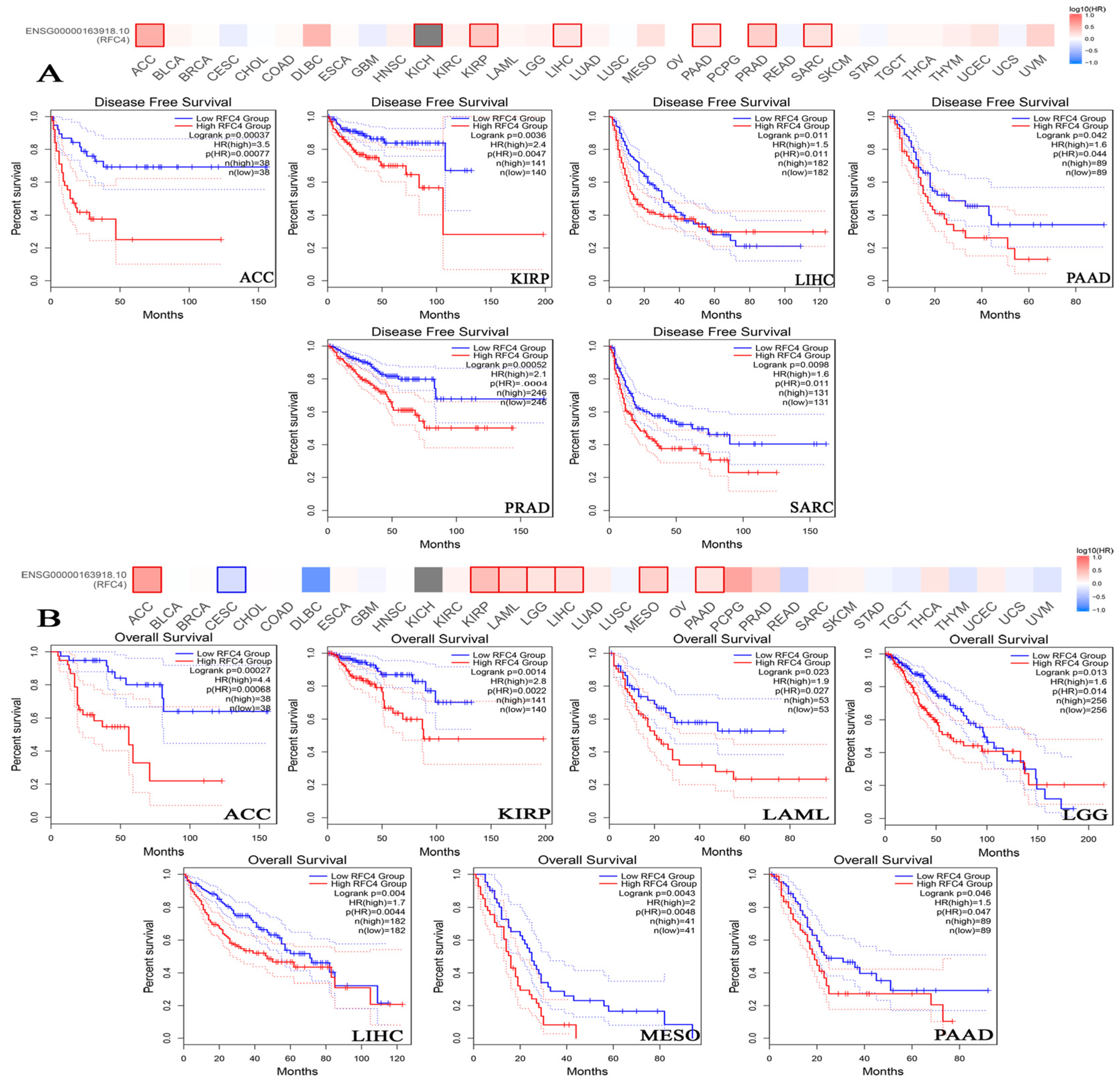
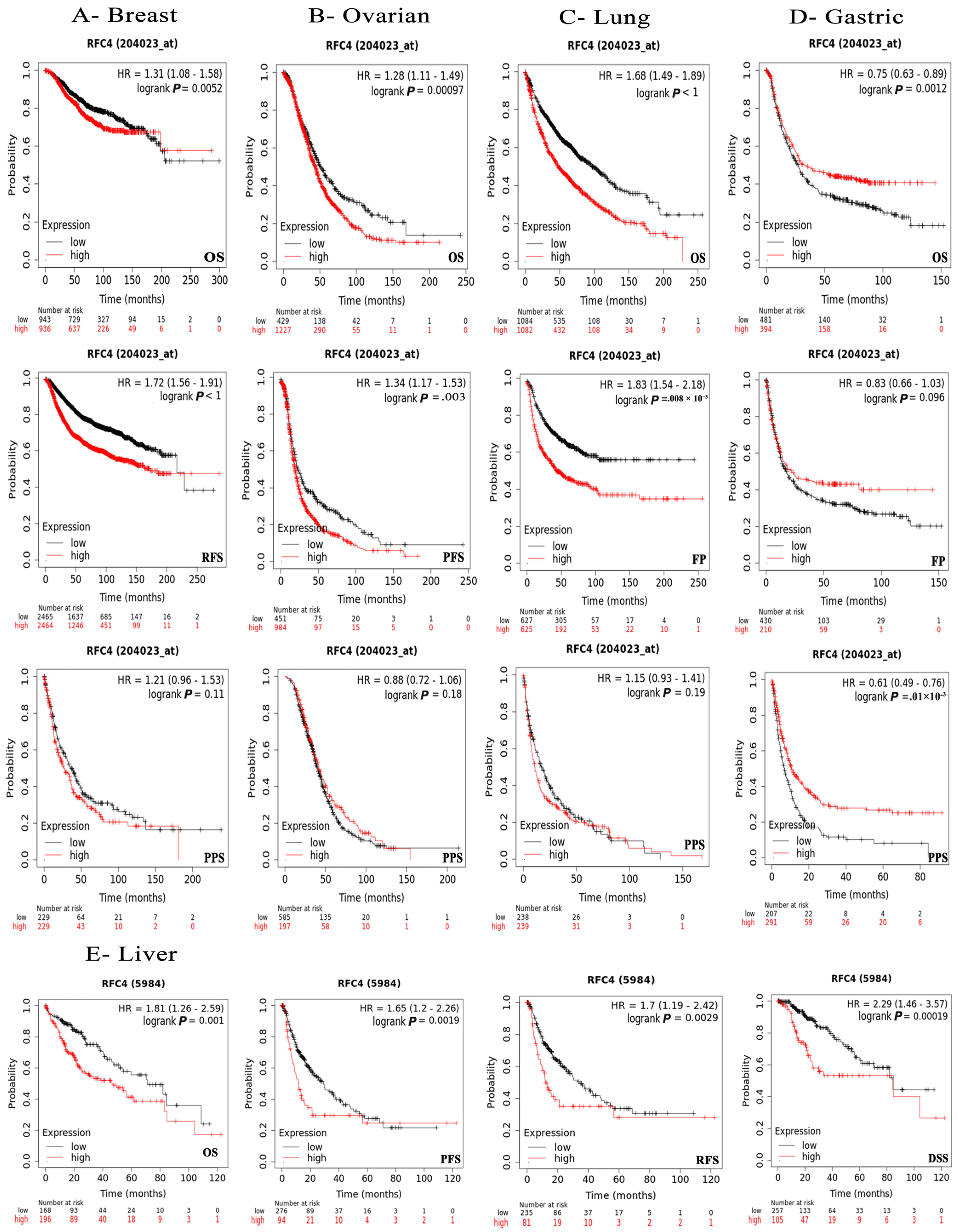
2.4. The Negative Correlation of Elevated RFC4 Levels with the Clinical Outcomes
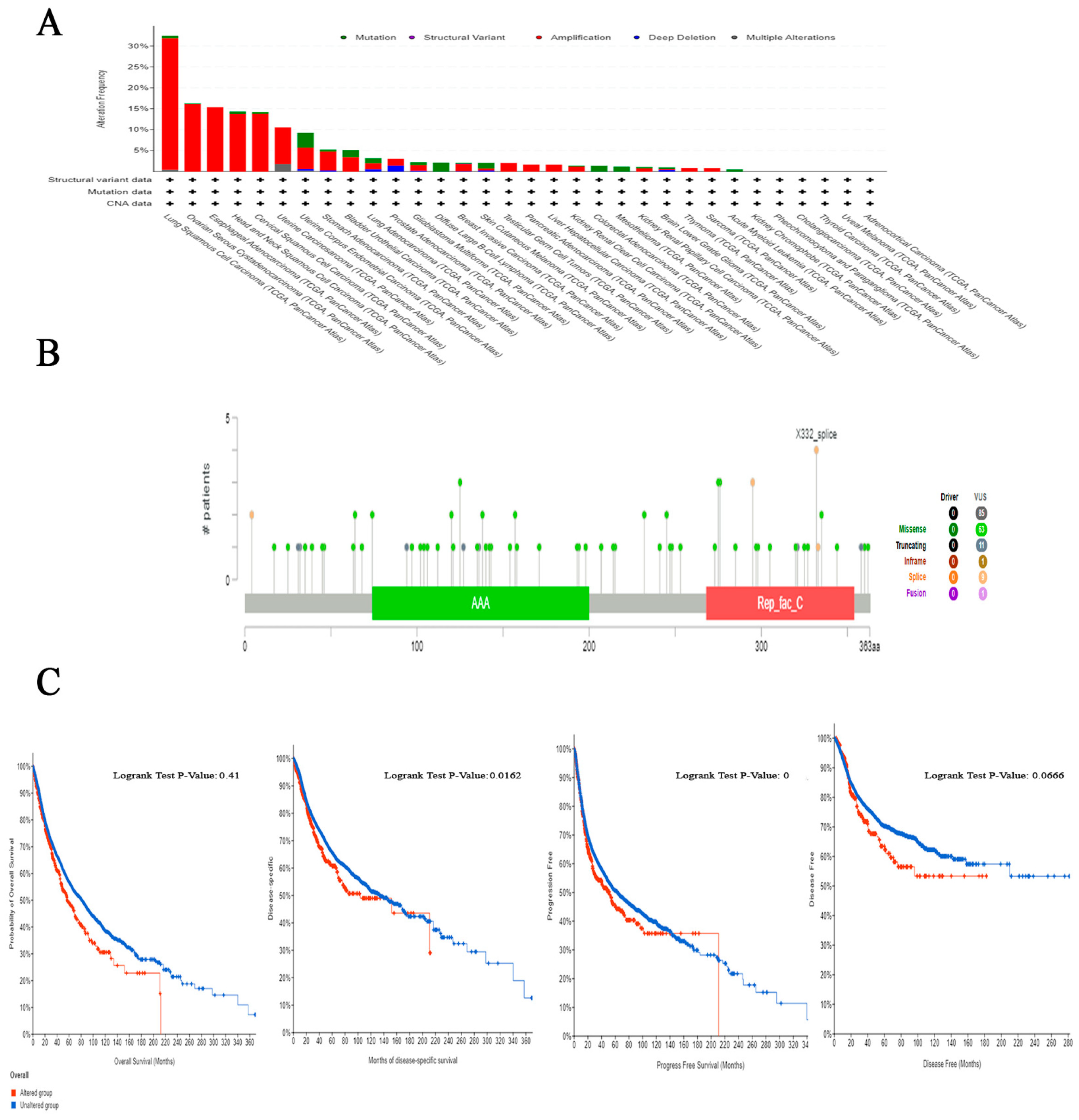
2.5. The Correlation between Genetic Alteration and Patient Outcome, Specifically Focusing on Predicting Poor Prognosis
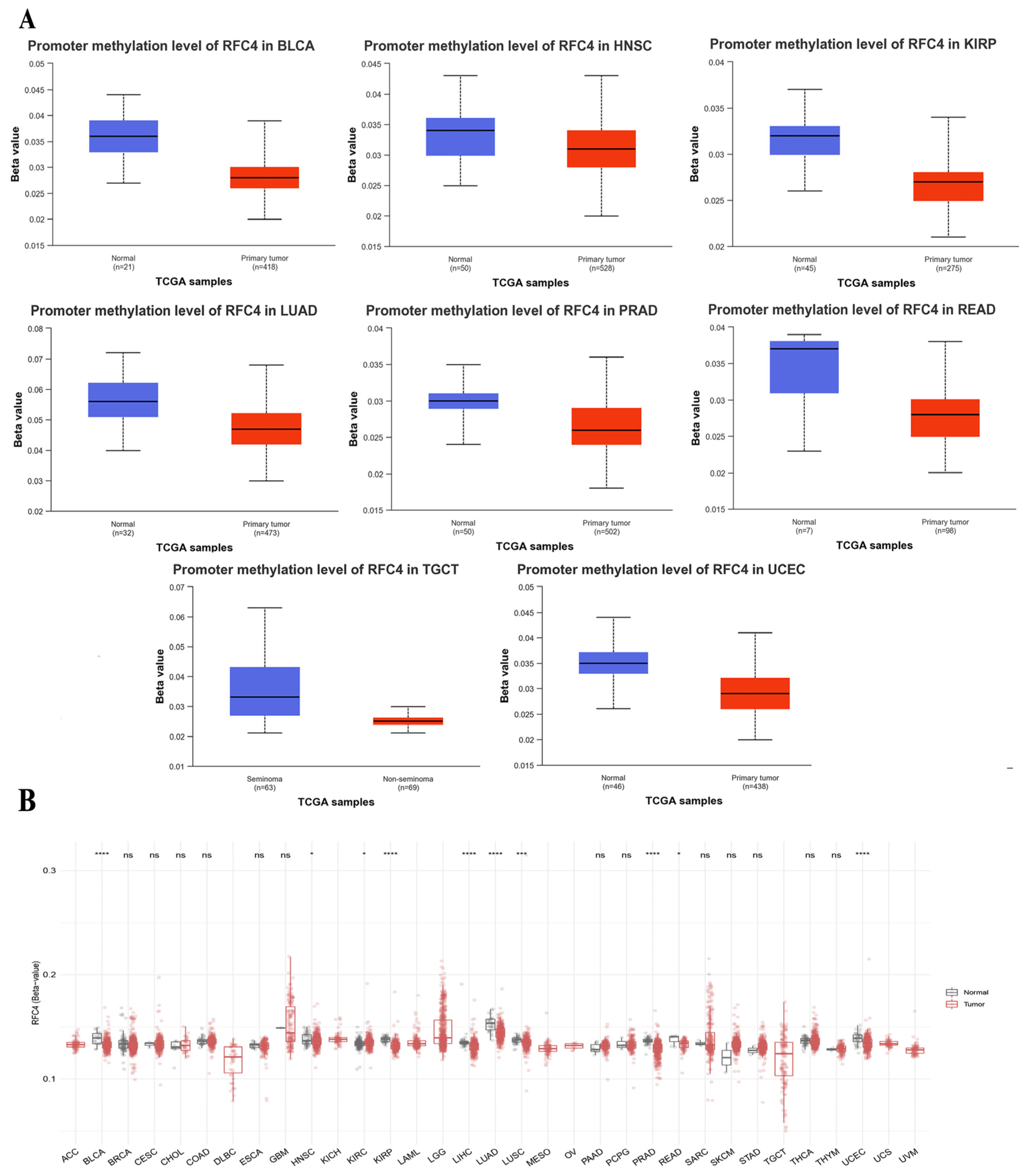
2.6. The Divergent Methylation Patterns of RFC4 in Various Human Cancers
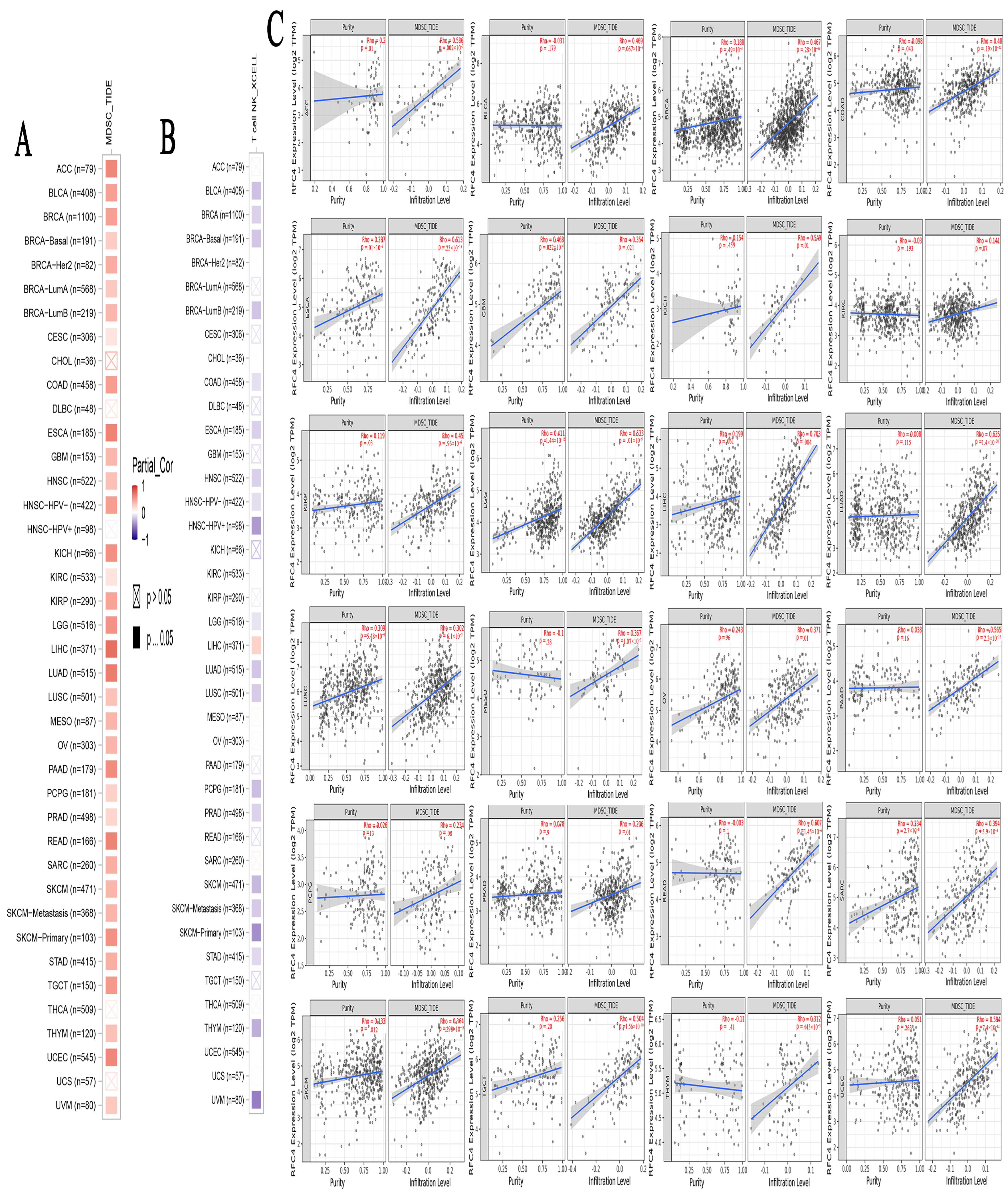
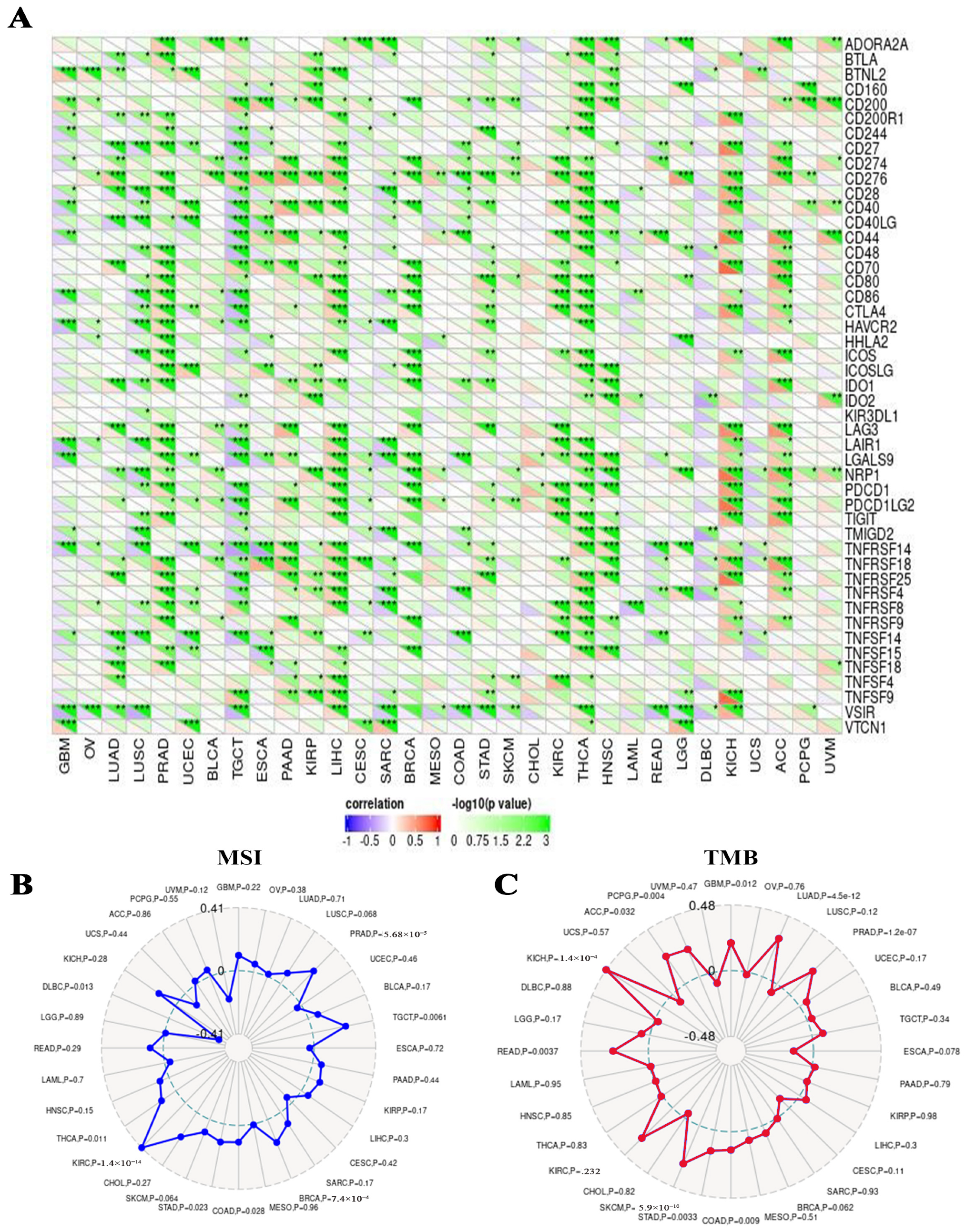
2.7. A Positive Link Was Seen between RFC4 Expression in Malignant Tissue and the Presence of Immunosuppressive Cells
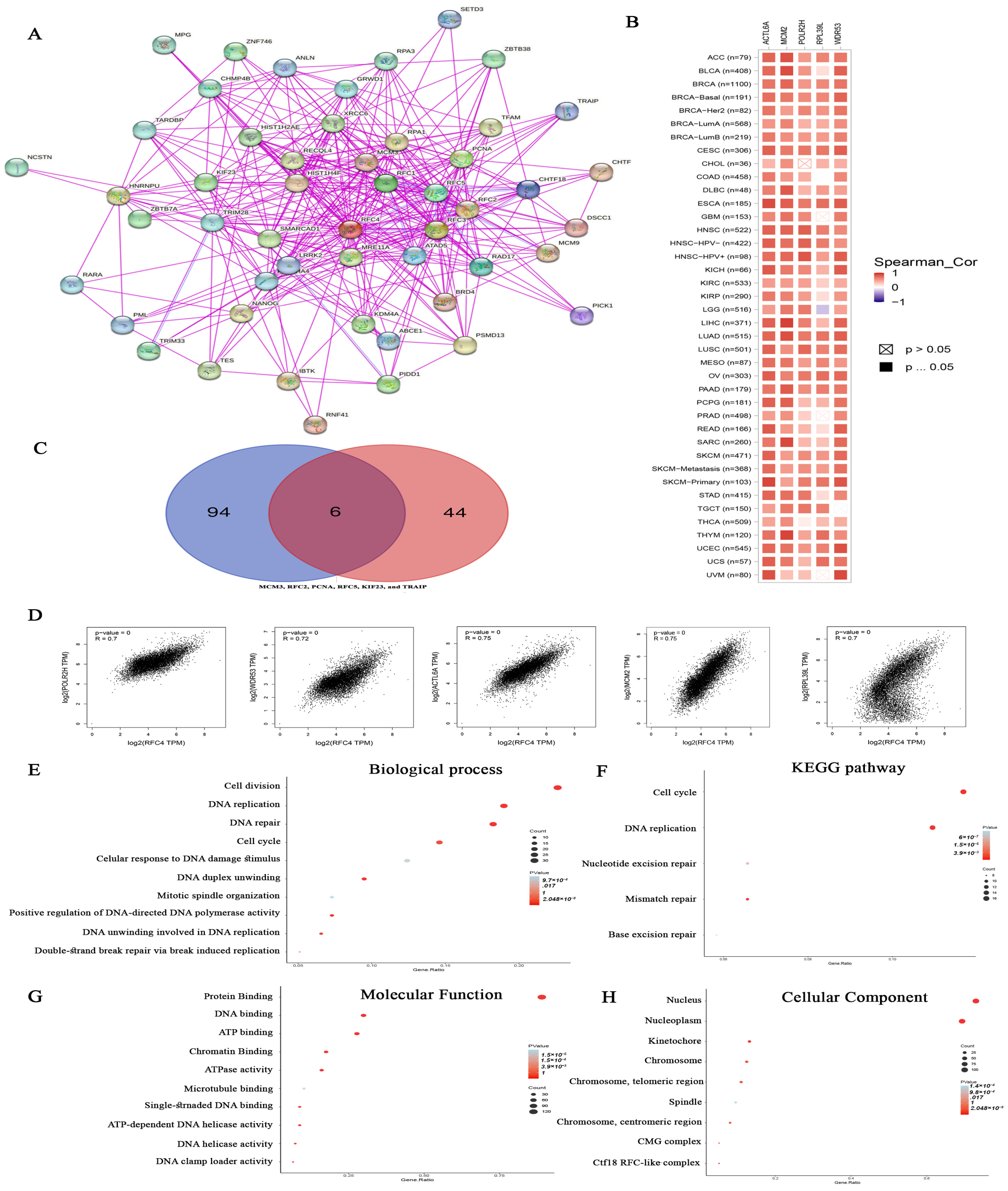
2.8. Analysis of Proteins Interacting and Correlated with RFC4
2.9. Molecular Docking
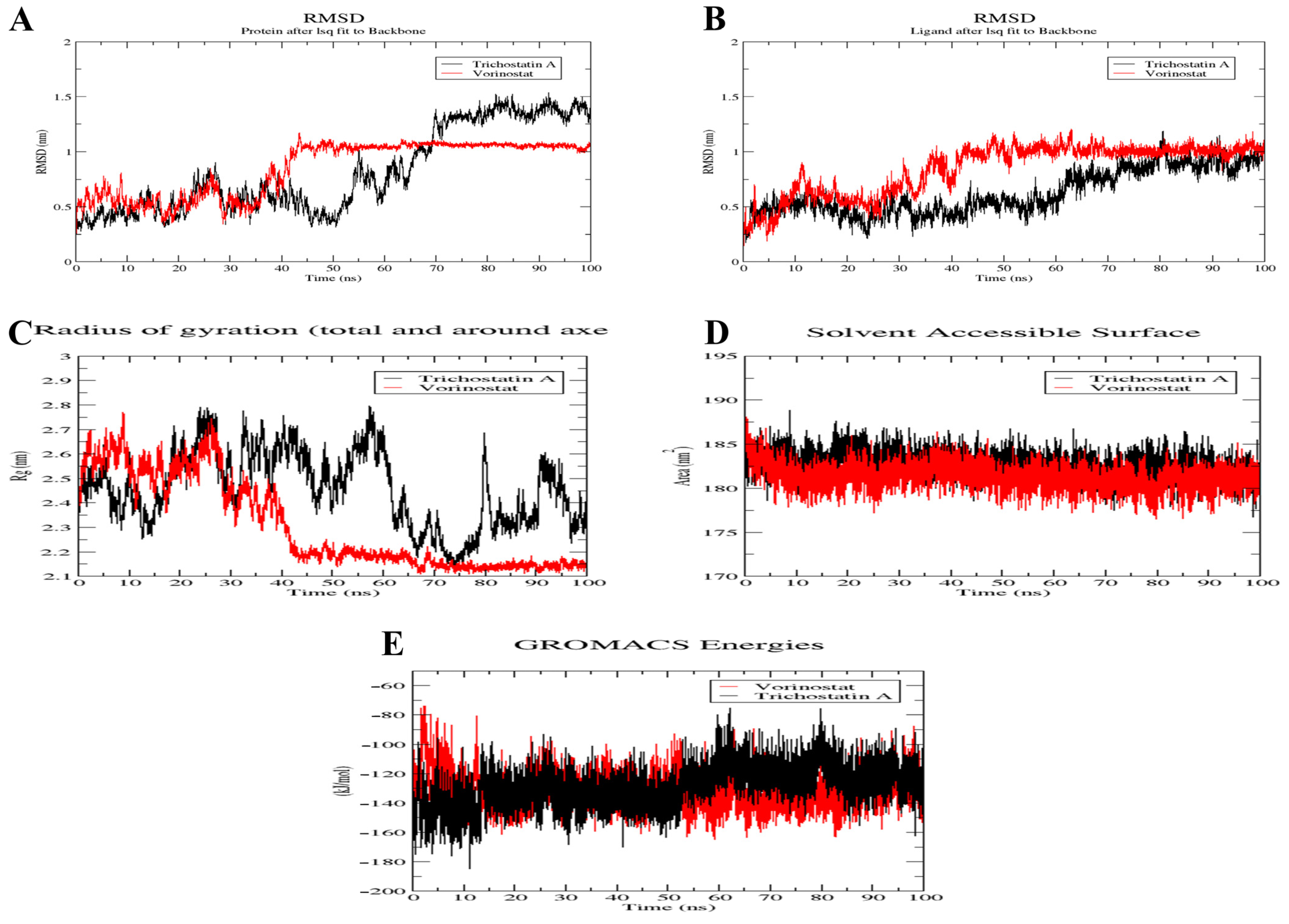
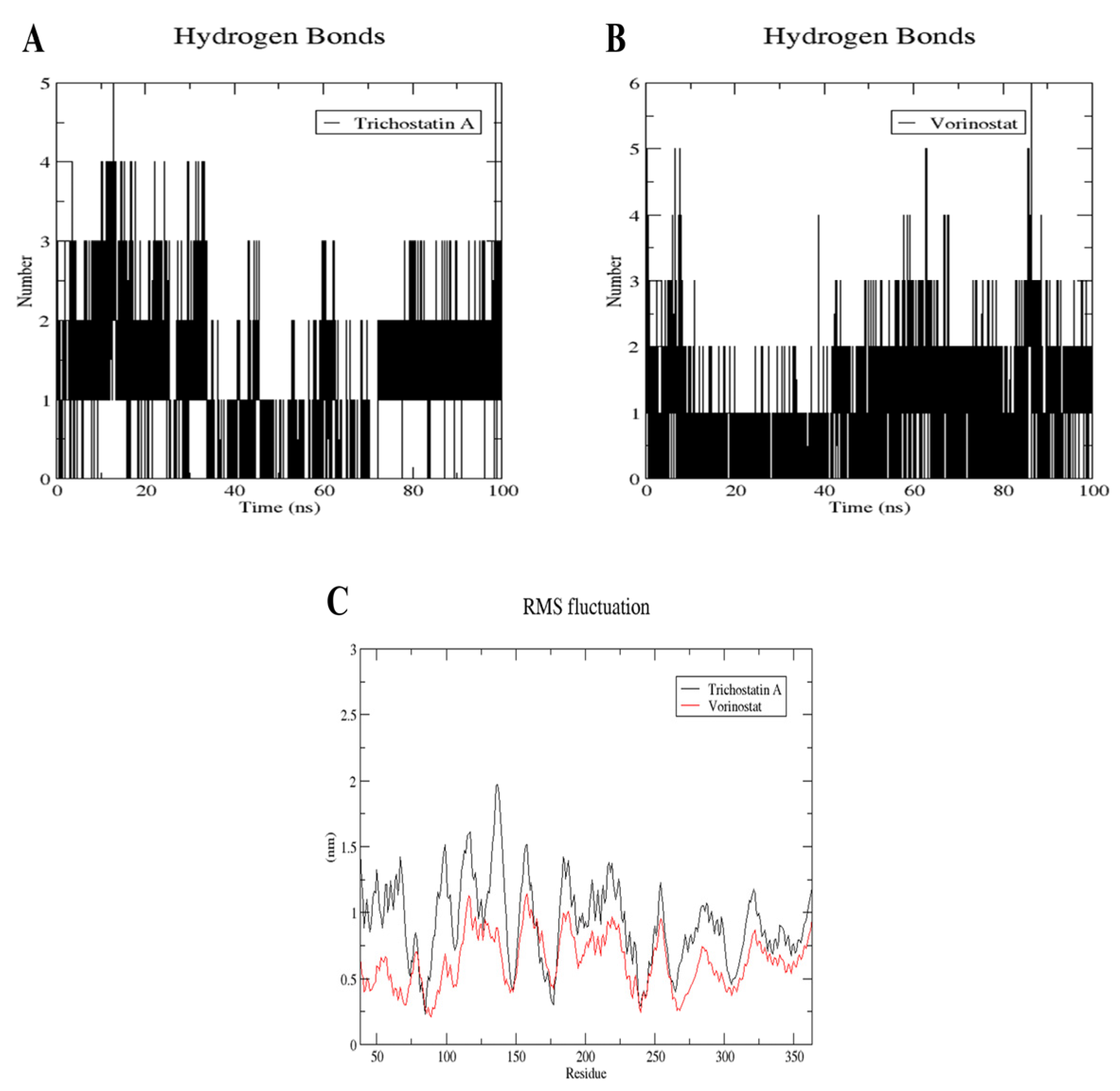
2.10. MD Simulation of Trichostatin A and Vorinostat on RFC4 Active Site
3. Discussion
4. Materials and Methods
4.1. RFC4 Differential Expression Analysis
4.2. Association between RFC4 and Tumor Grade and Stage
4.3. Assessment of Differential RFC4 Protein Levels
4.4. Survival Prognosis Analysis
4.5. The Connection between RFC4 Genetic Alteration and Patient Survival
4.6. Epigenetic Modulation of RFC4 under Tumor Conditions
4.7. The Impact of Modified RFC4 on the Infiltration and Functionality of Various Immune Components
4.8. RFC4 Enrichment Analysis
4.9. Molecular Docking
4.10. MD Simulation of Trichostatin A and Vorinostat on RFC4 Active Site
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soltan, M.A.; Eldeen, M.A.; Eid, R.A.; Alyamani, N.M.; Alqahtani, L.S.; Albogami, S.; Jafri, I.; Park, M.N.; Alsharif, G.; Fayad, E.; et al. A pan-cancer analysis reveals CHD1L as a prognostic and immunological biomarker in several human cancers. Front. Mol. Biosci. 2023, 10, 1017148. [Google Scholar] [CrossRef] [PubMed]
- Boutelle, A.M.; Attardi, L.D. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021, 31, 298–310. [Google Scholar] [CrossRef]
- Soltan, M.A.; Eldeen, M.A.; Sajer, B.H.; Abdelhameed, R.F.A.; Al-Salmi, F.A.; Fayad, E.; Jafri, I.; Ahmed, H.E.M.; Eid, R.A.; Hassan, H.M.; et al. Integration of Chemoinformatics and Multi-Omics Analysis Defines ECT2 as a Potential Target for Cancer Drug Therapy. Biology 2023, 12, 613. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.O.; Lee, M.; Chung, Y.J. AS-CMC: A pan-cancer database of alternative splicing for molecular classification of cancer. Sci. Rep. 2022, 12, 21074. [Google Scholar] [CrossRef]
