Structural Characterization of TRAF6 N-Terminal for Therapeutic Uses and Computational Studies on New Derivatives
Abstract
:1. Introduction
2. Results
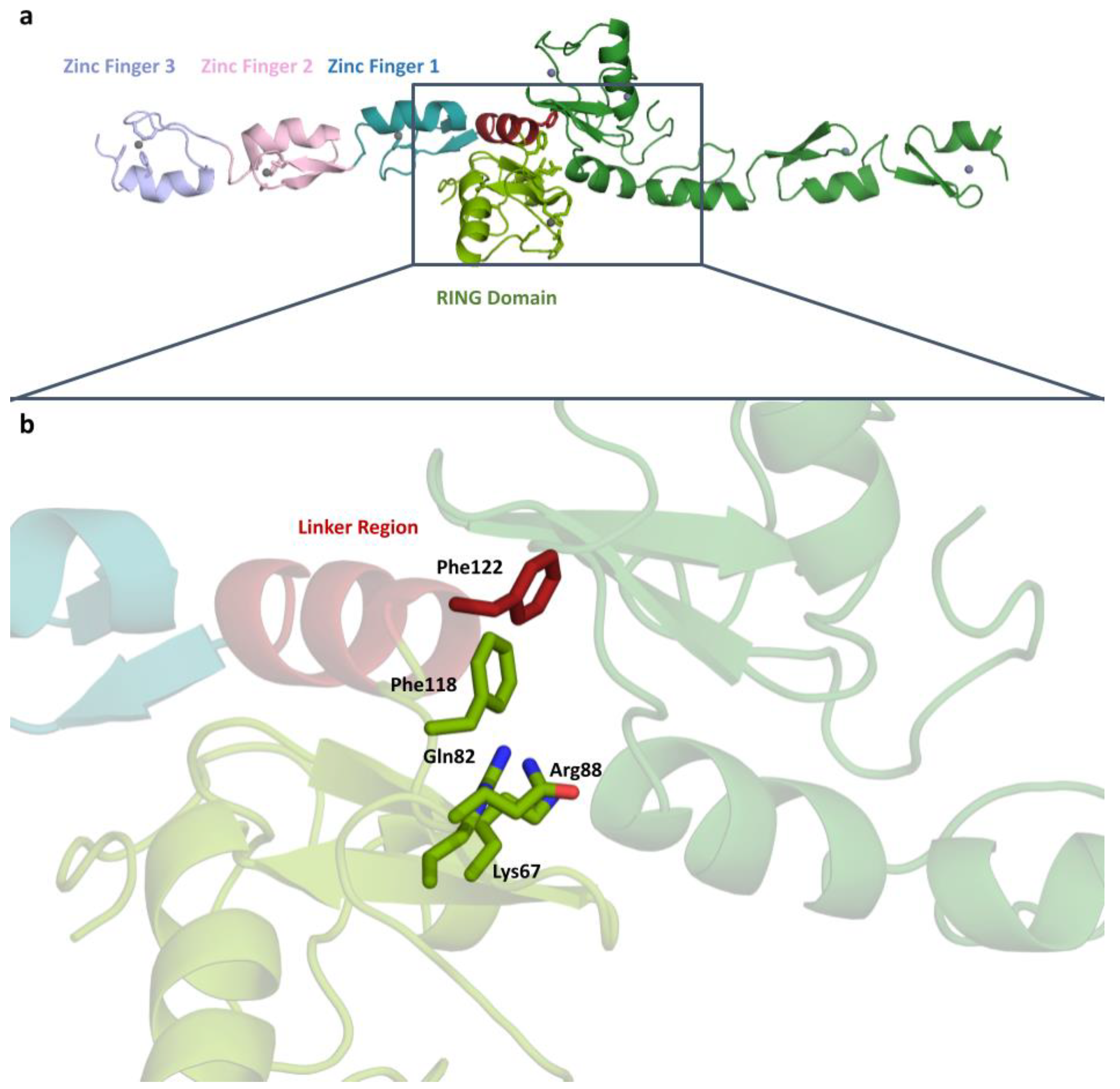
2.1. TRAF6 N-Terminal Structure at 3.2 Å Resolution
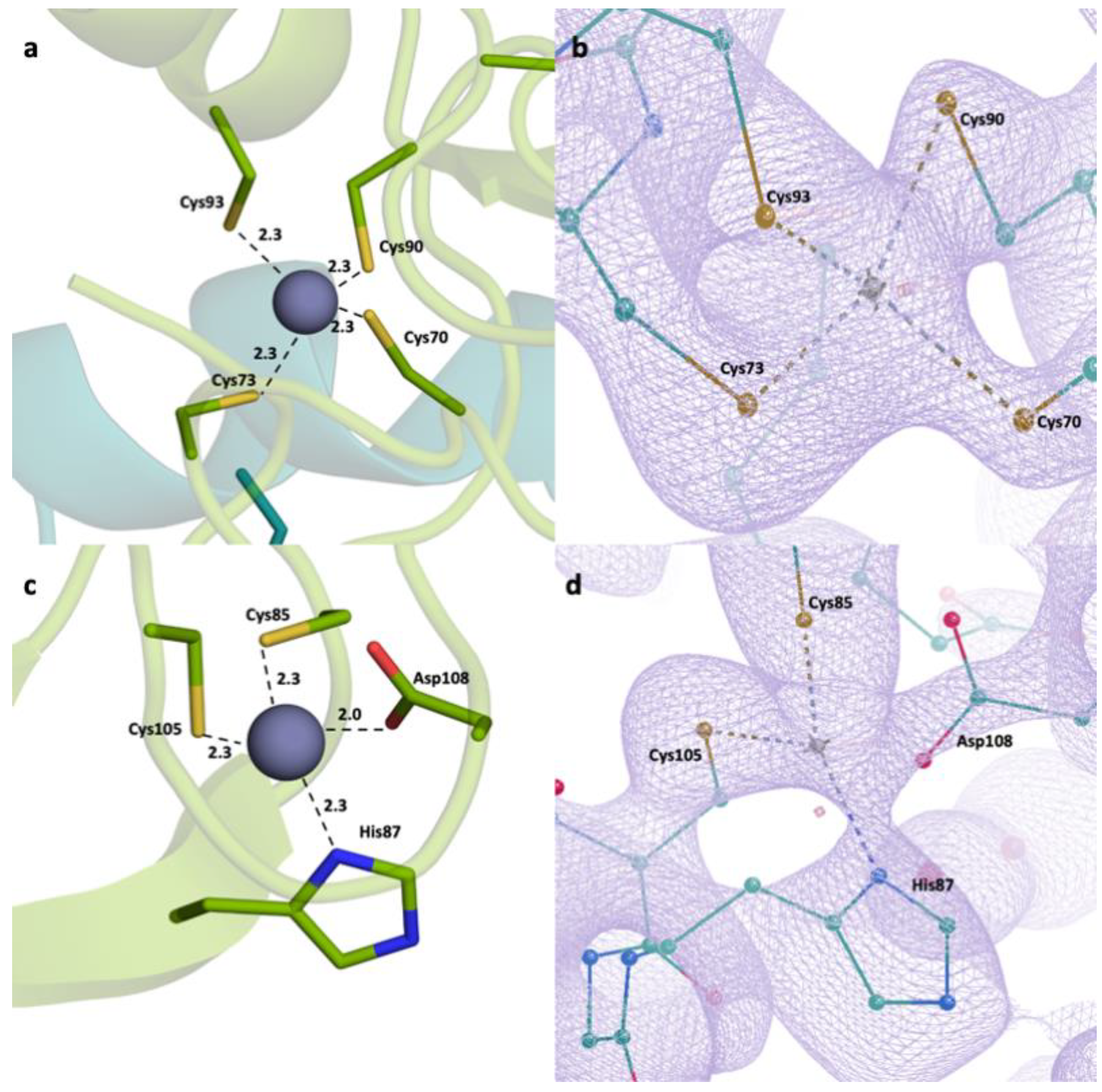
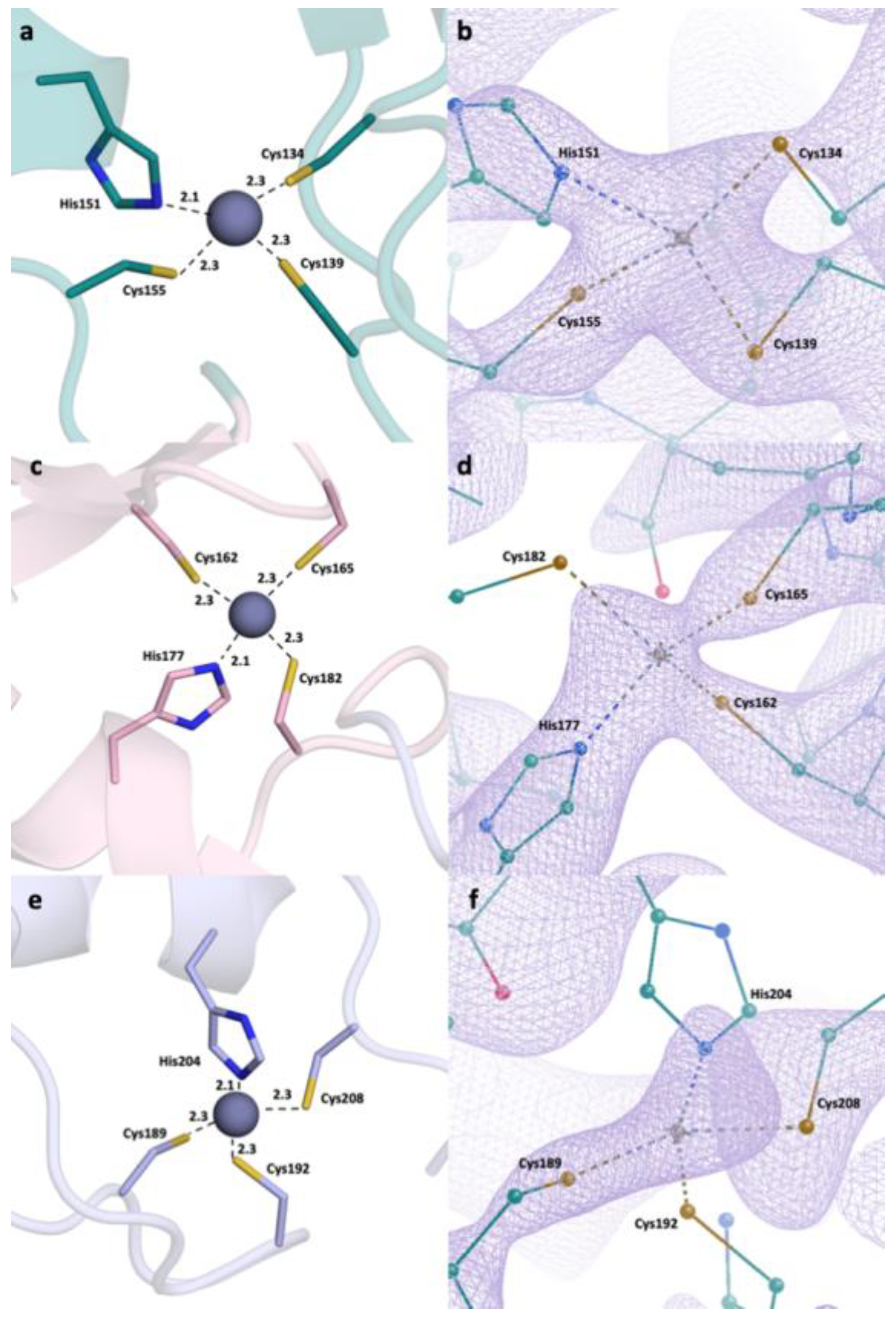
2.2. Detailed Analysis of RING Domain and Zinc Fingers
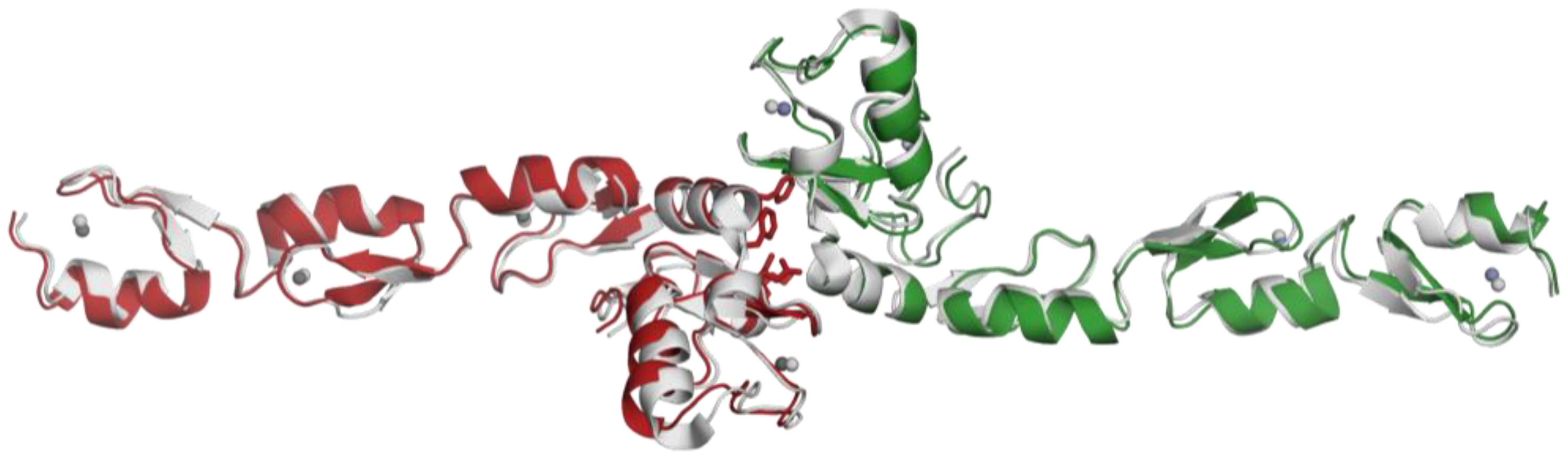
