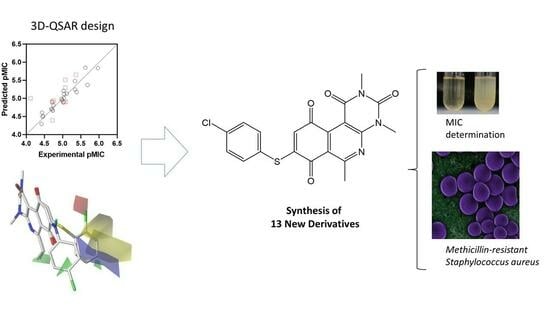
QSAR Studies, Synthesis, and Biological Evaluation of New Pyrimido-Isoquinolin-Quinone Derivatives against Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results and Discussion
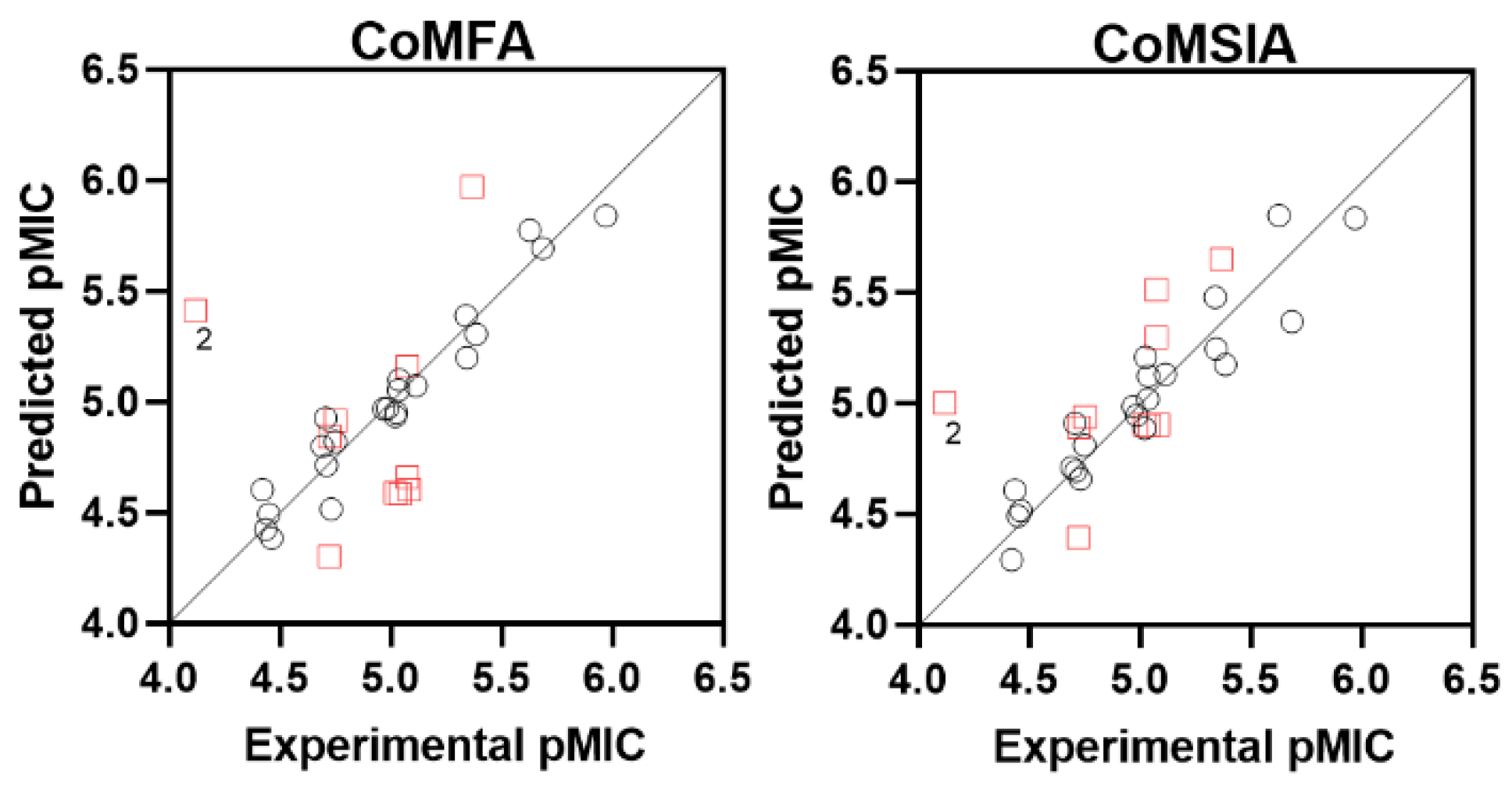
2.1. CoMFA/CoMSIA Studies
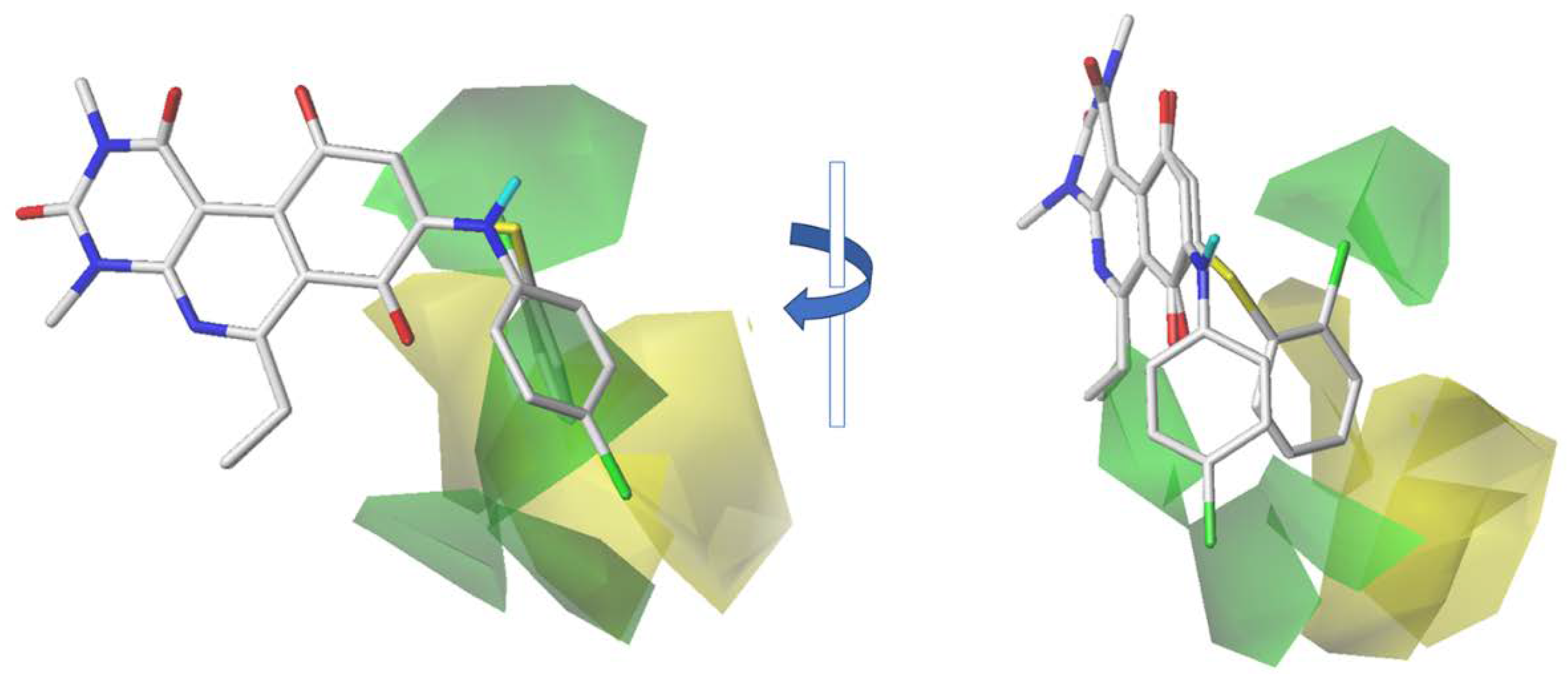
2.2. CoMFA Contour Maps
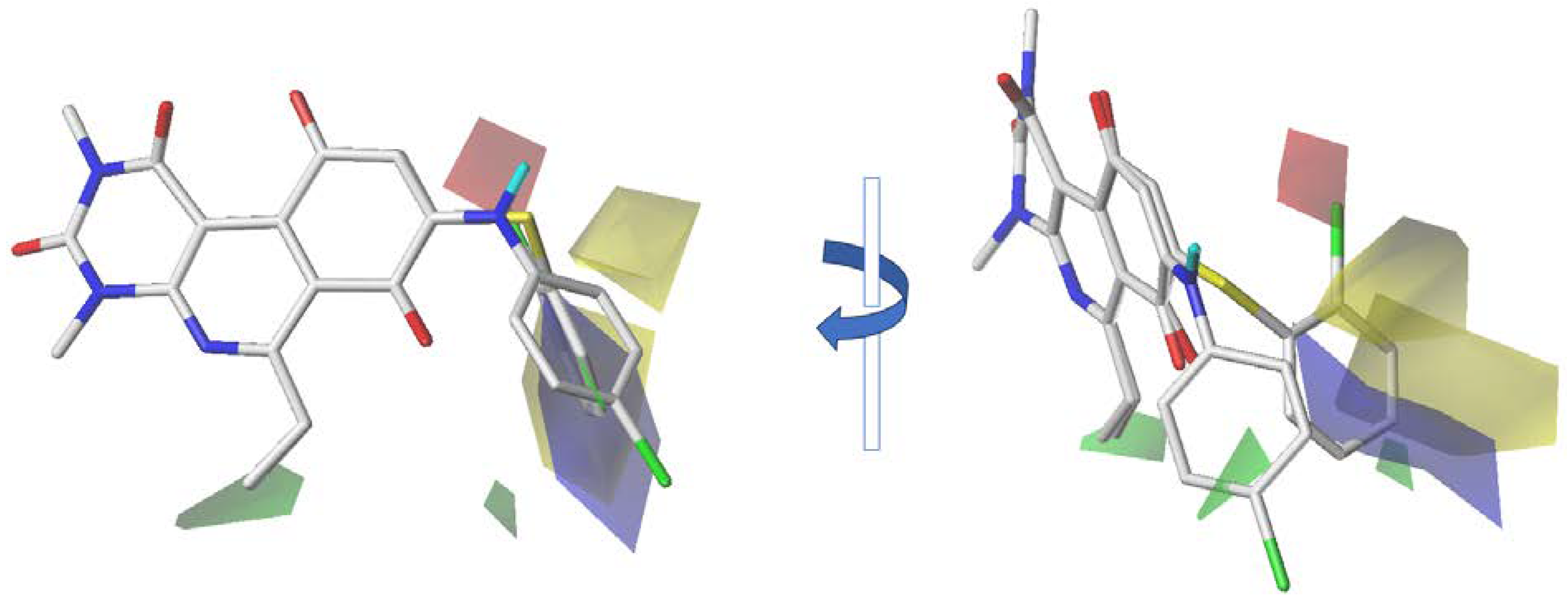
2.3. CoMSIA Contour Maps
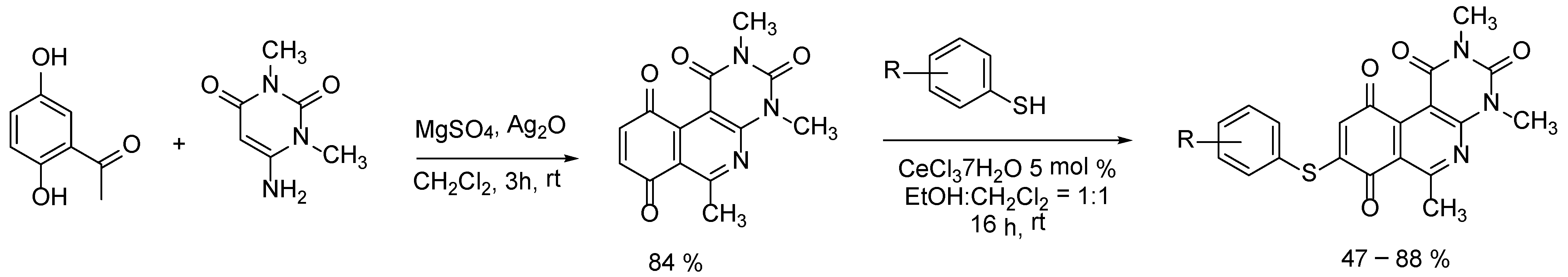
2.4. Design and Synthesis of New Derivatives
2.5. Antibacterial Activity Evaluation
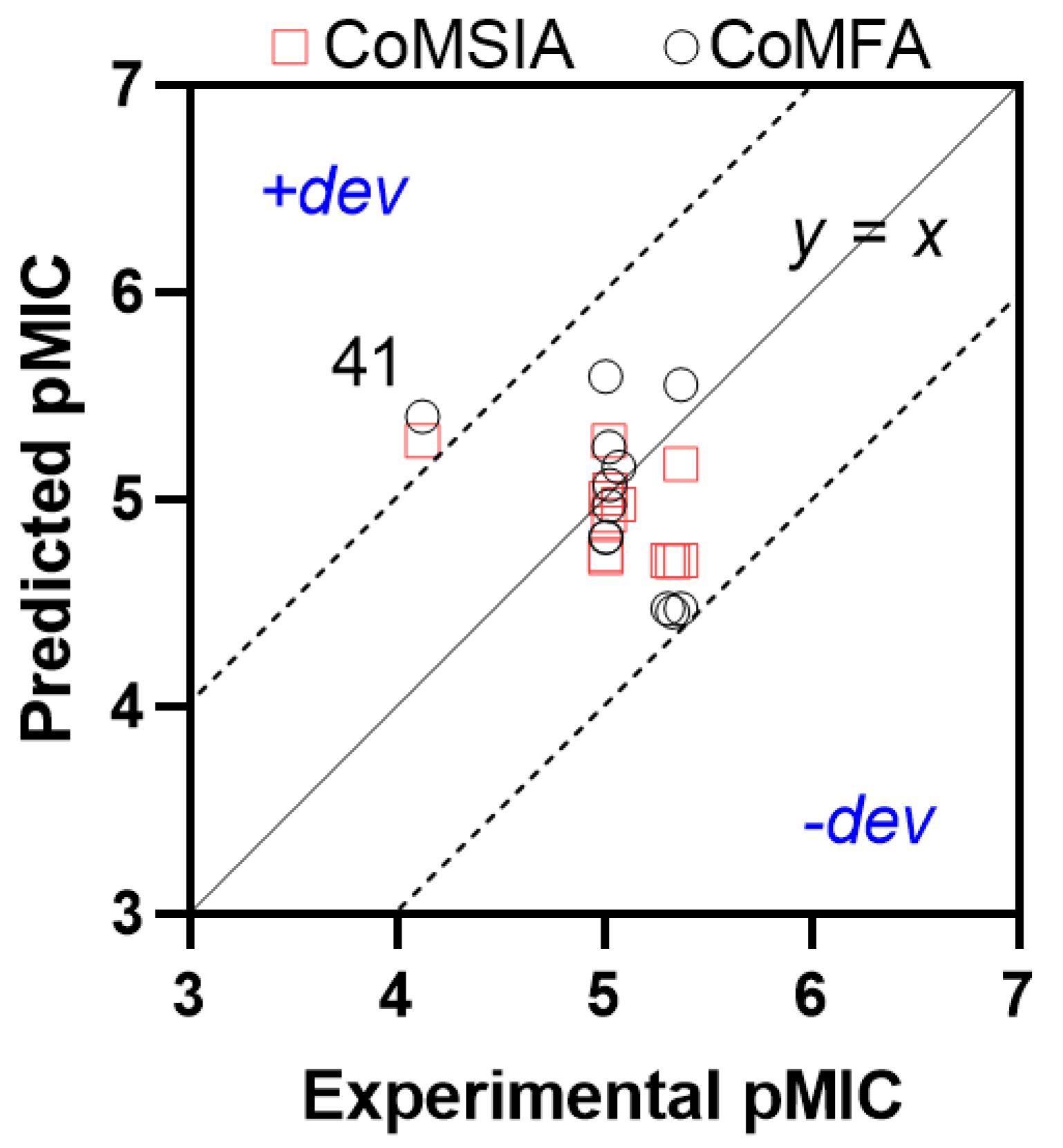
2.6. QSAR Model Challenge
3. Materials and Methods
3.1. QSAR Studies
3.2. Chemistry
3.3. Chemical Synthesis and Structural Characterization for Compounds
3.3.1. Synthesis of 2,4,6-Trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (Quinone Core, QC)
