Abstract
A novel series of hybrid compounds comprising quinazolin-4-one and 3-cyanopyridin-2-one structures has been developed, with dual inhibitory actions on both EGFR and BRAFV600E. These hybrid compounds were tested in vitro against four different cancer cell lines. Compounds 8, 9, 18, and 19 inhibited cell proliferation significantly in the four cancer cells, with GI50 values ranging from 1.20 to 1.80 µM when compared to Doxorubicin (GI50 = 1.10 µM). Within this group of hybrids, compounds 18 and 19 exhibited substantial inhibition of EGFR and BRAFV600E. Molecular docking investigations provided confirmation that compounds 18 and 19 possess the capability to inhibit EGFR and BRAFV600E. Moreover, computational ADMET prediction indicated that most of the newly synthesized hybrids have low toxicity and minimal side effects.
1. Introduction
Cancer is still a heavily researched public health issue in modern civilization. Despite intensive research, it remains a significant cause of death in wealthy countries [1,2,3]. Cancer’s danger rests in its capacity to infiltrate and destroy normal tissues and organs, causing them to malfunction. Furthermore, cancer can spread to other body regions, complicating treatment [4,5,6]. Cytotoxic therapy has long been considered the gold standard in cancer treatment [7]. However, due to inconsistencies in treatment outcomes and inadequate safety, new approaches to cancer treatment have evolved [8,9,10]. The development and widespread clinical testing of precisely targeted anti-cancer tools such as therapeutic antibodies [11], tyrosine kinase inhibitors (TKIs) [12,13,14], micro-RNA therapy [15], oncolytic viruses [16], and gene-editing treatments [17] have significantly expanded the arsenal of weapons available to combat various tumor types.
Central to these advancements is the Epidermal Growth Factor Receptor (EGFR), also known as HER1, a transmembrane receptor belonging to the ErbB family alongside HER2, HER3, and HER4 [18,19]. EGFR, a pivotal tyrosine kinase receptor, plays indispensable roles in various physiological processes, including cell cycle regulation [20], differentiation [21], and reorganization of the cytoskeleton [22]. It is frequently overexpressed in several cancer types, where it participates in activities such as cell proliferation, migration, invasion, and angiogenesis [23,24]. Mutations in EGFR are frequent in diseases such as non-small cell lung cancer, head and neck cancer, and colorectal cancer [25,26,27].
Conversely, the BRAF gene encodes BRAF kinase, a cytosolic serine/threonine kinase crucial in cell signaling, growth, and survival pathways [28,29]. BRAF mutations are commonly seen in melanoma, thyroid cancer, and colorectal cancer [30,31].
Nitrogen-based heterocycle molecules are a useful source of necessary building blocks for developing novel bioactive chemicals. N-Heterocyclic components are found in over three-quarters of the medications approved by the Food and Drug Administration (FDA) [32]. Quinazoline and quinazolin-4-one are crucial nitrogen-based heterocycles that have been extensively researched in numerous research activities revealing their adaptive biological effects [33,34]. These chemically basic compounds possess a wide range of medicinal properties, such as anti-cancer [33,35], anti-tubercular [36], anti-inflammatory [37], and antimicrobial activities [38]. Many quinazoline-based anti-cancer agents were granted FDA approval and are in clinical use for cancer management, such as Erlotinib (I) (Figure 1), demonstrating remarkable potency in inhibiting growth factor receptor tyrosine kinases, particularly targeting the EGFR receptor, and featuring quinazoline scaffolds [39]. Its anti-cancer effectiveness stems from its ability to hinder intracellular phosphorylation of tyrosine kinases at the ATP binding site of the receptor, block JAK2V617F—a mutant variant of JAK2, and initiate pathways that lead to apoptotic cell death [40]. Despite its notable efficacy, specificity, and favorable safety record, patients frequently acquire resistance to this treatment within 8–12 months of initiating therapy due to mutations occurring in the ATP binding site of the EGFR kinase domain [41]. In our recent publication, compound (II) with a quinazolin-4-one nucleus had robust antitumor activity against four cancer cell lines, as well as significant dual EGFR and BRAFV600E inhibitory action (IC50 = 0.11 µM and 0.65 µM, respectively) (Figure 1) [33].
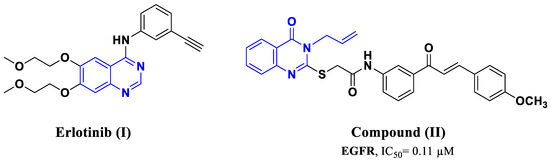
Figure 1.
Structure of compounds I and II.
Cyanopyridine derivatives have exhibited favorable characteristics in terms of antimicrobial [42], antioxidant [43], antibiotic [44], anti-inflammatory [45], anticonvulsant [46], as well as anti-cancer potential [47,48]. Because of their potential to interact with a variety of biological targets, including tubulin [49], the survivin protein [50,51], and PIM-1 Kinase [52,53], these compounds have received much interest for their anti-cancer characteristics. In our recent publication, 3-cyanopyridone/pyrazoline hybrid III (Figure 2) exhibited impressive dual inhibitory efficacy against EGFR and BRAF V600E (IC50 = 68, 65 nM, respectively), with remarkable GI50 values of 25 nM. Like Erlotinib, compound III displayed a potent inhibitor especially against both cancer cell proliferation and BRAFV600E [54]. In another study, compound IV (Figure 2) demonstrated significant efficacy in inhibiting cancer cell proliferation (with a GI50 value of 0.72 µM) and promising inhibitory potential against BRAFV600E (with an IC50 value of 58 nM), outperforming Erlotinib (with an IC50 value of 65 nM) [47].
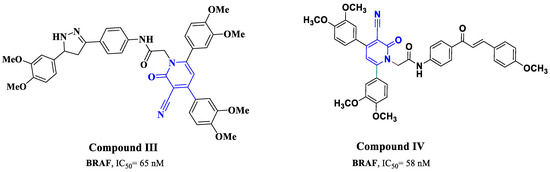
Figure 2.
Structure of compounds III and IV.