- Ramos, M.; Geistlinger, L.; Oh, S.; Schiffer, L.; Azhar, R.; Kodali, H.; de Bruijn, I.; Gao, J.; Carey, V.J.; Morgan, M.; et al. Multiomic Integration of Public Oncology Databases in Bioconductor. JCO Clin. Cancer Inform. 2020, 4, 958–971. [Google Scholar] [CrossRef]
- Hassan, R.M.; Elanany, M.G.; Mostafa, M.M.; Yousef, R.H.A.; Salem, S.T. Whole genome characterization of methicillin resistant Staphylococcus aureus in an Egyptian Tertiary Care Hospital. J. Microbiol. Immunol. Infect. = Wei Mian Yu Gan Ran Za Zhi 2023, 56, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Eid, R.A.; Soltan, M.A.; Eldeen, M.A.; Shati, A.A.; Dawood, S.A.; Eissa, M.; Zaki, M.S.A.; Algahtani, M.; Theyab, A.; Abdel-Daim, M.M.; et al. Assessment of RACGAP1 as a Prognostic and Immunological Biomarker in Multiple Human Tumors: A Multiomics Analysis. Int. J. Mol. Sci. 2022, 23, 14102. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Wu, K.; Yi, M.; Qin, S.; Chu, Q.; Zheng, X.; Wu, K. The efficacy and safety of combination of PD-1 and CTLA-4 inhibitors: A meta-analysis. Exp. Hematol. Oncol. 2019, 8, 26. [Google Scholar] [CrossRef]
- Eid, R.A.; Eldeen, M.A.; Soltan, M.A.; Al-Shraim, M.; Aldehri, M.; Alqahtani, L.S.; Alsharif, G.; Albogami, S.; Jafri, I.; Fayad, E.; et al. Integrative analysis of WDR12 as a potential prognostic and immunological biomarker in multiple human tumors. Front. Genet. 2022, 13, 1008502. [Google Scholar] [CrossRef]
- Yao, N.Y.; O’Donnell, M. The RFC clamp loader: Structure and function. In The Eukaryotic Replisome: A Guide to Protein Structure and Function; Subcellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2012; Volume 62, pp. 259–279. [Google Scholar] [CrossRef]