3.3.2. General Procedure (A) for Synthesis of 8-Thioaryl-pyrimidoisoquinolinequinones Derivatives (33–45)
8-((4-Chloro-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (33)
8-((4-Bromo-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (34)
2,4,6-Trimethyl-8-((4-methyl-phenyl)thio)pyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (35)
8-((3-Chloro-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (36)
8-((3-Bromo-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (37)
8-((3-Fluoro-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (38)
8-(3-Methoxy-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (39)
2,4,6-Trimethyl-8-((3-methyl-phenyl)thio)pyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (40)
8-((2-Chloro-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (41)
8-((2-Bromo-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (42)
8-(2-Methoxy-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (43)
2,4,6-Trimethyl-8-((3-methyl-phenyl)thio)pyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (44)
8-((2-Fluoro-phenyl)thio)-2,4,6-trimethylpyrimido[4,5-c]isoquinoline-1,3,7,10(2H,4H)-tetraone (45)
3.4. Evaluation of Antibacterial Activity
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Global Research Agenda for Antimicrobial Resistance in Human Health. Available online: https://www.who.int/publications/m/item/global-research-agenda-for-antimicrobial-resistance-in-human-health (accessed on 11 October 2023).
- Walsh, T.R.; Gales, A.C.; Laxminarayan, R.; Dodd, P.C. Antimicrobial Resistance: Addressing a Global Threat to Humanity. PLoS Med. 2023, 20, e1004264. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Frieden, T. Antibiotic Resistance Threats in the United States; United States Department of Health and Human Services: Washington, DC, USA, 2013.
- Kongnakorn, T.; Tichy, E.; Kengkla, K.; Kanokwanvimol, N.; Suthipinijtham, P.; Phuripakathorn, C.; Al Taie, A. Economic Burden of Antimicrobial Resistance and Inappropriate Empiric Treatment in Thailand. Antimicrob. Steward. Healthc. Epidemiol. 2023, 3, E109. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance: Global Report on Surveillance. Available online: https://www.who.int/publications/i/item/9789241564748 (accessed on 15 October 2023).
- CDC. The Biggest Antibiotic-Resistant Threats in the U.S. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 11 October 2023).