A challenging issue that has arisen during EGFR inhibitor therapy is the emergence of the BRAFV600E mutation as a potential resistance mechanism [55]. Inhibiting BRAF can also activate EGFR, reviving tumor growth [56,57,58]. Consequently, a strategic approach involving the dual inhibition of EGFR and BRAFV600E has been employed to circumvent these complexities. Several clinical trials, including patients with metastatic colorectal cancer with the BRAFV600E mutation, revealed that the combination of BRAFV600E and EGFR inhibitors was clinically successful [55,59,60].
In our ongoing research to develop antiproliferative hybrids inspired by the biological properties of compounds I–IV, we hypothesized that these hybrids might possess dual EGFR/BRAFV600E pharmacophoric elements. As shown in Figure 3, the quinazolin-4-one moiety (as seen in compounds I and II), spacer, and 3-cyanopyridin-2-one moiety (as seen in compounds III and IV) were identified as important components. As a result, combining these pharmacophores into a single compact structure can lead to the development of potent antiproliferative drugs with EGFR/BRAFV600E inhibitory actions. This approach may provide benefits such as reduced drug resistance development and fewer associated side effects.
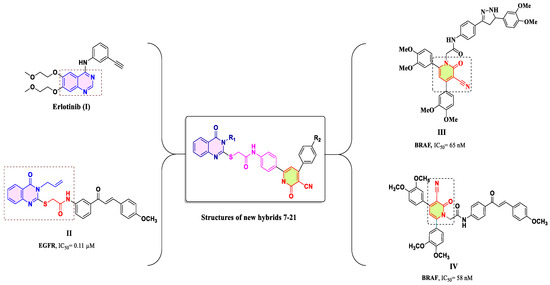
Figure 3.
Rational design of newly designed hybrids 7–21.
As a result, a novel series of quinazolin-4-one/3-cyanopyridin-2-one hybrids (7–21, Figure 3) were developed and synthesized as dual EGFR/BRAFV600E inhibitors with antiproliferative activity. The newly synthesized compounds were evaluated against a panel of four human cancer cell lines. The most potent compounds were then investigated as EGFR and BRAFV600E inhibitors. Finally, molecular docking studies were carried out to evaluate their binding mechanisms and interactions inside the active sites of molecular targets.
2. Results and Discussion
2.1. Chemistry
The reaction sequences shown in Scheme 1 were used to prepare compounds 7–21. Anthranilic acid 1 was reacted with appropriate ethyl/phenyl/allyl isothiocyanates (2a–c) in refluxing ethanol in the presence of triethylamine (TEA) as a base catalyst to generate 2-mercapto-3-substituted-(3H)-quinazolin-4-ones (3a–c) [33,35]. N-(4-Acetylphenyl)-2-bromoacetamide 4 was synthesized in high yield according to the previously described procedure [61] by treating p-aminoacetophenone with bromoacetyl bromide in water and methylene chloride bilayer solvent in the presence of potassium carbonate. The structure of product 4 was confirmed by the reported melting point [61]. Consequently, refluxing compounds 3a–c with N-(4-acetylphenyl)-2-bromoacetamide 4 in acetonitrile in the presence of TEA for 8–12 h, and the corresponding intermediates 5a–c were prepared in 75–85% yield. The melting points of compounds 5a–c were confirmed by a previously reported study [35].
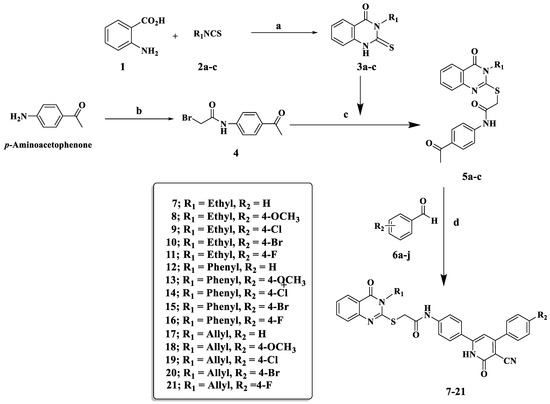
Scheme 1.
Synthesis of quinazolin-4-one/3-cyano-2-pyridone hybrids (7–21); reagents and reaction condition: (a) TEA/EtOH reflux 1.5 h; (b) BrCOCH2Br, CH2Cl2, K2CO3, H2O; (c) CH3CN, TEA reflux 8 h; (d) ethyl cyanoacetate, ammonium acetate, fusion 120–130° for 1.5 h.
The synthesis of quinazolin-4-one/3-cyano-2-pyridone hybrids 7–21 was carried out by a one-pot, four-component reaction of equal amounts of acylated quinazolin-4(3H)-one 5a–c, ethyl cyanoacetate, suitably substituted benzaldehyde, and an excess of ammonium acetate in absolute ethanol with vigorous stirring at 120–130 °C for 1.5 h to obtain final conjugates 7–21. Regarding the 1HNMR of compound 10, it exhibited a singlet signal at δ: 4.26 ppm, illustrating the presence of methylene group of SCH2 protons as well as the presence of a singlet signal at δ: 6.77 ppm corresponding to cyanopyridine H-5. Furthermore, the NH group of the 2-pyridone ring appeared as a singlet at δ: 12.75 ppm. In the 13C NMR of compound 10, a signal appeared at δ: 115.26 ppm corresponding to the nitrile group, and a signal at δ: 160.21 ppm related to the carbonyl group of the 2-pyridone ring.
2.2. Biology
2.2.1. In Vitro Anti-Cancer Activity
Cytotoxicity Assay in Non-Tumorigenic MCF-10A Cells
To evaluate the potential cytotoxic effects of the synthesized compounds on non-cancerous cells, we utilized the human mammary gland epithelial cell line, MCF-10A [62]. Compounds 7–21 were incubated with MCF-10A cells for a period of 4 days. After this incubation period, the cell survival was assessed through the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [9]. Importantly, at a concentration of 50 µM, the investigated compounds did not manifest any cytotoxic effects on MCF-10A cells. For most of the compounds tested, the cell viability remained high, surpassing 86% Table 1.

Table 1.
Cytotoxicity assay of compounds 7–21 and Doxorubicin in non-tumorigenic MCF-10A cells.