- Schmidt, S.L.; Gomes, X.V.; Burgers, P.M. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J. Biol. Chem. 2001, 276, 34784–34791. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.L.; Pautz, A.L.; Burgers, P.M. ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J. Biol. Chem. 2001, 276, 34792–34800. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Brill, S.J. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 3725–3737. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Feng, L.; Wei, J.; Wang, G.; Wu, L. Knockdown of RFC4 inhibits the cell proliferation of nasopharyngeal carcinoma in vitro and in vivo. Front. Med. 2023, 17, 132–142. [Google Scholar] [CrossRef]
- Liu, L.; Tao, T.; Liu, S.; Yang, X.; Chen, X.; Liang, J.; Hong, R.; Wang, W.; Yang, Y.; Li, X.; et al. An RFC4/Notch1 signaling feedback loop promotes NSCLC metastasis and stemness. Nat. Commun. 2021, 12, 2693. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.C.; Yue, X.; Zhang, R.X.; Liu, T.Y.; Pan, Z.Z.; Yang, M.J.; Lu, Z.H.; Wang, Z.Y.; Peng, J.H.; Le, L.Y.; et al. Genome-wide RNAi Screening Identifies RFC4 as a Factor That Mediates Radioresistance in Colorectal Cancer by Facilitating Nonhomologous End Joining Repair. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4567–4579. [Google Scholar] [CrossRef]
- Zhou, Y.; Hingorani, M.M. Impact of individual proliferating cell nuclear antigen-DNA contacts on clamp loading and function on DNA. J. Biol. Chem. 2012, 287, 35370–35381. [Google Scholar] [CrossRef]
- He, Y.; Hu, S.; Zhong, J.; Cheng, A.; Shan, N. Identification of significant genes signatures and prognostic biomarkers in cervical squamous carcinoma via bioinformatic data. PeerJ 2020, 8, e10386. [Google Scholar] [CrossRef]
- Gisatulin, M.; Dobricic, V.; Zühlke, C.; Hellenbroich, Y.; Tadic, V.; Münchau, A.; Isenhardt, K.; Bürk, K.; Bahlo, M.; Lockhart, P.J.; et al. Clinical spectrum of the pentanucleotide repeat expansion in the RFC1 gene in ataxia syndromes. Neurology 2020, 95, e2912–e2923. [Google Scholar] [CrossRef]
- Arai, M.; Kondoh, N.; Imazeki, N.; Hada, A.; Hatsuse, K.; Matsubara, O.; Yamamoto, M. The knockdown of endogenous replication factor C4 decreases the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Liver Int. 2009, 29, 55–62. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, D.; Li, F.; Zhao, Z.; Liu, X.; Gao, D.; Zhang, Y.; Li, H. Identification of biomarkers for acute leukemia via machine learning-based stemness index. Gene 2021, 804, 145903. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Pan, C.; Zhang, T.; Ni, M.; Wang, W.; Zhang, S.; Chen, Y.; Wang, J.; Fang, Q. Nrf2 overexpression increases the resistance of acute myeloid leukemia to Cytarabine by inhibiting replication factor C4. Cancer Gene Ther. 2022, 29, 1773–1790. [Google Scholar] [CrossRef] [PubMed]