- World Bank. Drug-Resistant Infections; World Bank: Washington, DC, USA, 2017. [Google Scholar]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.R. Bad Bugs, No Drugs: No ESCAPE Revisited. Clin. Infect. Dis. 2009, 49, 992–993. [Google Scholar] [CrossRef]
- Global Action Plan on Antimicrobial Resistance. Microbe Wash. DC 2015, 10, 354–355.
- Campanini-Salinas, J.; Andrades-Lagos, J.; Mella-Raipan, J.; Vasquez-Velasquez, D. Novel Classes of Antibacterial Drugs in Clinical Development, a Hope in a Post-Antibiotic Era. Curr. Top. Med. Chem. 2018, 18, 1188–1202. [Google Scholar] [CrossRef]
- Andrei, S.; Valeanu, L.; Chirvasuta, R.; Stefan, M.-G. New FDA Approved Antibacterial Drugs: 2015–2017. Discoveries 2018, 6, e81. [Google Scholar] [CrossRef]
- Piddock, L.J.V. The Crisis of No New Antibiotics—What Is the Way Forward? Lancet Infect. Dis. 2012, 12, 249–253. [Google Scholar] [CrossRef]
- WHO. Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 17 October 2023).
- Vuong, C.; Yeh, A.J.; Cheung, G.Y.C.; Otto, M. Investigational Drugs to Treat Methicillin-Resistant Staphylococcus aureus. Expert Opin. Investig. Drugs 2016, 25, 73–93. [Google Scholar] [CrossRef]
- Klein, E.; Smith, D.L.; Laxminarayan, R. Hospitalizations and Deaths Caused by Methicillin-Resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 2007, 13, 1840–1846. [Google Scholar] [CrossRef]
- Li, Z.; Zhuang, H.; Wang, G.; Wang, H.; Dong, Y. Prevalence, Predictors, and Mortality of Bloodstream Infections Due to Methicillin-Resistant Staphylococcus aureus in Patients with Malignancy: Systemic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 74. [Google Scholar] [CrossRef]
- David, L.; Brata, A.M.; Mogosan, C.; Pop, C.; Czako, Z.; Muresan, L.; Ismaiel, A.; Dumitrascu, D.I.; Leucuta, D.C.; Stanculete, M.F.; et al. Artificial Intelligence and Antibiotic Discovery. Antibiotics 2021, 10, 1376. [Google Scholar] [CrossRef]
- Fourches, D. Reaction: Molecular Modeling for Novel Antibacterials. Chem 2017, 3, 13–14. [Google Scholar] [CrossRef]
- Campanini-Salinas, J.; Andrades-Lagos, J.; Gonzalez Rocha, G.; Choquesillo-Lazarte, D.; Bollo Dragnic, S.; Faúndez, M.; Alarcón, P.; Silva, F.; Vidal, R.; Salas-Huenuleo, E.; et al. A New Kind of Quinonic-Antibiotic Useful against Multidrug-Resistant S. Aureus and E. faecium Infections. Molecules 2018, 23, 1776. [Google Scholar] [CrossRef] [PubMed]
- Campanini-Salinas, J.; Andrades-Lagos, J.; Hinojosa, N.; Moreno, F.; Alarcón, P.; González-Rocha, G.; Burbulis, I.E.; Vásquez-Velásquez, D. New Quinone Antibiotics against Methicillin-Resistant S. aureus. Antibiotics 2021, 10, 614. [Google Scholar] [CrossRef] [PubMed]