Antiproliferative Activity
The antiproliferative properties of synthesized compounds are essential in assessing their therapeutic potential. In this study, we embarked on a detailed evaluation of compounds 7–21, focusing on their ability to hinder cell proliferation across diverse cancer cell lines. The chosen cell lines represent a spectrum of human cancers, which are pancreatic cancer (Panc-1), breast cancer (MCF-7), colon cancer (HT-29), and epithelial cancer (A-549). This variety ensures that the synthesized compounds’ efficacy is tested against different cancer types, giving a holistic picture of their potential use. For a rigorous assessment, the widely accepted MTT assay [63,64] was employed. Using Doxorubicin, a well-established drug, as the reference standard ensures a benchmark for the synthesized compounds, allowing a contextual understanding of their efficacy. A first glance at the results in Table 2 showcases the promising antiproliferative potential of the synthesized compounds. With GI50 values spanning 1.40 µM to 9.40 µM, these compounds exhibited commendable inhibitory effects compared to the potent Doxorubicin with a GI50 of 1.10 µM. Among the series, compounds 8, 9, and 18–21 emerged as the frontrunners. Their antiproliferative activity was strikingly high, especially in compound 18 with a GI50 value tantamount to Doxorubicin, thereby indicating its impressive potency. The antiproliferative activity seems to hinge significantly on the type of substituent at the N-3 position of the quinazoline moiety. Compound 18, with an allyl group, showed a pronounced effect, whereas its counterparts with ethyl or phenyl groups, i.e., compounds 8 and 13, were less potent. This suggests the paramount role of allyl group in enhancing the antiproliferative activity, a crucial insight for future design improvements. Diving deeper into the structure–activity relationship, the position-4 substitution on the phenyl group of the pyridine-2-one moiety emerges as a critical determinant. Compounds 19, 20, and 21, bearing halogens, suggest a clear tolerance and even preference for halogens in antiproliferative activity. Interestingly, the order of efficacy based on the halogens is Cl > Br > F. Unsubstituted derivatives, namely compounds 7, 12, and 17, showcased relatively lower potency. This signifies the relevance of the substituents at position-4 of the phenyl group, stressing their importance in future design considerations.

Table 2.
Antiproliferative activity of compounds 7–21 and Doxorubicin.
In synthesizing compounds for antiproliferative purposes, the interplay between different functional groups and substituents becomes apparent. This detailed evaluation of the synthesized compounds offers valuable insights, not just in understanding their current efficacy but also in directing future compound designs and modifications for even enhanced therapeutic potential.
EGFR Inhibitory Activity
The most effective antiproliferative hybrids, 8, 9, 18, and 19, were tested for EGFR-TK inhibitory activity as a possible target for their antiproliferative activity [65]. Table 3 shows the IC50 values for each compound against Erlotinib, the reference drug.

Table 3.
Effects of compounds 8, 9, 18, 19, and Erlotinib on EGFR and BRAFV600E.
Compounds 8, 9, 18, and 19 displayed promising EGFR inhibitory activity, with IC50 values ranging from 110 nM to 190 nM compared to Erlotinib (IC50 = 80 ± 5 nM). As EGFR inhibitors, the investigated compounds were less effective than Erlotinib. The most potent antiproliferative agent, compound 18 (R1 = allyl, R2 = OCH3), was also the most potent EGFR inhibitor, with an IC50 value of 110 ± 10 nM, 1.4-fold less potent than Erlotinib. Compound 19 (R1 = allyl, R2 = Cl) was the second most active compound, with an IC50 value of 140 ± 11 nM, 1.8-fold less potent than Erlotinib. These findings add to the body of evidence supporting the significance of the methoxy group in these compounds’ antiproliferative effect. Finally, compounds 8 (R1 = ethyl, R2 = OCH3) and 9 (R1 = ethyl, R2 = Cl) were the least effective EGFR inhibitors, with IC50 values of 160 ± 12 nM and 190 ± 15 nM, respectively. These findings indicate that the EGFR may be a potential target for the antiproliferative activity of the investigated compounds, notably compounds 18 and 19.
BRAFV600E Inhibitory Activity
An in vitro study was carried out to evaluate the activity of the newly synthesized compounds 8, 9, 18, and 19 against BRAFV600E [59]. Table 3 shows the IC50 values for the tested compounds and Erlotinib, which was chosen as the reference drug. Table 2 shows that the investigated compounds had good inhibitory efficacy, with IC50 values ranging from 140 nM to 290 nM compared to Erlotinib (IC50 = 60 ± 5 nM). Compounds 18 (R1 = allyl, R2 = OCH3) and 19 (R1 = allyl, R2 = Cl) had the most potent inhibitory activities against BRAFV600E, with IC50 values of 140 ± 10 nM and 190 ± 12 nM, respectively, being 2- and 3-fold less potent than Erlotinib. According to the findings of this study, the tested compounds 18 and 19 had dual inhibitory effects against both EGFR and BRAFV600E, suggesting that they could be used as potential targets for antiproliferative action.
2.3. In Silico Studies
2.3.1. Docking Study
In this investigative study, we conducted a computational docking analysis to unveil the binding interactions involving the highly in vitro active compounds 18 and 19 with the tyrosine kinase receptors EGFR and BRAFV600E. We employed the Discovery Studio software, a computational tool designed for such analyses [66,67,68]. To streamline our inquiry, we obtained the X-ray crystallography structures of the EGFR and BRAFV600E tyrosine kinases. Specifically, the structure of EGFR was co-crystallized with Erlotinib as a reference structure (PDB: 1M17) [69]. Similarly, the BRAFV600E structure was co-crystallized with Vemurafenib as a reference (PDB: 3OG7) [70].
To validate the reliability of the EGFR docking procedure, we conducted a re-docking experiment using the co-crystallized Erlotinib. This was performed within the active site of EGFR, resulting in a S score of −8.69 kcal/mol. The root-mean-square deviation (RMSD) value, a structural similarity measure, was calculated to be 1.72 Å. A significant hydrogen bond interaction was established, specifically between the pyrimidine nitrogen of Erlotinib and the amino acid Met769, with a bond length of 2.29 Å. This interaction emphasized the pivotal role of Met769 in stabilizing the ligand within the binding site Figure 4.