- Breit, S.; Stanulla, M.; Flohr, T.; Schrappe, M.; Ludwig, W.D.; Tolle, G.; Happich, M.; Muckenthaler, M.U.; Kulozik, A.E. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood 2006, 108, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Chen, Q.; Shimada, K.; Tang, M.; Li, H.; Gurumurthy, A.; Khoury, J.D.; Xu, B.; Huang, S.; Qiu, Y. Histone deacetylase inhibitor targets CD123/CD47-positive cells and reverse chemoresistance phenotype in acute myeloid leukemia. Leukemia 2019, 33, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Xu, Y.; Lu, L.; Jiao, Y.; Liu, J.; Wang, L.; Zhao, H. Identification of key candidate genes and small molecule drugs in cervical cancer by bioinformatics strategy. Cancer Manag. Res. 2018, 10, 3533–3549. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Fang, L.; Luo, Y.; Yang, Z.; Liao, Y.; Cui, J.; Huang, M.; Yang, Z.; Huang, Y.; Fan, X.; et al. Levels of human replication factor C4, a clamp loader, correlate with tumor progression and predict the prognosis for colorectal cancer. J. Transl. Med. 2014, 12, 320. [Google Scholar] [CrossRef]
- Zhang, J.; Meng, S.; Wang, X.; Wang, J.; Fan, X.; Sun, H.; Ning, R.; Xiao, B.; Li, X.; Jia, Y.; et al. Sequential gene expression analysis of cervical malignant transformation identifies RFC4 as a novel diagnostic and prognostic biomarker. BMC Med. 2022, 20, 437. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Xie, X. RFC4 promotes the progression and growth of Oral Tongue squamous cell carcinoma in vivo and vitro. J. Clin. Lab. Anal. 2021, 35, e23761. [Google Scholar] [CrossRef]
- Zabady, S.; Mahran, N.; Soltan, M.A.; Alaa Eldeen, M.; Eid, R.A.; Albogami, S.; Fayad, E.; Matboli, M.; Habib, E.K.; Hasanin, A.H.; et al. Cyanidin-3-Glucoside Modulates hsa_circ_0001345/miRNA106b/ATG16L1 Axis Expression as a Potential Protective Mechanism against Hepatocellular Carcinoma. Curr. Issues Mol. Biol. 2022, 44, 1677–1687. [Google Scholar] [CrossRef]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Byers, L.A.; Wang, J.; Nilsson, M.B.; Fujimoto, J.; Saintigny, P.; Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012, 2, 798–811. [Google Scholar] [CrossRef]