2.3. Structural Alignment with the Reference Protein
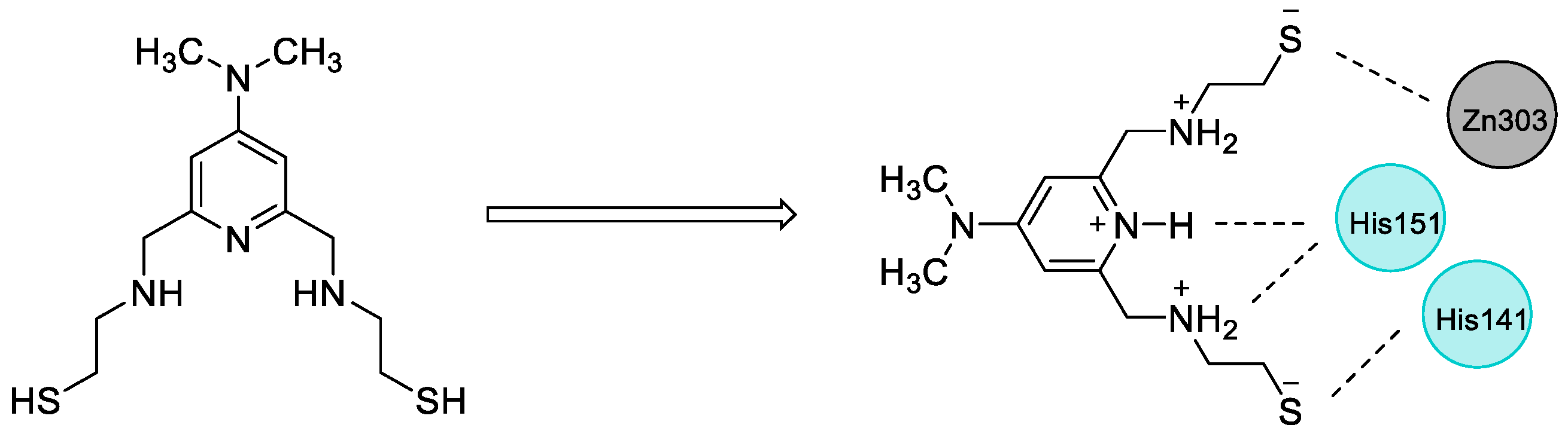
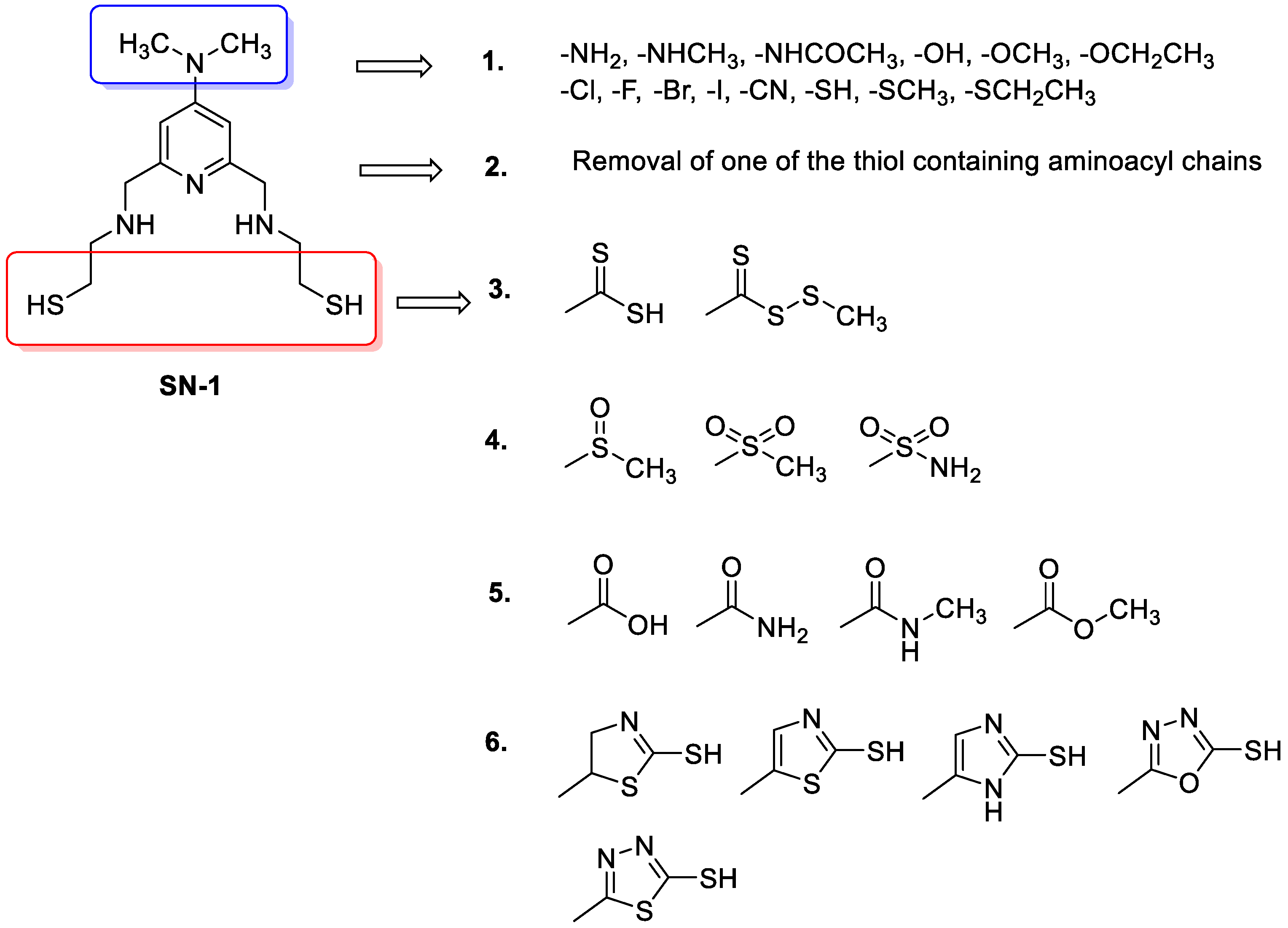
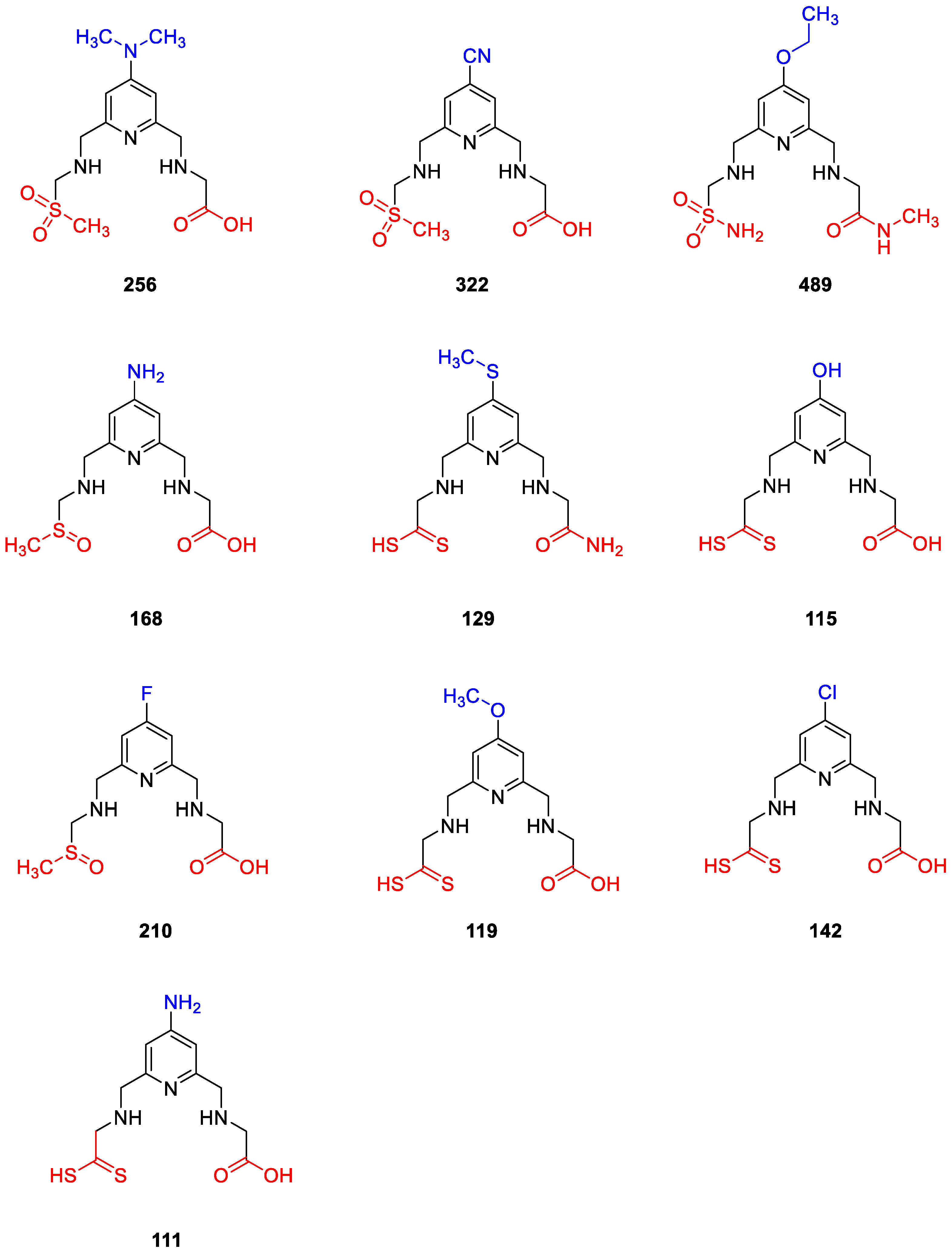
2.4. Design of New Compounds
- Replacement of dimethylamino groups with amine, methylamine, acetamido, hydroxyl, methoxy, ethoxy, fluoro, chloro, bromo, iodo, cyano, thiol, thiomethyl, thioethyl groups;
- Removal of one of the thiols containing aminoacyl chains;
- Replacement of thiol group to methyl/ethyl (dithioperoxo)thioate, dithiocarbamates in aminoacyl chains;
- Replacement of thiol group to methylsulfinyl, methylsulfonyl, and sulfonamide groups in aminoacyl chains;
- Replacement of thiol group to amide, carboxylic acid, and ester groups in aminoacyl chains;
- Replacement of thiol group to thiol-substituted thiazoline, thiazole, imidazole, oxadiazole, thiadiazole, and pyridine rings in aminoacyl chains.