- Andrades-Lagos, J.; Campanini-Salinas, J.; Pedreros-Riquelme, A.; Mella, J.; Choquesillo-Lazarte, D.; Zamora, P.P.; Pessoa-Mahana, H.; Burbulis, I.; Vásquez-Velásquez, D. Design, Synthesis, and Structure–Activity Relationship Studies of New Quinone Derivatives as Antibacterial Agents. Antibiotics 2023, 12, 1065. [Google Scholar] [CrossRef]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W., Jr. Computational Methods in Drug Discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative Molecular Field Analysis (CoMFA). 1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Klebe, G.; Abraham, U.; Mietzner, T. Molecular Similarity Indices in a Comparative Analysis (CoMSIA) of Drug Molecules to Correlate and Predict Their Biological Activity. J. Med. Chem. 1994, 37, 4130–4146. [Google Scholar] [CrossRef]
- Wendt, B.; Cramer, R.D. Challenging the Gold Standard for 3D-QSAR: Template CoMFA versus X-Ray Alignment. J. Comput. Aided Mol. Des. 2014, 28, 803–824. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.-S.; Huang, R.-B.; Wei, Y.-T.; Du, L.-Q.; Chou, K.-C. Multiple Field Three Dimensional Quantitative Structure–Activity Relationship (MF-3D-QSAR). J. Comput. Chem. 2008, 29, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-L.; Wang, F.; Shi, X.-X.; Jia, C.-Y.; Wu, F.-X.; Hao, G.-F.; Yang, G.-F. Cloud 3D-QSAR: A Web Tool for the Development of Quantitative Structure–Activity Relationship Models in Drug Discovery. Brief. Bioinform. 2021, 22, bbaa276. [Google Scholar] [CrossRef] [PubMed]
- Daré, J.K.; Freitas, M.P. Is Conformation Relevant for QSAR Purposes? 2D Chemical Representation in a 3D-QSAR Perspective. J. Comput. Chem. 2022, 43, 917–922. [Google Scholar] [CrossRef]
- Cronin, M.T.D.; Schultz, T.W. Pitfalls in QSAR. Theochem 2003, 622, 39–51. [Google Scholar] [CrossRef]
- Walesch, S.; Birkelbach, J.; Jézéquel, G.; Haeckl, F.P.J.; Hegemann, J.D.; Hesterkamp, T.; Hirsch, A.K.H.; Hammann, P.; Müller, R. Fighting Antibiotic Resistance—Strategies and (Pre)Clinical Developments to Find New Antibacterials. EMBO Rep. 2023, 24, e56033. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Valderrama, J.A.; Leiva, H.; Rodríguez, J.A.; Theoduloz, C.; Schmeda-Hirshmann, G. Studies on Quinones. Part 43: Synthesis and Cytotoxic Evaluation of Polyoxyethylene-Containing 1,4-Naphthoquinones☆. Bioorg. Med. Chem. 2008, 16, 3687–3693. [Google Scholar] [CrossRef]
- Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Tenth Edition. Available online: https://clsi.org/media/1632/m07a10_sample.pdf (accessed on 15 October 2023).
- Cabezas, D.; Mellado, G.; Espinoza, N.; Gárate, J.A.; Morales, C.; Castro-Alvarez, A.; Matos, M.J.; Mellado, M.; Mella, J. In Silico Approaches to Develop New Phenyl-Pyrimidines as Glycogen Synthase Kinase 3 (GSK-3) Inhibitors with Halogen-Bonding Capabilities: 3D-QSAR CoMFA/CoMSIA, Molecular Docking and Molecular Dynamics Studies. J. Biomol. Struct. Dyn. 2023, 1–10. [Google Scholar] [CrossRef] [PubMed]