- Shen, H.; Che, K.; Cong, L.; Dong, W.; Zhang, T.; Liu, Q.; Du, J. Diagnostic and prognostic value of blood samples for KRAS mutation identification in lung cancer: A meta-analysis. Oncotarget 2017, 8, 36812–36823. [Google Scholar] [CrossRef]
- Jakob, J.A.; Bassett, R.L., Jr.; Ng, C.S.; Curry, J.L.; Joseph, R.W.; Alvarado, G.C.; Rohlfs, M.L.; Richard, J.; Gershenwald, J.E.; Kim, K.B.; et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012, 118, 4014–4023. [Google Scholar] [CrossRef]
- Anglim, P.P.; Alonzo, T.A.; Laird-Offringa, I.A. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: An update. Mol. Cancer 2008, 7, 81. [Google Scholar] [CrossRef]
- Romero-Garcia, S.; Prado-Garcia, H.; Carlos-Reyes, A. Role of DNA Methylation in the Resistance to Therapy in Solid Tumors. Front. Oncol. 2020, 10, 1152. [Google Scholar] [CrossRef]
- Shao, C.; Sun, W.; Tan, M.; Glazer, C.A.; Bhan, S.; Zhong, X.; Fakhry, C.; Sharma, R.; Westra, W.H.; Hoque, M.O.; et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin. Cancer Res. 2011, 17, 4320–4330. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Soes, S.; Daugaard, I.L.; Sorensen, B.S.; Carus, A.; Mattheisen, M.; Alsner, J.; Overgaard, J.; Hager, H.; Hansen, L.L.; Kristensen, L.S. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience 2014, 1, 367–374. [Google Scholar] [CrossRef]
- Peng, M.; Mo, Y.; Wang, Y.; Wu, P.; Zhang, Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; Li, X.; et al. Neoantigen vaccine: An emerging tumor immunotherapy. Mol. Cancer 2019, 18, 128. [Google Scholar] [CrossRef]
- Chang, E.; Pelosof, L.; Lemery, S.; Gong, Y.; Goldberg, K.B.; Farrell, A.T.; Keegan, P.; Veeraraghavan, J.; Wei, G.; Blumenthal, G.M.; et al. Systematic Review of PD-1/PD-L1 Inhibitors in Oncology: From Personalized Medicine to Public Health. Oncologist 2021, 26, e1786–e1799. [Google Scholar] [CrossRef] [PubMed]