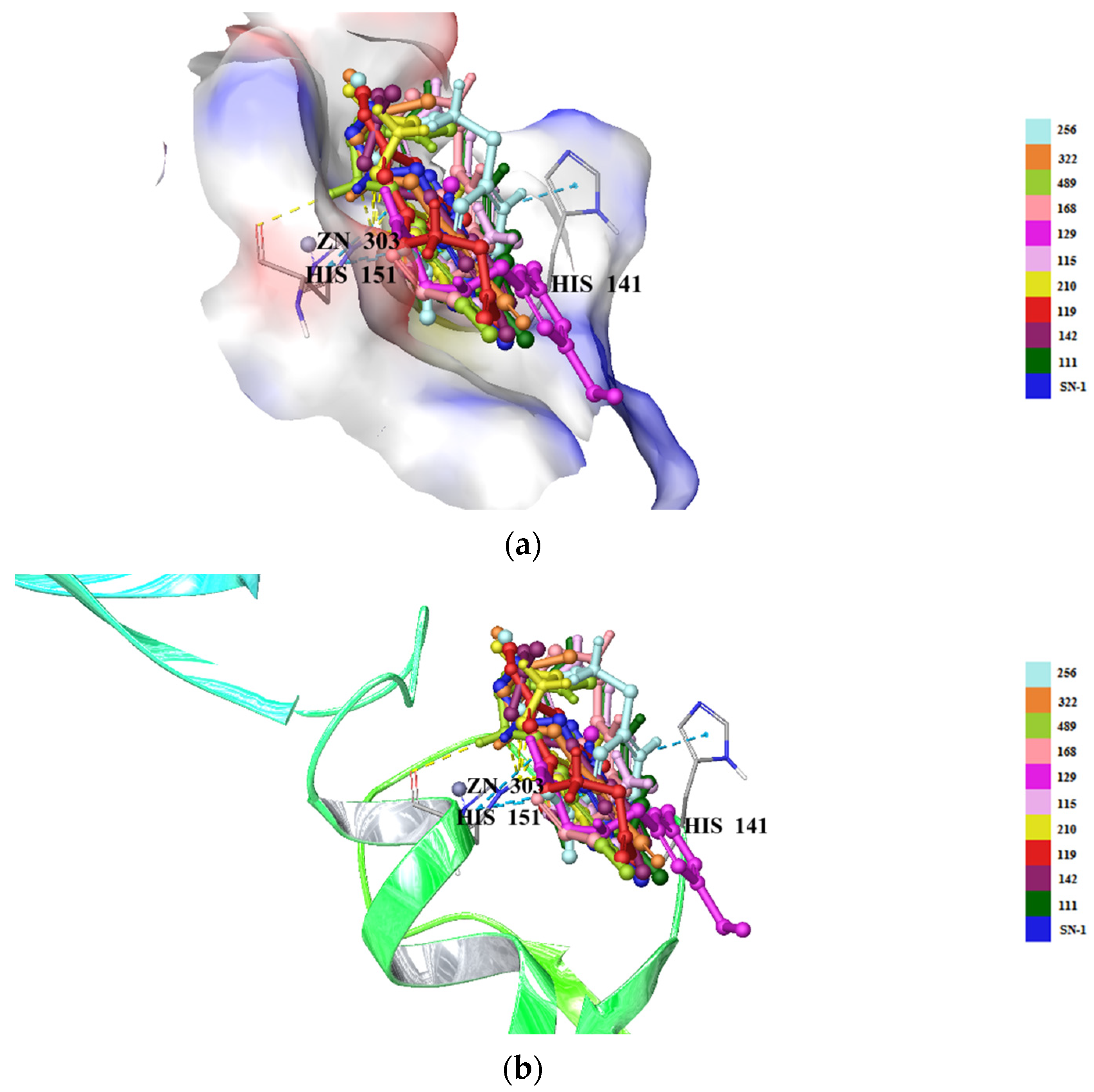
2.5. Molecular Docking Studies for New Compounds
2.6. MD Simulations
2.7. ADME Prediction of New Compounds
3. Discussion
4. Materials and Methods
4.1. Transformation and Expression
4.2. Purification
4.3. Crystallization
4.4. Crystal Harvesting and Delivery
4.5. Data Collection and Data Reduction
4.6. Structure Determination and Refinement
4.7. Molecular Docking Studies
4.8. MD Simulations
4.9. In Silico ADME Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Wang, Y.C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 53. [Google Scholar] [CrossRef]
- Gibson, T.J.; Postma, J.P.; Brown, R.S.; Argos, P. A model for the tertiary structure of the 28 residue DNA-binding motif (‘zinc finger’) common to many eukaryotic transcriptional regulatory proteins. Protein Eng. 1988, 2, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Lv, Y.; Liu, Y.; Qiao, S.; Wang, H. Zinc Finger Proteins in Neuro-Related Diseases Progression. Front. Neurosci. 2021, 15, 760567. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Hao, Y.; Song, Z.; Fan, Y.; Li, S. TRIM37 negatively regulates inflammatory responses induced by virus infection via controlling TRAF6 ubiquitination. Biochem. Biophys. Res. Commun. 2021, 556, 87–92. [Google Scholar] [CrossRef]
- Li, J.; Liu, N.; Tang, L.; Yan, B.; Chen, X.; Zhang, J.; Peng, C. The relationship between TRAF6 and tumors. Cancer Cell Int. 2020, 20, 429. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Li, P.T.; Luo, Z.H.; Zhang, Z.P.; Wang, Y.L.; Zou, P.F. Novel Findings in Teleost TRAF4, a Protein Acts as an Enhancer in TRIF and TRAF6 Mediated Antiviral and Inflammatory Signaling. Front. Immunol. 2022, 13, 944528. [Google Scholar] [CrossRef]
- Lamothe, B.; Campos, A.D.; Webster, W.K.; Gopinathan, A.; Hur, L.; Darnay, B.G. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J. Biol. Chem. 2008, 283, 24871–24880. [Google Scholar] [CrossRef]
- He, X.; Li, Y.; Li, C.; Liu, L.J.; Zhang, X.D.; Liu, Y.; Shu, H.B. USP2a negatively regulates IL-1β- and virus-induced NF-κB activation by deubiquitinating TRAF6. J. Mol. Cell Biol. 2013, 5, 39–47. [Google Scholar] [CrossRef]
- Lalani, A.I.; Zhu, S.; Gokhale, S.; Jin, J.; Xie, P. TRAF molecules in inflammation and inflammatory diseases. Curr. Pharmacol. Rep. 2018, 4, 64–90. [Google Scholar] [CrossRef]
- Bradley, J.R.; Pober, J.S. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 2001, 20, 6482–6491. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.H.; Wan, D.H.; Gu, Z.H.; Deng, X.X.; Weng, S.P.; Yu, X.Q.; He, J.G. Litopenaeus vannamei tumor necrosis factor receptor-associated factor 6 (TRAF6) responds to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection and activates antimicrobial peptide genes. Dev. Comp. Immunol. 2011, 35, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.C.; Lee, J.; Choi, Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015, 266, 72–92. [Google Scholar] [CrossRef]
- Inoue, J.I.; Ishida, T.; Tsukamoto, N.; Kobayashi, N.; Naito, A.; Azuma, S.; Yamamoto, T. Tumor necrosis factor receptor-associated factor (TRAF) family: Adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000, 254, 14–24. [Google Scholar] [CrossRef]
- Yamamoto, M.; Gohda, J.; Akiyama, T.; Inoue, J.I. TNF receptor-associated factor 6 (TRAF6) plays crucial roles in multiple biological systems through polyubiquitination-mediated NF-κB activation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2021, 97, 145–160. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Signaling to NF-kappaB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef]
- Park, M.H.; Hong, J.T. Roles of NF-κB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, A.; Rahmani, F.; Ferns, G.A.; Ryzhikov, M.; Avan, A.; Hassanian, S.M. Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene 2020, 726, 144132. [Google Scholar] [CrossRef]