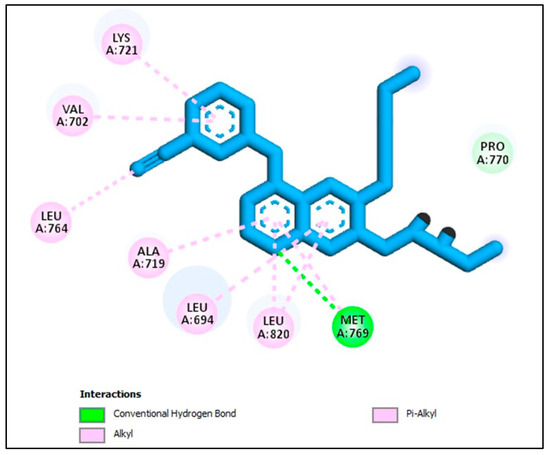
Figure 4.
Docking representation model of co-crystallized Erlotinib within the binding site of EGFR (PDB ID: 1M17).
The molecular modelling and interactions presented in the docking studies of compounds 18, 19, and the quinazoline-single moiety 5C with the EGFR receptor provide a profound insight into the mechanisms and intricacies of drug-receptor binding. The conjugation of quinazolin-4-one and 3-cyanopyridin-2-one motifs into a singular entity in compounds 18 and 19 and their subsequent interactions illuminate the significance of strategic molecular design for targeted receptor engagement. A critical observation from the provided data is the pronounced binding efficacy of the conjugated hybrids, 18 and 19, despite the evident absence of direct interactions of their quinazoline ring with the receptor amino acids. This absence might initially seem counterintuitive, given the potent nature of quinazoline as a pharmacophore. However, the enhanced activity of these hybrids can be attributed to their encompassing and vital interactions with pivotal EGFR amino acids. Such interactions not only anchor the molecules within the active site, but they also likely disrupt native functions of the receptor, rendering it inactive (Figure 5). The diversity and multiplicity of the interactions exhibited by compounds 18 and 19 emphasize the wisdom behind the conjugation of the two moieties. Whereas each moiety is capable of specific interactions, their combination seems to exploit a broader range of binding possibilities, ensuring a comprehensive engagement with the receptor. This intricate binding landscape, especially the interactions with crucial amino acids like MET A:769, ALA A:719, and VAL A:702, indicates a highly strategic alignment within the receptor active site, which could explain their potent in vitro activity. Compound 18 establishes a more diverse binding pattern within the active site of EGFR compared to compound 19. This diversity is evident in its broader interaction landscape, especially its π-sigma and π-alkyl interactions with pivotal amino acids like PHE A:771 Figure 5. These extensive interactions not only stabilize the compound within the active site but could also enhance its inhibitory effect on the receptor. Furthermore, compound 18′s computed docking score of −7.35 kcal/mol, which is higher than the −6.85 kcal/mol score of compound 19, provides a quantitative measure of its superior binding affinity (Figure 5). The constrained binding pattern of compound 19, in contrast, might limit its binding stability and, consequently, its therapeutic efficacy. Whereas compound 19 still exhibits valuable interactions and remains a potential EGFR inhibitor, compound 18, with its multifaceted engagement and superior docking score, stands out as the more potent entity in the series. On the contrary, the quinazoline-single moiety 5c, although demonstrating a series of interactions, lacks the diversity and depth of engagements witnessed in the hybrids (Figure 6). This difference is likely due to the absence of the 3-cyanopyridin-2-one motif, which, when present in the hybrids, amplifies their binding profiles. The lower S score for 5c compared to the hybrids further supports this notion (Figure 6).
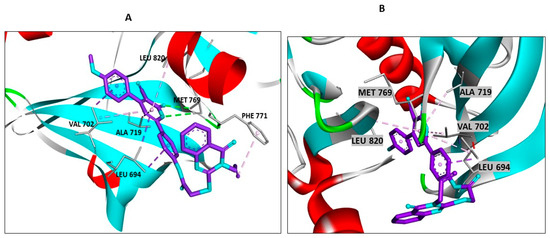
Figure 5.
Docking representation models of compounds 18 and 19 within the binding site of EGFR (PDB ID: 1M17); (A) 3D-docked model of compound 18; (B) 3D-docked model of compound 19.
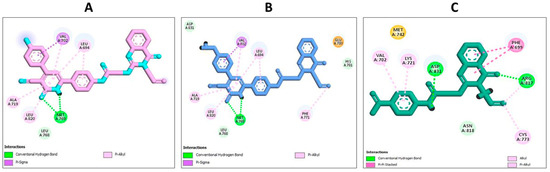
Figure 6.
Docking representation models of compound 18, 19, and 5c within the binding site of EGFR (PDB ID: 1M17); (A) 2D-docked model of compound 19; (B) 2D-docked model of compound 18; (C) 2D-docked model of compound 5c.
Similarly, to validate the docking protocol concerning BRAFV600E, we conducted a re-docking experiment utilizing the co-crystallized Vemurafenib. This endeavor took place at active site of BRAFV600E, resulting in an impressive S score of −11.99 kcal/mol. To gauge the extent of structural conformity, we computed a root-mean-square deviation (RMSD) value of 1.55 Å. Substantial hydrogen bond interactions were established, particularly between Vemurafenib and the amino acids Cys532 and Asn580, firmly anchoring its position within the binding site (Figure 7).
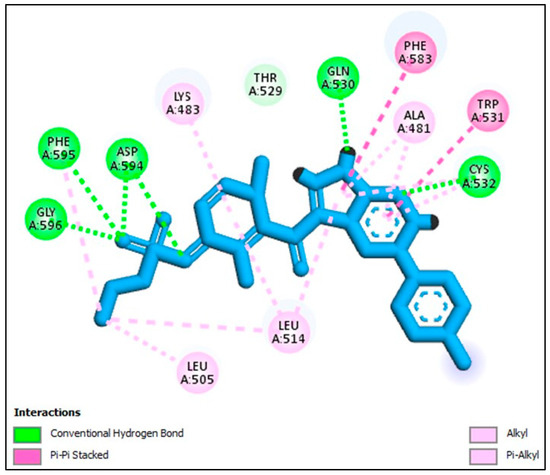
Figure 7.
Docking representation model of co-crystallized Vemurafenib within the binding site of BRAF (PDB ID: 3OG7).