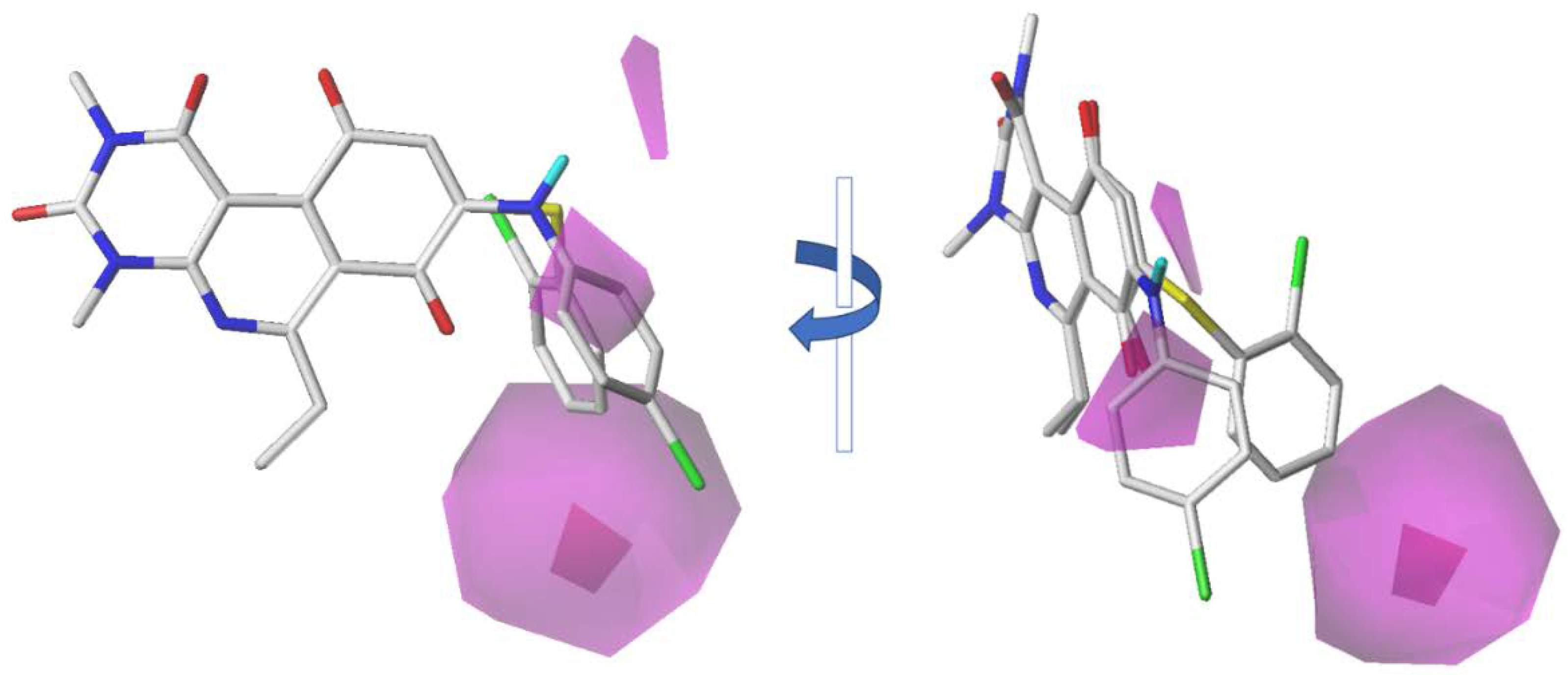
| Model | N | q2 | r2 | SEE | F | Contribution | ||
|---|---|---|---|---|---|---|---|---|
| Steric | Electrostatic | Acceptor | ||||||
| CoMFA | 5 | 0.660 | 0.938 | 0.286 | 48.017 | 1 | ||
| CoMSIA | 5 | 0.596 | 0.895 | 0.312 | 27.276 | 0.269 | 0.504 | 0.227 |
| CoMFA | CoMSIA | |||
|---|---|---|---|---|
| Random | q2 | N | q2 | N |
| 1 | −0.166 | 1 | −0.08 | 1 |
| 2 | −0.193 | 1 | 0.095 | 1 |
| 3 | −0.287 | 1 | −0.696 | 1 |
| 4 | −0.398 | 1 | −0.047 | 1 |
| 5 | −0.219 | 1 | −0.234 | 1 |
| 6 | −0.017 | 9 | −0.339 | 1 |
| 7 | −0.246 | 1 | −0.142 | 3 |
| 8 | −0.199 | 1 | 0.108 | 1 |
| 9 | −0.12 | 1 | −0.319 | 1 |
| 10 | −0.088 | 1 | 0.041 | 4 |
| Average | −0.193 | −0.161 | ||
| Molecule | Exp. pMIC | CoMFA | CoMSIA | ||
|---|---|---|---|---|---|
| Pred. pMIC | Residual | Pred. pMIC | Residual | ||
| 1 | 4.707 | 4.926 | −0.22 | 4.909 | −0.20 |
| 2 * | 4.120 | 5.412 | −1.29 | 5.001 | −0.88 |
| 3 | 5.340 | 5.388 | −0.05 | 5.478 | −0.14 |
| 4 | 5.687 | 5.692 | −0.01 | 5.367 | 0.32 |
| 5 | 5.023 | 4.934 | 0.09 | 4.887 | 0.14 |
| 6 | 5.039 | 5.098 | −0.06 | 5.122 | −0.08 |
| 7 | 5.027 | 4.950 | 0.08 | 5.207 | −0.18 |
| 8 | 5.344 | 5.199 | 0.15 | 5.244 | 0.10 |
| 9 | 5.386 | 5.306 | 0.08 | 5.175 | 0.21 |
| 10 * | 5.023 | 4.588 | 0.43 | 4.904 | 0.12 |
| 11 | 4.437 | 4.425 | 0.01 | 4.608 | −0.17 |
| 12 * | 4.726 | 4.843 | −0.12 | 4.890 | −0.16 |
| 13 * | 5.043 | 4.585 | 0.46 | 4.898 | 0.15 |
| 14 * | 5.085 | 4.604 | 0.48 | 4.903 | 0.18 |
| 15 | 4.463 | 4.384 | 0.08 | 4.514 | −0.05 |
| 16 | 4.423 | 4.604 | −0.18 | 4.293 | 0.13 |
| 17 | 4.733 | 4.515 | 0.22 | 4.657 | 0.08 |
| 18 | 4.451 | 4.492 | −0.04 | 4.491 | −0.04 |
| 19 * | 4.753 | 4.915 | −0.16 | 4.937 | −0.18 |
| 20 | 4.751 | 4.819 | −0.07 | 4.809 | −0.06 |
| 21 | 5.037 | 5.053 | −0.02 | 5.017 | 0.02 |
| 22 * | 5.075 | 4.660 | 0.42 | 5.296 | −0.22 |
| 23 | 5.628 | 5.774 | −0.15 | 5.847 | −0.22 |
| 24 | 5.973 | 5.838 | 0.13 | 5.834 | 0.14 |
| 25 | 4.970 | 4.968 | 0.00 | 4.984 | −0.01 |
| 26 | 4.986 | 4.970 | 0.02 | 4.945 | 0.04 |
| 27 | 5.115 | 5.072 | 0.04 | 5.130 | −0.02 |
| 28 * | 5.369 | 5.969 | −0.60 | 5.648 | −0.28 |
| 29 * | 5.076 | 5.160 | −0.08 | 5.512 | −0.44 |
| 30 | 4.692 | 4.798 | −0.11 | 4.709 | −0.02 |
| 31 * | 4.724 | 4.301 | 0.42 | 4.392 | 0.33 |
| 32 | 4.711 | 4.714 | 0.00 | 4.692 | 0.02 |
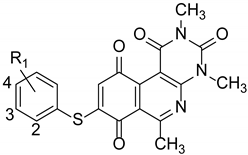 | ||||||
|---|---|---|---|---|---|---|
| Compounds | MIC (µg/mL) | |||||
| Label | R1 | MRSA (ATCC 43300) | MSSA (ATCC 29213) | E. faecalis (ATCC 29212) | E. coli (ATCC 25922) | P. aeruginosa (ATCC 27853) |