- Middleton, A.J.; Budhidarmo, R.; Das, A.; Zhu, J.; Foglizzo, M.; Mace, P.D.; Day, C.L. The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1. Nat. Commun. 2017, 8, 1788. [Google Scholar] [CrossRef]
- Qi, Y.; Pradipta, A.R.; Li, M.; Zhao, X.; Lu, L.; Fu, X.; Wei, J.; Hsung, R.P.; Tanaka, K.; Zhou, L. Cinchonine induces apoptosis of HeLa and A549 cells through targeting TRAF6. J. Exp. Clin. Cancer Res. 2017, 36, 35. [Google Scholar] [CrossRef]
- Khusbu, F.Y.; Zhou, X.; Roy, M.; Chen, F.Z.; Cao, Q.; Chen, H.C. Resveratrol induces depletion of TRAF6 and suppresses prostate cancer cell proliferation and migration. Int. J. Biochem. Cell Biol. 2020, 118, 105644. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Luo, L.; Wei, J.; Liu, Y.; Haque, N.; Huang, H.; Qi, Y.; Huang, Z. Identification of a new TRAF6 inhibitor for the treatment of hepatocellular carcinoma. Int. J. Biol. Macromol. 2021, 182, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Guangwei, Z.; Zhibin, C.; Qin, W.; Chunlin, L.; Penghang, L.; Ruofan, H.; Hui, C.; Hoffman, R.M.; Jianxin, Y. TRAF6 regulates the signaling pathway influencing colorectal cancer function through ubiquitination mechanisms. Cancer Sci. 2022, 113, 1393–1405. [Google Scholar] [CrossRef]
- Zhao, X.; Ren, L.; Wang, X.; Han, G.; Wang, S.; Yao, Q.; Qi, Y. Benzoyl-xanthone derivative induces apoptosis in MCF-7 cells by binding TRAF6. Exp. Ther. Med. 2022, 23, 181. [Google Scholar] [CrossRef]
- Bai, S.; Zha, J.; Zhao, H.; Ross, F.P.; Teitelbaum, S.L. Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J. Biol. Chem. 2008, 283, 30861–30867. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Li, J.W.; Qin, Y.H.; Zhai, H.; Feng, B.; Li, H.; Zhang, N.N.; Yang, C.S. Expression of TRAF6 in peripheral blood B cells of patients with myasthenia gravis. BMC Neurol. 2022, 22, 302. [Google Scholar] [CrossRef]
- Semmler, S.; Gagné, M.; Garg, P.; Pickles, S.R.; Baudouin, C.; Hamon-Keromen, E.; Destroismaisons, L.; Khalfallah, Y.; Chaineau, M.; Caron, E.; et al. TNF receptor-associated factor 6 interacts with ALS-linked misfolded superoxide dismutase 1 and promotes aggregation. J. Biol. Chem. 2020, 295, 3808–3825. [Google Scholar] [CrossRef]
- Huang, H.; Xia, A.; Sun, L.; Lu, C.; Liu, Y.; Zhu, Z.; Wang, S.; Cai, J.; Zhou, X.; Liu, S. Pathogenic Functions of Tumor Necrosis Factor Receptor-Associated Factor 6 Signaling Following Traumatic Brain Injury. Front. Mol. Neurosci. 2021, 14, 629910. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cao, D.L.; Ma, L.J.; Gao, Y.J. TRAF6 Contributes to CFA-Induced Spinal Microglial Activation and Chronic Inflammatory Pain in Mice. Cell Mol. Neurobiol. 2022, 42, 1543–1555. [Google Scholar] [CrossRef]
- Masperone, L.; Codrich, M.; Persichetti, F.; Gustincich, S.; Zucchelli, S.; Legname, G. The E3 Ubiquitin Ligase TRAF6 Interacts with the Cellular Prion Protein and Modulates Its Solubility and Recruitment to Cytoplasmic p62/SQSTM1-Positive Aggresome-Like Structures. Mol. Neurobiol. 2022, 59, 1577–1588. [Google Scholar] [CrossRef]
- Koga, R.; Radwan, M.O.; Ejima, T.; Kanemaru, Y.; Tateishi, H.; Ali, T.F.S.; Ciftci, H.I.; Shibata, Y.; Taguchi, Y.; Inoue, J.I.; et al. A Dithiol Compound Binds to the Zinc Finger Protein TRAF6 and Suppresses Its Ubiquitination. ChemMedChem 2017, 12, 1935–1941. [Google Scholar] [CrossRef]
- Radwan, M.O.; Koga, R.; Hida, T.; Ejima, T.; Kanemaru, Y.; Tateishi, H.; Okamoto, Y.; Inoue, J.I.; Fujita, M.; Otsuka, M. Minimum structural requirements for inhibitors of the zinc finger protein TRAF6. Bioorg. Med. Chem. Lett. 2019, 29, 2162–2167. [Google Scholar] [CrossRef]
- Guven, O.; Ciftci, H.; DeMirci, H. Tumor Necrosis Factor Receptor Associated Factor 6 (TRAF6) N-terminal Domain. PDB Entry—8HZ2. 2023. Available online: https://doi.org/10.2210/pdb8HZ2/pdb (accessed on 8 November 2023).