Hybrid 18, with a docking score of −7.85 kcal/mol, showcases more pronounced interactions with the receptor. From the visualization, hybrid 18 forms strong conventional hydrogen bonds with residues like CYS A:532, ASN A:580, and SER A:536 (Figure 8). These interactions, particularly with CYS A:532, are of utmost importance, considering the pivotal role cysteine residues often play in the active sites of various proteins. The compound aromatic regions also engage in multiple pi interactions, further reinforcing its grip within the active site. Alkyl and pi-alkyl interactions with residues such as ALA A:481, LYS A:483, and VAL A:471 solidify its binding affinity, potentially leading to its potent in vitro activity (Figure 8). On the other hand, hybrid 19, despite having a commendable docking score of −7.53 kcal/mol, falls slightly short when compared to hybrid 18. The visual representation suggests robust interactions, but the comparative difference in docking scores indicates that hybrid 18 might have a slightly superior binding affinity or forms more favorable interactions, translating to enhanced inhibitory potential. However, with a docking score of −6.33 kcal/mol, Erlotinib interaction is somewhat less optimal than the scores observed for hybrids 18 and 19 (Figure 8). One reason for this could be the extent and depth of interactions Erlotinib forms with the receptor. Whereas it does form important bonds with vital residues, it may not span as diverse a landscape as the hybrids, especially hybrid 18 (Figure 8). These findings closely aligned with dual EGFR/BRAFV600E in vitro inhibition assay outcomes. In conclusion, the findings presented in this study underscore the notable inhibitory potential of compound 18 against the BRAFV600E kinase. This propensity is underpinned by meaningful binding interactions observed within the active site. This observation resonates with the outcomes obtained from the rigorous BRAFV600E in vitro inhibition assay, lending credibility to the predictive capabilities of our computational approach.
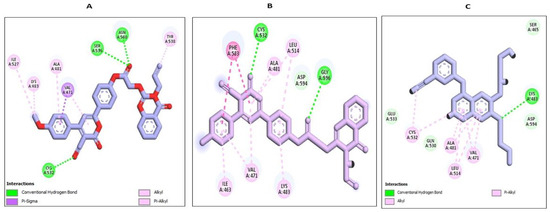
Figure 8.
Docking representation models of compound 18, 19, and Erlotinib within the binding site of BRAF (PDB ID: 3OG7); (A) 2D-docked model of compound 18; (B) 2D-docked model of compound 19; (C) 2D-docked model of compound Erlotinib.
Additionally, our investigation unveils a compelling avenue for potential dual inhibition, as indicated by supplementary simulations targeting both EGFR and BRAFV600E; specifically, compounds 18 and 19 promise to function as dual inhibitors.
The insights garnered from this study offer a foundational comprehension of the inhibitory implications embedded within the investigated compounds. This understanding, in turn, sets the stage for subsequent experimental investigations and optimization endeavors within the dynamic realm of drug discovery. Describing the complex interactions between these compounds and the target kinases is an important guide for developing targeted hit compounds. These compounds could interfere with pathways linked to cancer signaling.
2.3.2. In Silico ADMET Studies
Considering the promising in vitro and silico docking results, we proceeded to conduct supplementary ADMET studies for the newly synthesized compounds. This choice was motivated by the intention to gain a more comprehensive insight into these significant activities [71]. During the ADMET investigations, we utilized Erlotinib as the established reference compound. Using Discovery Studio 4.0, we conducted predictions for the ADMET descriptors of all synthesized compounds. The expected descriptors are outlined in Table 4.

Table 4.
Predicted ADMET for synthesized qunazolin-4-one/3-cyanopyrid-2-one hybrids 7–21.
A significant number of the hybrid compounds exhibited a modest predicted level of intestinal absorption (absorption level = 2). This positioning designates them as promising candidates for localized treatment of gastrointestinal tumors or as potential contenders for intravenous administration. Notably, most of these novel compounds displayed low solubility (ADME aqueous solubility level = 2).
Furthermore, the AlogP98 tool aided in predicting the logP value based on the molecular structure of each compound. This approach allows researchers to estimate how a compound will partition between a nonpolar solvent (octanol) and water. Notably, most hybrid compounds demonstrated AlogP98 values ranging from 5 to 6, indicating their heightened lipophilicity. This characteristic contributes to their reduced aqueous solubility. However, there is a potential solution in the form of Cyclodextrin Complexation, a method that improves solubility by encapsulating these hybrid molecules within the hydrophobic cavity of cyclodextrin molecules.
During the ADMET assessment, all the newly synthesized hybrids were categorized at a blood–brain barrier (BBB) level of 4, effectively hindering their ability to penetrate across the BBB. Importantly, the drug bioavailability was interconnected with the fundamental property of 2D polar surface area (ADMET 2D PSA).
Utilizing the calculated 2D polar surface area (PSA 2D) and Atom-based Log P98 (A log P98) properties, the outcomes have been visualized through a 2D ADMET plot (Figure 9). It is worth noting that molecules with a PSA less than 145 generally exhibit characteristics of low bioavailability and passive absorption [72]. Using a 2D chemical structure as input, the cytochrome P450 2D6 (CYP2D6) model predicts the potential for inhibiting the CYP2D6 enzyme. This enzyme, located in the liver, plays a critical role in metabolizing a wide range of substrates, thereby significantly contributing to various drug–drug interaction scenarios [73]. As a result, experimenting to evaluate CYP2D6 inhibition becomes crucial within the regulatory protocols followed during drug discovery and development [74]. Each evaluated hybrid compound was predicted to display non-inhibitory behavior towards CYP2D6. As a result, the risk of inducing liver dysfunction after administering these hybrids is minimal.
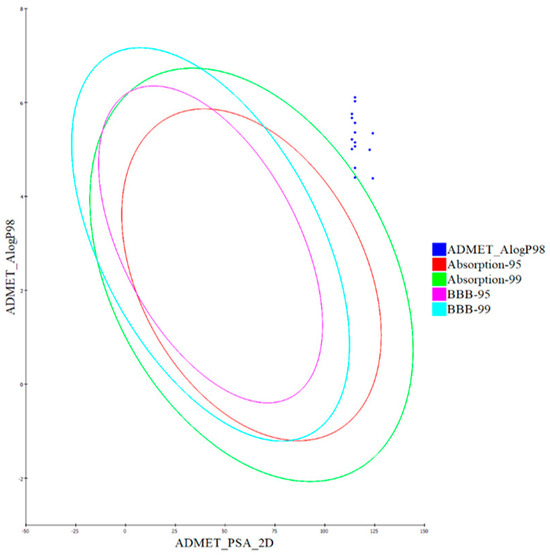
Figure 9.