- Condamine, T.; Ramachandran, I.; Youn, J.I.; Gabrilovich, D.I. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu. Rev. Med. 2015, 66, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ma, X.; Zhu, C.; Liu, L.; Wang, G.; Yuan, X. The Role of Myeloid-Derived Suppressor Cells in Patients with Solid Tumors: A Meta-Analysis. PLoS ONE 2016, 11, e0164514. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.A.; Seo, H.; Kim, I.K.; Jeon, I.; Kang, C.Y. Roles of NKT cells in cancer immunotherapy. Arch. Pharm. Res. 2019, 42, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.J.; Choi, J.E.; Exley, M.A. Novel Approaches to Exploiting Invariant NKT Cells in Cancer Immunotherapy. Front. Immunol. 2018, 9, 384. [Google Scholar] [CrossRef]
- ZHAO, Z.; Li, W.; Zhang, X.; Ge, M.; Song, C. Correlation between TMB and MSI in patients with solid tumors. J. Clin. Oncol. 2020, 38, e15169. [Google Scholar] [CrossRef]
- Diaz, L.A.; Marabelle, A.; Delord, J.-P.; Shapira-Frommer, R.; Geva, R.; Peled, N.; Kim, T.W.; Andre, T.; Cutsem, E.V.; Guimbaud, R.; et al. Pembrolizumab therapy for microsatellite instability high (MSI-H) colorectal cancer (CRC) and non-CRC. J. Clin. Oncol. 2017, 35, 3071. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, S.; Chen, W.; Lin, Y.; Zhang, Q.; Huang, D. A Bioinformatics-Based Screening and Analysis of Key Genes in Hepatocellular Carcinoma. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Eid, R.A.; Alaa Edeen, M.; Shedid, E.M.; Kamal, A.S.S.; Warda, M.M.; Mamdouh, F.; Khedr, S.A.; Soltan, M.A.; Jeon, H.W.; Zaki, M.S.A.; et al. Targeting Cancer Stem Cells as the Key Driver of Carcinogenesis and Therapeutic Resistance. Int. J. Mol. Sci. 2023, 24, 1786. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2013, 424, 53–65. [Google Scholar] [CrossRef]
- Tran, M.; Yeh, K.T.; Chuang, Y.M.; Hsu, P.Y.; Low, J.T.; Kumari, H.; Lee, Y.T.; Chen, Y.C.; Huang, W.H.; Jin, H.; et al. Methylomic analysis identifies C11orf87 as a novel epigenetic biomarker for GI cancers. PLoS ONE 2021, 16, e0250499. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, D.; Lu, C. The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics Chromatin 2019, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Dunn, G.P.; Schreiber, R.D. Cancer immunosurveillance and immunoediting: The roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 2006, 90, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Sharma, N.; Gupta, K.B.; Dhiman, M. Role of immune system in tumor progression and carcinogenesis. J. Cell. Biochem. 2018, 119, 5028–5042. [Google Scholar] [CrossRef] [PubMed]