- Gul, M.; Ayan, E.; Destan, E.; Johnson, J.A.; Shafiei, A.; Kepceoglu, A.; Yilmaz, M.; Ertem, F.B.; Yapici, I.; Tosun, B.; et al. Rapid and efficient ambient temperature X-ray crystal structure determination at Turkish Light Source. Sci. Rep. 2023, 13, 8123. [Google Scholar] [CrossRef]
- Yin, Q.; Lin, S.C.; Lamothe, B.; Lu, M.; Lo, Y.C.; Hura, G.; Zheng, L.; Rich, R.L.; Campos, A.D.; Myszka, D.G.; et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 2009, 16, 658–666. [Google Scholar] [CrossRef]
- Otsuka, M.; Fujita, M.; Aoki, T.; Ishii, S.; Sugiura, Y.; Yamamoto, T.; Inoue, J. Novel zinc chelators with dual activity in the inhibition of the kappa B site-binding proteins HIV-EP1 and NF-kappa. Br. J. Med. Chem. 1995, 38, 3264–3270. [Google Scholar] [CrossRef]
- Otsuka, M.; Fujita, M.; Sugiura, Y.; Yamamoto, T.; Inoue, J.; Maekawa, T.; Ishii, S. Synthetic inhibitors of regulatory proteins involved in the signaling pathway of the replication of human immunodeficiency virus 1. Bioorg. Med. Chem. 1997, 5, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Otsuka, M.; Sugiura, Y. Metal-chelating inhibitors of a zinc finger protein HIV-EP1. Remarkable potentiation of inhibitory activity by introduction of SH groups. J. Med. Chem. 1996, 39, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Ejima, T.; Hirota, M.; Mizukami, T.; Otsuka, M.; Fujita, M. An anti-HIV-1 compound that increases steady-state expression of apoplipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G. Int. J. Mol. Med. 2011, 28, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Radwan, M.O.; Hamasaki, A.; Ejima, A.; Obata, E.; Koga, R.; Tateishi, H.; Okamoto, Y.; Fujita, M.; Nakao, M.; et al. A novel inhibitor of farnesyltransferase with a zinc site recognition moiety and a farnesyl group. Bioorg. Med. Chem. Lett. 2017, 27, 3862–3866. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, H.; Tateishi, M.; Radwan, M.O.; Masunaga, T.; Kawatashiro, K.; Oba, Y.; Oyama, M.; Inoue-Kitahashi, N.; Fujita, M.; Okamoto, Y.; et al. A new inhibitor of ADAM17 composed of a zinc-binding dithiol moiety and a specificity pocket-binding appendage. Chem. Pharm. Bull. 2021, 69, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Ece, A. Computer-aided drug design. BMC Chem. 2023, 17, 26. [Google Scholar] [CrossRef]
- Güleç, Ö.; Türkeş, C.; Arslan, M.; Demir, Y.; Dincer, B.; Ece, A.; Beydemir, Ş. Novel beta-lactam substituted benzenesulfonamides: In vitro enzyme inhibition, cytotoxic activity and in silico interactions. J. Biomol. Struct. Dyn. 2023, 1–19. [Google Scholar] [CrossRef]
- Çelik Onar, H.; Özden, E.M.; Taslak, H.D.; Gülçin, İ.; Ece, A.; Erçağ, E. Novel coumarin-chalcone derivatives: Synthesis, characterization, antioxidant, cyclic voltammetry, molecular modelling and biological evaluation studies as acetylcholinesterase, α-glycosidase, and carbonic anhydrase inhibitors. Chem. Biol. Interact. 2023, 383, 110655. [Google Scholar] [CrossRef]
- Schrödinger Release 2016-2: QikProp, Schrödinger, LLC.: New York, NY, USA, 2016.
- SwissADME. Available online: http://www.swissadme.ch (accessed on 30 August 2023).
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Dou, Y.; Tian, X.; Zhang, J.; Wang, Z.; Chen, G. Roles of TRAF6 in Central Nervous System. Curr. Neuropharmacol. 2018, 16, 1306–1313. [Google Scholar] [CrossRef]
- Min, Y.; Kim, M.J.; Lee, S.; Chun, E.; Lee, K.Y. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy 2018, 14, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Bai, L.; Chen, W.; Xu, S. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin. Ther. Targets 2010, 14, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschella, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Tang, Y.-D.; Zhai, J.; Zheng, C. The RING finger protein family in health and disease. Signal Transduct. Target. Ther. 2022, 7, 300. [Google Scholar] [CrossRef]
- Ertem, F.B.; Guven, O.; Buyukdag, C.; Gocenler, O.; Ayan, E.; Yuksel, B.; Gul, M.; Usta, G.; Cakılkaya, B.; Johnson, J.A.; et al. Protocol for structure determination of SARS-CoV-2 main protease at near-physiological-temperature by serial femtosecond crystallography. STAR Protoc. 2022, 3, 101158. [Google Scholar] [CrossRef]
- Garman, E.F.; Owen, R.L. Cryocooling and radiation damage in macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 32–47. [Google Scholar] [CrossRef]
- Atalay, N.; Akcan, E.K.; Gul, M.; Ayan, E.; Destan, E.; Ertem, F.B.; Tokay, N.; Çakilkaya, B.; Nergiz, Z.; Karakadioğlu, G.; et al. Cryogenic X-ray crystallographic studies of biomacromolecules at Turkish Light Source “Turkish DeLight”. Turk. J. Biol. 2022, 47, 1–13. [Google Scholar] [CrossRef]
- Rigaku. CrysAlisPro Software System, Version 1.171.42.35a. 2021. Rigaku Oxford Diffraction. Available online: https://www.rigaku.com (accessed on 8 November 2023).
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 2.5.2, Schrödinger, LLC.: New York, NY, USA, 2023.
- Schrödinger Release 2016-2, Schrödinger, LLC.: New York, NY, USA, 2016.
- Ciftci, H.; Sever, B.; Ayan, E.; Can, M.; DeMirci, H.; Otsuka, M.; TuYuN, A.F.; Tateishi, H.; Fujita, M. Identification of New L-Heptanoylphosphatidyl Inositol Pentakisphosphate Derivatives Targeting the Interaction with HIV-1 Gag by Molecular Modelling Studies. Pharmaceuticals 2022, 15, 1255. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2023-3, Schrödinger, LLC.: New York, NY, USA, 2023.