The expected ADMET study of quinazolin-4-one/3-cyanopyrid-2-one hybrids 7–21.
The plasma protein binding model determines whether a substance will bind robustly (>90%) to carrier proteins in the blood. Notably, a substantial binding to plasma proteins (>90%) was expected for the majority of hybrid compounds, as detailed in Table 4.
2.3.3. In Silico Toxicity Predictions
To predict toxicity, we utilized the constructed and validated models within the Discovery Studio software for the synthesized compounds [75]. The rodent carcinogenicity test carried out by the FDA evaluates the capacity of a chemical structure to induce cancer in rats. On the other hand, the prediction of rat maximum tolerated dose (MTD) estimates the highest dose of a chemical substance that can be administered to rats without resulting in adverse effects [76]. In assessing toxicity for a chemical compound, the prediction of rat oral LD50 is utilized to forecast the acute median lethal dose (LD50) in rats after the compound is administered orally [77]. As part of the Draize test, ocular irritancy analysis is utilized to determine the potential of a particular compound to cause eye irritation and assess the degree of severity of this irritation [78]. In evaluations based on rabbits, investigations into skin irritancy assess the probability of a substance causing skin irritation and the potential severity of such effects. As per the in silico assessments outlined in Table 5, most compounds displayed low toxicity and minimal adverse effects.

Table 5.
The predicted toxicity properties of the synthesized compounds.
Additionally, all the hybrid compounds subjected to testing were forecasted to have non-carcinogenic attributes, aligning with the preliminary findings of the FDA rodent carcinogenicity assessment. The majority of the evaluated compounds exhibited higher rat oral LD50 values compared to the value of Erlotinib.
Furthermore, the predictive models suggested that all hybrids are anticipated to not irritate in the context of skin irritancy testing, and the majority of them are projected to induce mild irritation in the context of ocular irritancy testing.
In conclusion, the ADMET investigations conducted in this study provide crucial insights into the potential effectiveness, safety, and pharmacokinetic behavior of the newly developed hybrids. The insights gained from these assessments hold immense significance in guiding the drug discovery and development trajectory. This wealth of information assists in identifying promising drug candidates that merit further evaluation and advancement in the testing and developmental stages.
In summation, the ADMET explorations undertaken in this investigation offer pivotal insights into the potential efficacy, safety profile, and pharmacokinetic attributes of the recently formulated hybrids. The discernments gleaned from these analyses carry substantial importance, serving as a compass to navigate the path of drug discovery and development. This reservoir of knowledge aids in pinpointing auspicious drug candidates warranting meticulous scrutiny and progression through subsequent testing and developmental phases.
2.4. Structure–Activity Relationship (SAR)
Based on the observed results, the structure–activity relationship of our novel quinazolin-4-one/3-cyanopyridin-2-one hybrids (7–21) is as follows.
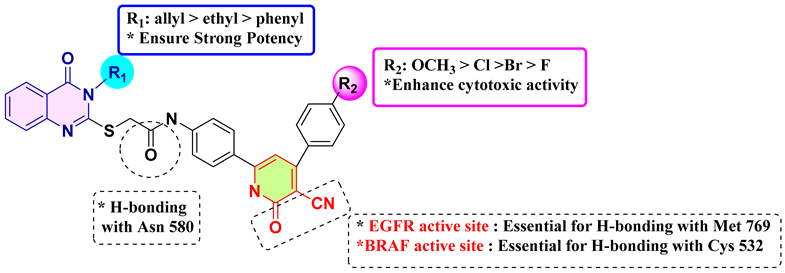
Finally, introducing substituents into the quinazolin-4-one and 3-cyanopyridin-2-one moieties significantly influences the efficacy of the newly synthesized hybrids 7–21. The highest levels of anti-proliferative activity were observed in 3-allyl quinazolin-4-one derivatives, with 3-ethyl and 3-phenyl derivatives displaying lower levels of activity in that order. The addition of specific functional groups, such as an electron-donating methoxy group at position 4 of the phenyl group in the 3-cyano-2-pyridone moiety (as seen in compound 18), appears to enhance both EGFR and BRAF inhibitory activity. The introduction of electron-withdrawing substituents at the para position of phenyl at position 4 of 3-cyano-2-pyridone seems to improve cytotoxicity in the order (4-Cl > 4-Br > 4-F). Moreover, the presence of a nitrile group (CN) at the C3 and carbonyl group of 3-cyano-2-pyridone moiety are essential for binding EGFR and BRAF active site. Moreover, both the pyridone nitrogen and the carbonyl oxygen atom within the 3-cyanopyrid-2-one core form hydrogen bonds with the backbone NH of Met769 located in the hinge region of the active site of EGFR kinase. On other hand, the nitrile group within the 3-cyanopyrid-2-one core is essential for forming a hydrogen bond with the key residue Cys 532 in the N-lobe, while the carbonyl oxygen atom of the amidic linkage is crucial for creating a hydrogen bond with Asn 580 within the catalytic loop active site of BRAFV600E.The strategic positioning of these functional groups is critical in defining these compounds’ antiproliferative efficacy.
3. Materials and Methods
3.1. Chemistry
General details: see Section A (Supplementary File). Compounds 3a–c and 5a–c were prepared according to previous studies [33,35].
General Procedures for the Synthesis of Compounds 7–21
A mixture of appropriate derivatives 5a–c (1 mmol) substituted benzaldehydes 6a–j (1 mmol), ethyl cyanoacetate (0.113 g, 1 mmol), and ammonium acetate (0.154 g, 2 mmol) were fused at 120–130 °C for 30 min. After cooling, the crude product was filtered off and crystallized from absolute ethanol [48,52].