- Solito, S.; Falisi, E.; Diaz-Montero, C.M.; Doni, A.; Pinton, L.; Rosato, A.; Francescato, S.; Basso, G.; Zanovello, P.; Onicescu, G.; et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood 2011, 118, 2254–2265. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, J.; Weng, L.; Tang, W.; Jin, S.; Ma, W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J. Hematol. Oncol. 2020, 13, 10. [Google Scholar] [CrossRef]
- Yu, S.; Ren, X.; Li, L. Myeloid-derived suppressor cells in hematologic malignancies: Two sides of the same coin. Exp. Hematol. Oncol. 2022, 11, 43. [Google Scholar] [CrossRef]
- Hao, Z.; Li, R.; Wang, Y.; Li, S.; Hong, Z.; Han, Z. Landscape of Myeloid-derived Suppressor Cell in Tumor Immunotherapy. Biomark. Res. 2021, 9, 77. [Google Scholar] [CrossRef]
- Metelitsa, L.S.; Naidenko, O.V.; Kant, A.; Wu, H.W.; Loza, M.J.; Perussia, B.; Kronenberg, M.; Seeger, R.C. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J. Immunol. 2001, 167, 3114–3122. [Google Scholar] [CrossRef]
- Kuylenstierna, C.; Björkström, N.K.; Andersson, S.K.; Sahlström, P.; Bosnjak, L.; Paquin-Proulx, D.; Malmberg, K.J.; Ljunggren, H.G.; Moll, M.; Sandberg, J.K. NKG2D performs two functions in invariant NKT cells: Direct TCR-independent activation of NK-like cytolysis and co-stimulation of activation by CD1d. Eur. J. Immunol. 2011, 41, 1913–1923. [Google Scholar] [CrossRef]
- McEwen-Smith, R.M.; Salio, M.; Cerundolo, V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol. Res. 2015, 3, 425–435. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Weiner, S.J.; Kollman, P.A.; Case, D.A.; Singh, U.C.; Ghio, C.; Alagona, G.; Profeta, S.; Weiner, P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA. Discovery Studio, v4.5; Dassault Systèmes BIOVIA: San Diego, CA, USA, 2008. [Google Scholar]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol. Inf. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Genheden, S.; Kongsted, J.; Soderhjelm, P.; Ryde, U. Nonpolar Solvation Free Energies of Protein-Ligand Complexes. J. Chem. Theory Comput. 2010, 6, 3558–3568. [Google Scholar] [CrossRef]


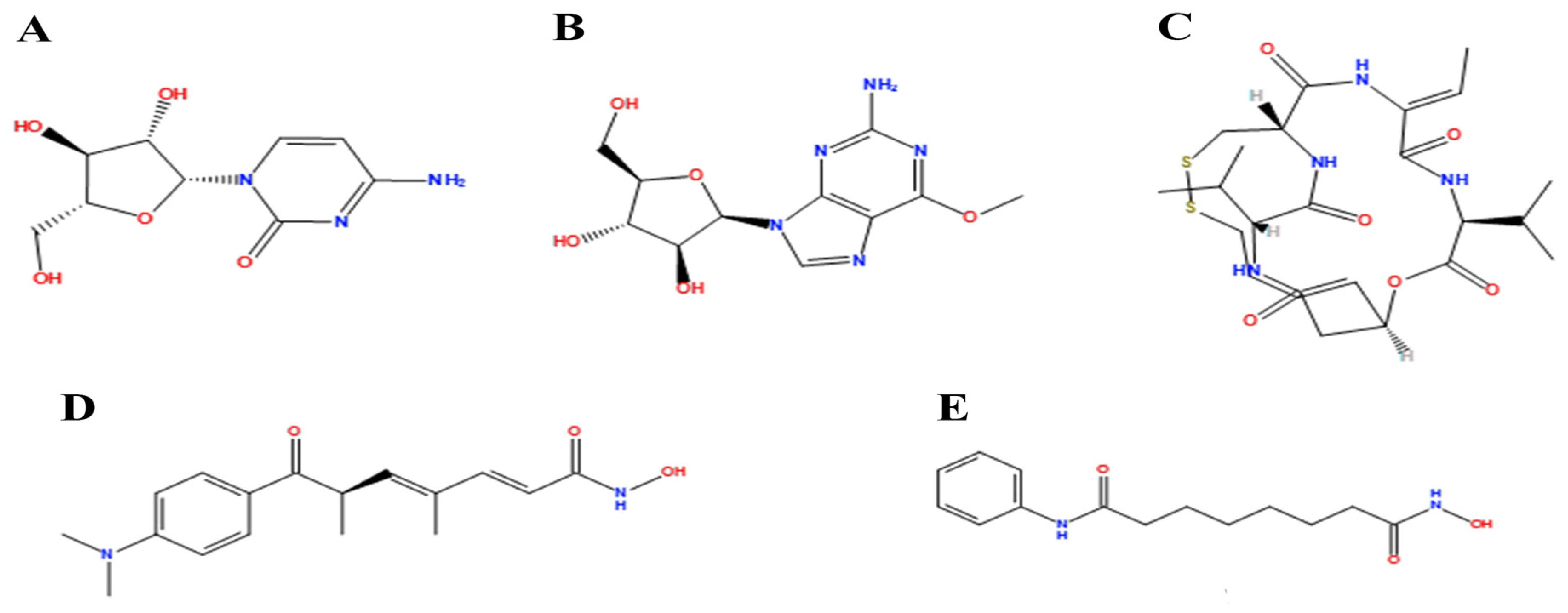