| 33 | 4-Cl | 2 | 2 | 4 | >32 | >32 |
| 34 | 4-Br | 2 | 2 | 4 | >32 | >32 |
| 35 | 4-Me | 2 | 4 | 4 | >32 | >32 |
| 36 | 3-Cl | 4 | 4 | 8 | >32 | >32 |
| 37 | 3-Br | 4 | 4 | 4 | >32 | >32 |
| 38 | 3-F | 4 | 4 | 4 | >32 | >32 |
| 39 | 3-OMe | 4 | 8 | 8 | >32 | >32 |
| 40 | 3-Me | 4 | 4 | 4 | >32 | >32 |
| 41 | 2-Cl | 32 | 32 | >32 | >32 | >32 |
| 42 | 2-Br | 2 | 4 | 4 | >32 | >32 |
| 43 | 2-OMe | 4 | 8 | 8 | >32 | >32 |
| 44 | 2-Me | 4 | 4 | 4 | >32 | >32 |
| 45 | 2-F | >32 | >32 | >32 | >32 | >32 |
| VAN | - | 1 | 1 | 2 | NT | NT |
| GEN | - | NT | NT | NT | 0.5 | 1 |
| Experimental pMIC | Predicted pMIC | ||||
|---|---|---|---|---|---|
| No. | CoMFA | Residual | CoMSIA | Residual | |
| 33 | 5.3303 | 4.456 | 0.87 | 4.699 | 0.63 |
| 34 | 5.3732 | 4.472 | 0.90 | 4.704 | 0.67 |
| 35 | 5.3090 | 4.472 | 0.84 | 4.704 | 0.61 |
| 36 | 5.0292 | 5.067 | −0.04 | 5.046 | −0.02 |
| 37 | 5.0722 | 5.155 | −0.08 | 4.974 | 0.10 |
| 38 | 5.0122 | 4.819 | 0.19 | 5.006 | 0.01 |
| 39 | 5.0247 | 4.970 | 0.05 | 4.923 | 0.10 |
| 40 | 5.0080 | 4.814 | 0.19 | 4.730 | 0.28 |
| 41 | 4.1262 | 5.397 | −1.27 | 5.280 | −1.15 |
| 42 | 5.3732 | 5.553 | −0.18 | 5.168 | 0.21 |
| 43 | 5.0247 | 5.250 | −0.23 | 5.280 | −0.26 |
| 44 | 5.0080 | 5.591 | −0.58 | 4.720 | 0.29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrades-Lagos, J.; Campanini-Salinas, J.; Sabadini, G.; Andrade, V.; Mella, J.; Vásquez-Velásquez, D. QSAR Studies, Synthesis, and Biological Evaluation of New Pyrimido-Isoquinolin-Quinone Derivatives against Methicillin-Resistant Staphylococcus aureus. Pharmaceuticals 2023, 16, 1621. https://doi.org/10.3390/ph16111621
Andrades-Lagos J, Campanini-Salinas J, Sabadini G, Andrade V, Mella J, Vásquez-Velásquez D. QSAR Studies, Synthesis, and Biological Evaluation of New Pyrimido-Isoquinolin-Quinone Derivatives against Methicillin-Resistant Staphylococcus aureus. Pharmaceuticals. 2023; 16(11):1621. https://doi.org/10.3390/ph16111621
Chicago/Turabian StyleAndrades-Lagos, Juan, Javier Campanini-Salinas, Gianfranco Sabadini, Victor Andrade, Jaime Mella, and David Vásquez-Velásquez. 2023. "QSAR Studies, Synthesis, and Biological Evaluation of New Pyrimido-Isoquinolin-Quinone Derivatives against Methicillin-Resistant Staphylococcus aureus" Pharmaceuticals 16, no. 11: 1621. https://doi.org/10.3390/ph16111621
APA StyleAndrades-Lagos, J., Campanini-Salinas, J., Sabadini, G., Andrade, V., Mella, J., & Vásquez-Velásquez, D. (2023). QSAR Studies, Synthesis, and Biological Evaluation of New Pyrimido-Isoquinolin-Quinone Derivatives against Methicillin-Resistant Staphylococcus aureus. Pharmaceuticals, 16(11), 1621. https://doi.org/10.3390/ph16111621