- Abascal, J.L.; Vega, C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 2005, 123, 234505. [Google Scholar] [CrossRef]
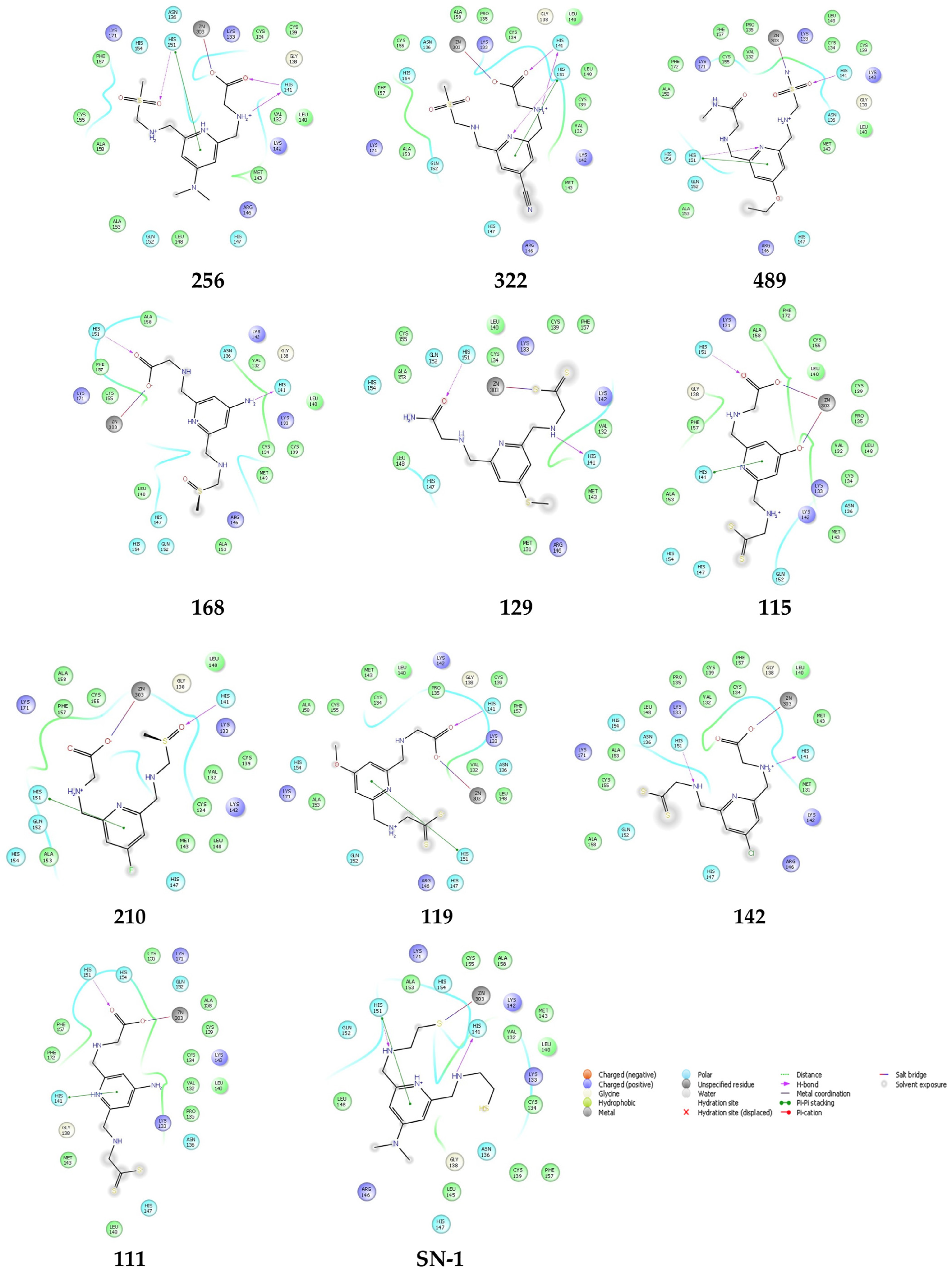
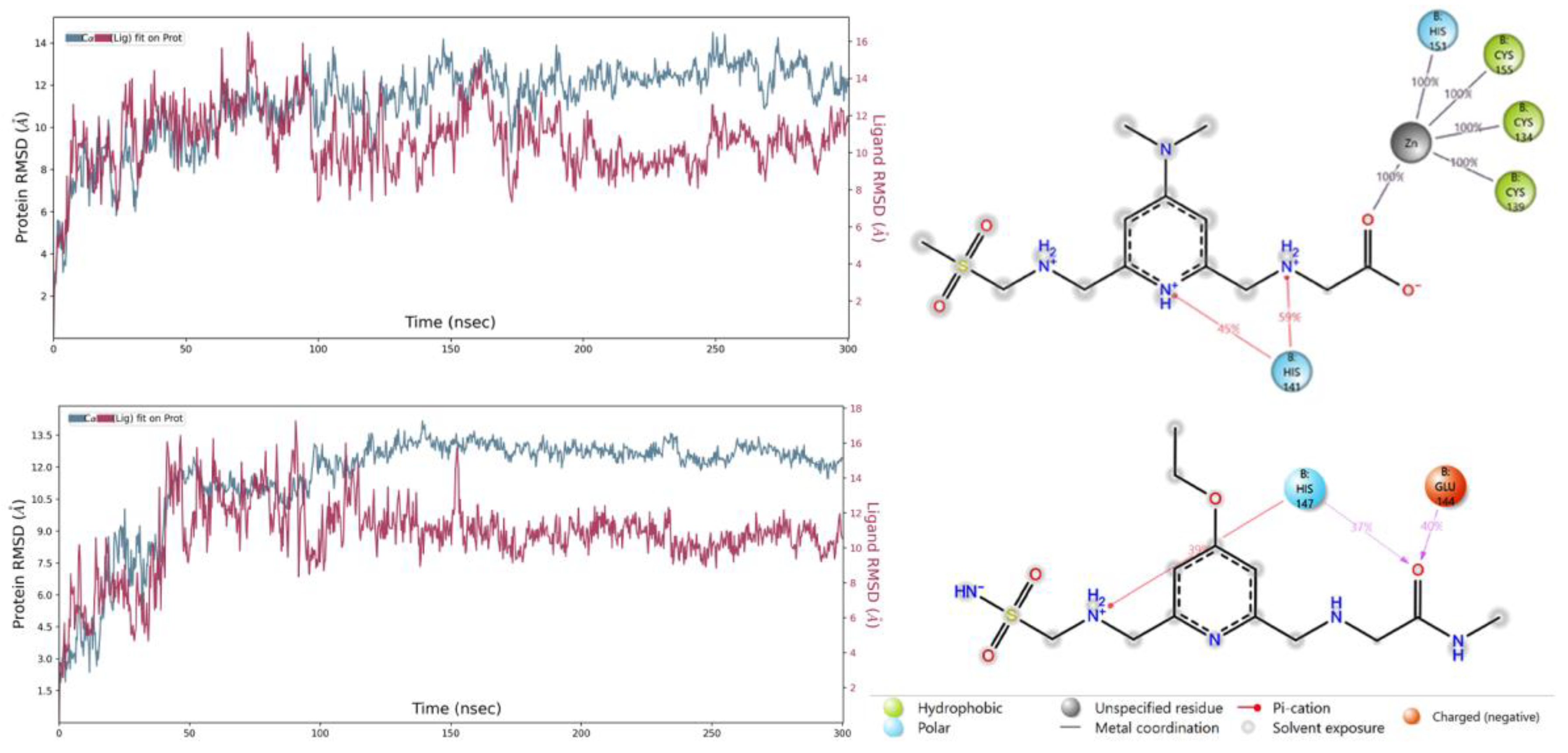
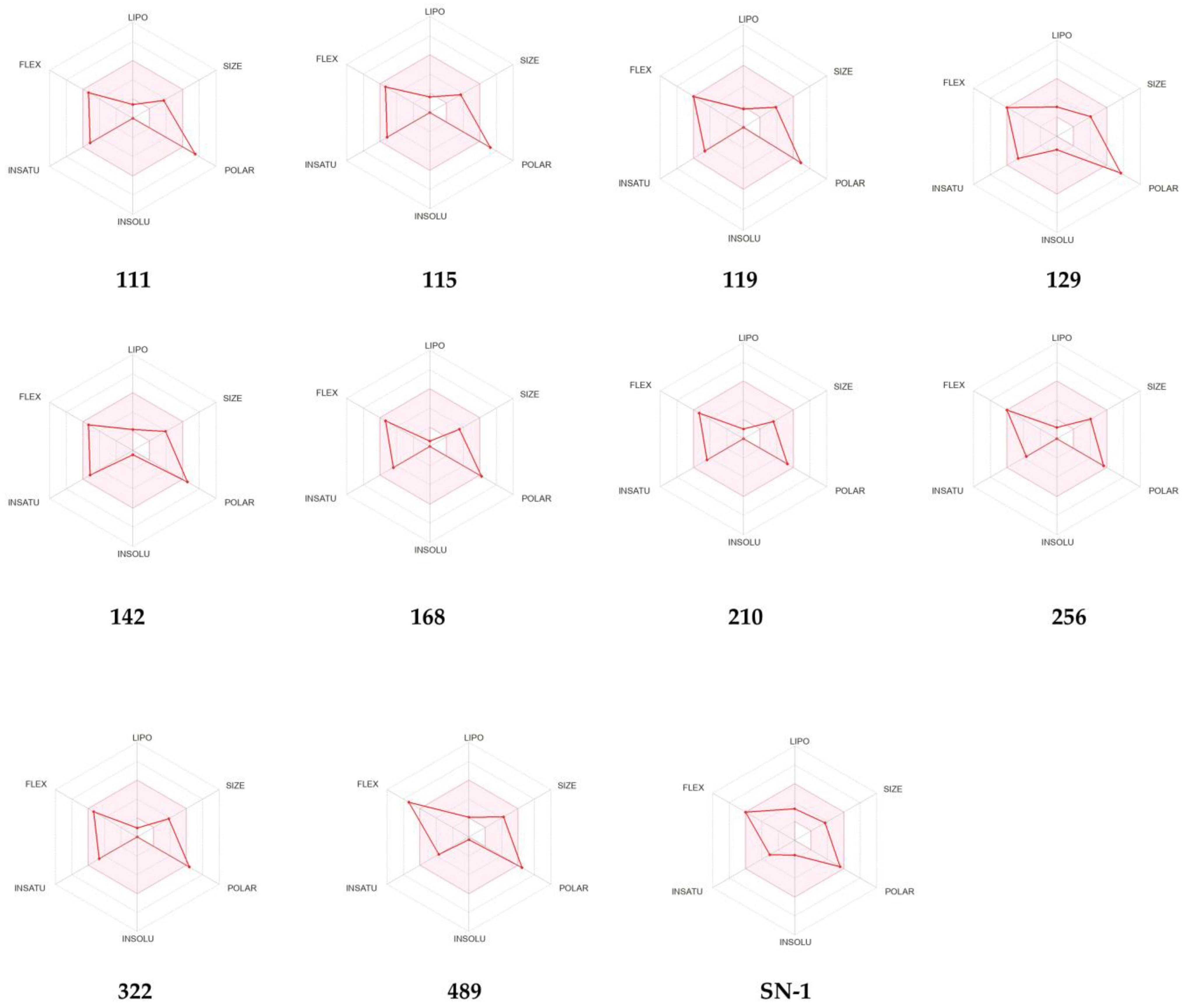
| Compound | 8HZ2 |
|---|---|
| Docking Score | |
| 256 | −6.889 |
| 322 | −6.747 |
| 489 | −6.259 |
| 168 | −5.698 |
| 129 | −5.647 |
| 115 | −5.578 |
| 210 | −5.570 |
| 119 | −5.532 |
| 142 | −5.526 |
| 111 | −5.522 |
| SN-1 | −7.335 |
| Compound | QPlogS | QPlogPo/w | QPlogBB | QPlogKhsa | Rule of Five |
|---|---|---|---|---|---|
| 111 | −1.331 | −1.943 | −1.128 | −0.978 | 0 |
| 115 | −1.187 | −1.972 | −0.805 | −0.965 | 0 |
| 119 | −1.822 | −0.885 | −0.427 | −0.798 | 0 |
| 129 | −0.174 | −0.024 | −0.462 | −0.942 | 0 |
| 142 | −2.235 | −0.552 | −0.211 | −0.730 | 0 |
| 168 | 0.638 | −3.322 | −1.680 | −1.184 | 0 |
| 210 | 0.258 | −2.265 | −0.872 | −1.059 | 0 |
| 256 | −1.368 | −2.015 | −1.648 | −0.957 | 0 |
| 322 | −1.024 | −3.418 | −1.717 | −1.194 | 0 |
| 489 | 0.345 | −1.981 | −2.084 | −1.254 | 0 |
| SN-1 | −2.049 | 2.396 | 0.624 | −0.244 | 0 |
| Dataset | TRAF6 |
|---|---|
| Wavelength (Å) | 1.54 |
| Resolution range | 20.58–3.231 (3.346–3.231) |
| Space group | P 1 |
| Unit cell | a = 45.893 Å b = 51.693 Å c = 54.302 Å α = 91.064° β = 112.116° γ = 108.43° |
| Total reflections | 6725 (639) |
| Unique reflections | 4506 (545) |
| Multiplicity | 1.5.(1.5) |
| Completeness (%) | 88.28 (79.80) |
| Mean I/sigma (I) | 7.48 (7.05) |
| Wilson B-factor | 31.96 |
| R-merge | 0.6108 (0.6305) |
| R-meas | 0.8208 (0.8537) |
| R-pim | 0.5435 (0.5705) |
| CC1/2 | 0.0159 (−0.0765) |
| CC * | 0.177 (−0.407) |
| Reflections used in refinement | 6129 (545) |
| Reflections used for R-free | 606 (56) |
| R-work | 0.2708 (0.3092) |
| R-free | 0.3750 (0.3958) |
| CC (work) | −0.007 (−0.107) |
| CC (free) | 0.066 (0.380) |