| Cpd. | Bonded Residues | Binding Energy (Kcal/mol) | 2D Interaction |
|---|---|---|---|
| Adenosine 5′-[gamma-thio]triphosphate | Val41 Tyr44 Arg45 Glu51 Gly81 Thr82 Gly83 Lys84 Thr85 Arg210 Arg239 | −8.1078 | 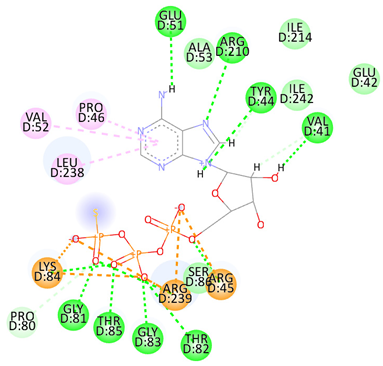 |
| Cytarabine | Arg45 Lys84 Thr85 Ser86 Arg239 | −5.5553 | 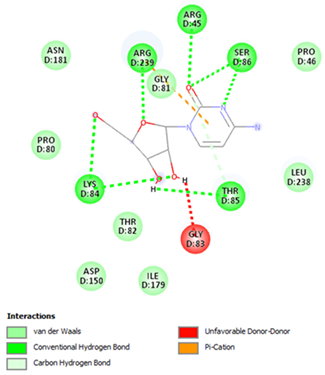 |
| Nelarabine | Arg45 | −6.2007 | 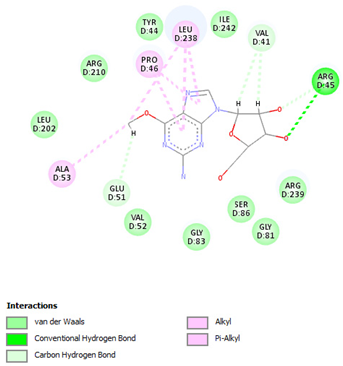 |
| Romidepsin | Lys84 Thr85 Glu151 Arg239 | −6.2108 | 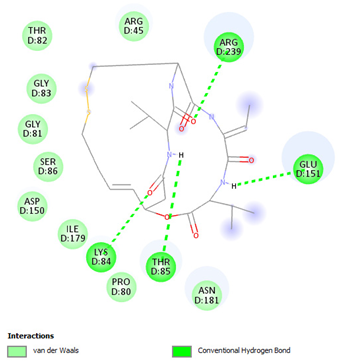 |
| Trichostatin A | Arg45 Glu51 Ala53 Ser86 | −7.4355 | Ar |
| Vorinostat | Gly83 Lys84 Thr85 Ser86 | −6.6417 | 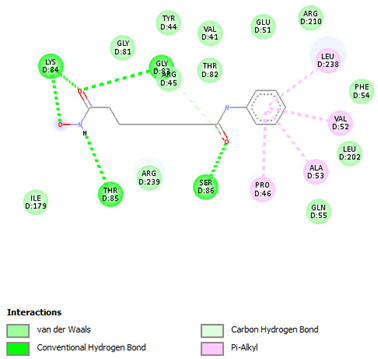 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaa Eldeen, M.; Mamdouh, F.; Abdulsahib, W.K.; Eid, R.A.; Alhanshani, A.A.; Shati, A.A.; Alqahtani, Y.A.; Alshehri, M.A.; Samir A. Zaki, M.; Soltan, M.A.; et al. Oncogenic Potential of Replication Factor C Subunit 4: Correlations with Tumor Progression and Assessment of Potential Inhibitors. Pharmaceuticals 2024, 17, 152. https://doi.org/10.3390/ph17020152
Alaa Eldeen M, Mamdouh F, Abdulsahib WK, Eid RA, Alhanshani AA, Shati AA, Alqahtani YA, Alshehri MA, Samir A. Zaki M, Soltan MA, et al. Oncogenic Potential of Replication Factor C Subunit 4: Correlations with Tumor Progression and Assessment of Potential Inhibitors. Pharmaceuticals. 2024; 17(2):152. https://doi.org/10.3390/ph17020152
Chicago/Turabian StyleAlaa Eldeen, Muhammad, Farag Mamdouh, Waleed K. Abdulsahib, Refaat A. Eid, Ahmad A. Alhanshani, Ayed A. Shati, Youssef A. Alqahtani, Mohammed A. Alshehri, Mohamed Samir A. Zaki, Mohamed A. Soltan, and et al. 2024. "Oncogenic Potential of Replication Factor C Subunit 4: Correlations with Tumor Progression and Assessment of Potential Inhibitors" Pharmaceuticals 17, no. 2: 152. https://doi.org/10.3390/ph17020152
APA StyleAlaa Eldeen, M., Mamdouh, F., Abdulsahib, W. K., Eid, R. A., Alhanshani, A. A., Shati, A. A., Alqahtani, Y. A., Alshehri, M. A., Samir A. Zaki, M., Soltan, M. A., & Noreldin, A. E. (2024). Oncogenic Potential of Replication Factor C Subunit 4: Correlations with Tumor Progression and Assessment of Potential Inhibitors. Pharmaceuticals, 17(2), 152. https://doi.org/10.3390/ph17020152