- N-(4-(5-Cyano-6-oxo-4-phenyl-1,6-dihydropyridin-2-yl)phenyl)-2-((3-ethyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetamide (7). White powder; yield (0.400 g, 75%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 1.33 (t, J = 7.0 Hz, 3H, N3-CH2-CH3), 4.14 (q, J = 7.1 Hz, 2H, N3-CH2-CH3), 4.27 (s, 2H, SCH2), 6.74 (s, 1H, pyridine-C5–H), 7.41–7.48 (m, 4H, Ar-H), 7.57–7.60 (m, 3H, Ar-H), 7.77 (d, J = 8.2 Hz, 3H, Ar-H), 7.92 (d, J = 8.5 Hz, 2H, Ar-H), 8.08 (d, J = 7.9 Hz, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.67 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 13.02, 36.78, 37.61, 91.52, 104.39, 118.53, 118.83, 118.88, 121.61, 124.00, 125.67, 125.99, 126.43, 127.50, 128.69, 130.37, 131.29, 131.77, 134.68, 135.36, 138.26, 141.76, 146.68, 155.88, 160.21, 166.36. Anal. Calcd. For C30H23N5O3S (533.61): C, 67.53; H, 4.34; N, 13.12; S, 6.01. Found: C, 67.64; H, 4.46; N, 13.15; S, 6.23.
- N-(4-(5-Cyano-4-(4-methoxyphenyl)-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((3-ethyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetamide (8). Yellow powder; yield (0.292 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 1.33 (t, J = 7.0 Hz, 3H, N3-CH2-CH3), 3.85 (s, 3H, OCH3), 4.14 (q, J = 7.1 Hz, 2H, N3-CH2-CH3), 4.27 (s, 2H, SCH2), 6.74 (s, 1H, pyridine-C5–H), 7.10–7.16 (m, 2H, Ar-H), 7.40–7.46 (m, 2H, Ar-H), 7.72–7.78 (m, 5H, Ar-H), 7.89 (s, 2H, Ar-H), 8.07 (dd, J = 8.2, 1.6 Hz, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.61 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 13.02, 36.78, 37.61, 55.61, 91.52, 104.39, 118.53, 118.83, 118.88, 121.61, 124.00, 125.67, 125.99, 126.43, 127.50, 128.69, 130.37, 131.29, 131.77, 134.68, 135.36, 138.26, 141.76, 146.68, 155.88, 160.21, 166.36. Anal. Calcd. For C31H25N5O4S (563.63): C, 66.06; H, 4.47; N, 12.43; S, 5.69. Found: C, 66.28; H, 4.67; N, 12.44; S, 5.71.
- N-(4-(4-(4-Chlorophenyl)-5-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((3-ethyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetamide (9). Yellow powder; yield (0.282 g, 78%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 1.33 (t, J = 7.0 Hz, 3H, N3-CH2-CH3), 4.13 (q, J = 7.1 Hz, 2H, N3-CH2-CH3), 4.26 (s, 2H, SCH2), 6.77 (s, 1H, pyridine-C5–H), 7.34–7.48 (m, 3H, Ar-H), 7.58–7.71 (m, 3H, Ar-H), 7.72–7.85 (m, 5H, Ar-H), 8.04 –8.09 (m, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.75 (s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 13.02, 36.78, 38.01, 91.52, 104.45, 115.26, 118.83, 118.87, 125.67, 125.99, 126.43, 127.50, 128.69, 130.37, 131.29, 131.77, 134.68, 135.36, 141.76, 146.68, 152.76, 155.88, 157.34,158.88, 159.35, 160.21, 166.36. Anal. Calcd. For C30H22ClN5O3S (568.05): C, 63.43; H, 3.90; N, 12.33; S, 5.64. Found: C, 63.45; H, 4.10; N, 12.35; S, 5.66.
- N-(4-(4-(4-Bromophenyl)-5-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((3-ethyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetamide (10). Yellow powder; yield (0.262 g, 78%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 1.33 (t, J = 7.0 Hz, 3H, N3-CH2-CH3), 4.13 (q, J = 7.1 Hz, 2H, N3-CH2-CH3), 4.26 (s, 2H, SCH2), 6.77 (s, 1H, pyridine-C5–H), 7.34–7.48 (m, 3H, Ar-H), 7.58–7.71 (m, 3H, Ar-H), 7.72–7.85 (m, 5H, Ar-H), 8.04–8.09 (m, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.75 (s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 13.02, 36.78, 38.01, 91.52, 104.45, 115.26, 118.83, 118.87, 125.67, 125.99, 126.43, 127.50, 128.69, 130.37, 131.29, 131.77, 134.68, 135.36, 141.76, 146.68, 152.76, 155.88, 157.34, 158.88, 159.35, 160.21, 166.36. Anal. Calcd. For C30H22BrN5O3S (612.50): C, 58.83; H, 3.62; N, 11.43; S, 5.23. Found: C, 58.90; H, 3.68; N, 11.48; S, 5.28.
- N-(4-(5-Cyano-4-(4-fluorophenyl)-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((3-ethyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)acetamide (11). Yellow powder; yield (0.298 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 1.33 (t, J = 7.0 Hz, 3H, N3-CH2-CH3), 4.13 (q, J = 7.1 Hz, 2H, N3-CH2-CH3), 4.26 (s, 2H, SCH2), 6.77 (s, 1H, pyridine-C5–H), 7.34–7.48 (m, 3H, Ar-H), 7.58–7.71 (m, 3H, Ar-H), 7.72–7.85 (m, 5H, Ar-H), 8.04–8.09 (m, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.75 (s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 13.02, 36.78, 38.01, 91.52, 104.45, 115.26, 118.83, 118.87, 125.67, 125.99, 126.43, 127.50, 128.69, 130.37, 131.29, 131.77, 134.68, 135.36, 141.76, 146.68, 152.76, 155.88, 157.34, 158.88, 159.35, 160.21, 166.36. Anal. Calcd. For C30H22FN5O3S (551.60): C, 65.32; H, 4.02; N, 12.70; S, 5.81. Found: C, 65.42; H, 4.22; N, 12.60; S, 5.85.