| Number of non-hydrogen atoms | 2537 |
| proteins | 2518 |
| ligands | 10 |
| solvent | 9 |
| Protein residues | 314 |
| RMS(bonds) | 0.008 |
| RMS(angles) | 1.07 |
| Ramachandran favored (%) | 90.00 |
| Ramachandran allowed (%) | 9.68 |
| Ramachandran outliers (%) | 0.32 |
| Rotamer outliers (%) | 12.24 |
| Clashscore | 19.22 |
| Average B-factor | 41.64 |
| macromolecules | 41.69 |
| ligands | 44.54 |
| solvent | 25.02 |
| Number of TLS groups | 13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guven, O.; Sever, B.; Başoğlu-Ünal, F.; Ece, A.; Tateishi, H.; Koga, R.; Radwan, M.O.; Demir, N.; Can, M.; Dilsiz Aytemir, M.; et al. Structural Characterization of TRAF6 N-Terminal for Therapeutic Uses and Computational Studies on New Derivatives. Pharmaceuticals 2023, 16, 1608. https://doi.org/10.3390/ph16111608
Guven O, Sever B, Başoğlu-Ünal F, Ece A, Tateishi H, Koga R, Radwan MO, Demir N, Can M, Dilsiz Aytemir M, et al. Structural Characterization of TRAF6 N-Terminal for Therapeutic Uses and Computational Studies on New Derivatives. Pharmaceuticals. 2023; 16(11):1608. https://doi.org/10.3390/ph16111608
Chicago/Turabian StyleGuven, Omur, Belgin Sever, Faika Başoğlu-Ünal, Abdulilah Ece, Hiroshi Tateishi, Ryoko Koga, Mohamed O. Radwan, Nefise Demir, Mustafa Can, Mutlu Dilsiz Aytemir, and et al. 2023. "Structural Characterization of TRAF6 N-Terminal for Therapeutic Uses and Computational Studies on New Derivatives" Pharmaceuticals 16, no. 11: 1608. https://doi.org/10.3390/ph16111608
APA StyleGuven, O., Sever, B., Başoğlu-Ünal, F., Ece, A., Tateishi, H., Koga, R., Radwan, M. O., Demir, N., Can, M., Dilsiz Aytemir, M., Inoue, J.-i., Otsuka, M., Fujita, M., Ciftci, H., & DeMirci, H. (2023). Structural Characterization of TRAF6 N-Terminal for Therapeutic Uses and Computational Studies on New Derivatives. Pharmaceuticals, 16(11), 1608. https://doi.org/10.3390/ph16111608