- N-(4-(5-Cyano-6-oxo-4-phenyl-1,6-dihydropyridin-2-yl)phenyl)-2-((4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetamide (12). Yellow powder; yield (0.298 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ: 4.11 (s, 2H, SCH2), 6.78 (s, 1H, pyridine-C5–H), 7.43–7.48 (m, 1H, Ar-H), 7.48–7.51 (m, 2H, Ar-H), 7.51–7.56 (m, 2H, Ar-H), 7.56–7.58 (m, 2H, Ar-H), 7.59–7.62 (m, 3H, Ar-H), 7.70–7.76 (m, 4H, Ar-H), 7.78–7.84 (m, 1H, Ar-H), 7.90 (d, J = 8.3 Hz, 2H, Ar-H), 8.08 (m, 1H, Ar-H), 10.67 (s, 1H, CONH), 12.71 (s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.43, 91.84, 104.34, 108.15, 116.84, 119.18, 119.67, 126.10, 126.38, 126.85, 128.39, 128.86, 129.04, 129.57, 129.85, 130.32, 130.66, 135.25, 135.90, 136.30, 138.07, 141.85, 143.68, 147.28, 157.06, 158.78, 160.90, 166.71. Anal. Calcd. For C34H23N5O3S (581.65): C, 70.21; H, 3.99; N, 12.04; S, 5.51. Found: C, 70.41; H, 3.95; N, 12.01; S, 5.50.
- N-(4-(5-Cyano-4-(4-methoxyphenyl)-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetamide (13). Yellow powder; yield (0.332 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 3.86 (s, 3H, OCH3), 4.13 (s, 2H, SCH2), 6.78 (s, 1H, pyridine-C5–H), 7.13 (d, J = 8.4 Hz, 2H, Ar-H), 7.44–7.58 (m, 4H, Ar-H), 7.62 (d, J = 5.7 Hz, 3H, Ar-H), 7.66–7.85 (m, 5H, Ar-H), 7.90 (d, J = 8.3 Hz, 2H, Ar-H), 8.09 (d, J = 7.9 Hz, 1H, Ar-H), 10.68 (s, 1H, CONH), 12.58 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 36.81, 55.22, 91.02, 112.65, 115.63, 118.17, 119.34, 120.04, 122.73, 123.64, 126.34, 126.55, 127.13, 129.17, 129.90, 130.05, 130.52, 130.85, 132.25, 135.44, 135.79, 136.25, 142.22, 147.58, 157.39, 160.59, 165.72. Anal. Calcd. For C35H25N5O4S (611.68): C, 68.73; H, 4.12; N, 11.45; S, 5.24. Found: C, 68.75; H, 4.17; N, 11.50; S, 5.25.
- N-(4-(4-(4-Chlorophenyl)-5-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetamide (14). Yellow powder; yield (0.309 g, 75%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.13 (s, 2H, SCH2), 6.82 (s, 1H, pyridine-C5–H), 7.47 (d, J = 7.5 Hz, 1H, Ar-H), 7.49–7.52 (m, 2H, Ar-H), 7.54 (d, J = 8.0 Hz, 1H, Ar-H), 7.57–7.63 (m, 3H, Ar-H), 7.63–7.68 (m, 2H, Ar-H), 7.72–7.79 (m, 4H, Ar-H), 7.79–7.84 (m, 1H, Ar-H), 7.91 (d, J = 8.3 Hz, 2H, Ar-H), 8.09 (dd, J = 8.0, 1.6 Hz, 1H, Ar-H), 10.68 (s, 1H, CONH), 12.76 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.82, 91.02, 104.65, 119.34, 120.04, 122.73, 123.64, 124.51, 126.34, 126.55, 127.13, 129.17, 129.90, 130.05, 130.52, 130.85, 132.25, 135.44, 135.79, 136.25, 142.22, 147.58, 157.39, 161.08, 166.86. Anal. Calcd. For C34H22ClN5O3S (616.09): C, 66.28; H, 3.60; N, 11.37; S, 5.20. Found: C, 66.30; H, 3.70; N, 11.36; S, 5.29.
- N-(4-(4-(4-Bromophenyl)-5-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetamide (15). Yellow powder; yield (0.296 g, 77%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.13 (s, 2H, SCH2), 6.80 (s, 1H, pyridine-C5–H), 7.47 (d, J = 7.5 Hz, 1H, Ar-H), 7.49–7.52 (m, 2H, Ar-H), 7.54 (d, J = 8.0 Hz, 1H, Ar-H), 7.57–7.63 (m, 3H, Ar-H), 7.63–7.68 (m, 2H, Ar-H), 7.72–7.79 (m, 4H, Ar-H), 7.79–7.84 (m, 1H, Ar-H), 7.91 (d, J = 8.3 Hz, 2H, Ar-H), 8.09 (dd, J = 8.0, 1.6 Hz, 1H, Ar-H), 10.68 (s, 1H, CONH), 12.76 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.82, 91.02, 104.65, 119.34, 120.04, 122.73, 123.64, 124.51, 126.34, 126.55, 127.13, 129.17, 129.90, 130.05, 130.52, 130.85, 132.25, 135.44, 135.79, 136.25, 142.22, 147.58, 157.39, 161.08, 166.86. Anal. Calcd. For C34H22BrN5O3S (660.55): C, 61.82; H, 12.10; N, 7.27; S, 4.85. Found: C, 61.84; H, 12.30; N, 7.47; S, 4.90.
- N-(4-(5-Cyano-4-(4-fluorophenyl)-6-oxo-1,6-dihydropyridin-2-yl)phenyl)-2-((4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio)acetamide (16). Yellow powder; yield (0.339 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ 4.11 (s, 2H, SCH2), 6.80 (s, 1H, pyridine-C5–H), 7.17–7.34 (m, 1H, Ar-H), 7.34–7.56 (m, 5H, Ar-H), 7.56 –7.68 (m, 3H, Ar-H), 7.68–7.77 (m, 3H, Ar-H), 7.77–7.84 (m, 2H, Ar-H), 7.85–8.02 (m, 2H, Ar-H), 8.08 (dd, J = 8.0, 1.6 Hz, 1H), 10.67 (s, 1H, CONH), 12.71 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.82, 91.02, 104.65, 119.34, 120.04, 122.73, 123.64, 124.51, 126.34, 126.55, 127.13, 129.17, 129.90, 130.05, 130.52, 130.85, 132.25, 135.44, 135.79, 136.25, 142.22, 147.58, 157.39, 161.08, 166.86. Anal. Calcd. For C34H22FN5O3S (599.64): C, 68.10; H, 3.70; N, 11.68; S, 5.35. Found: C, 68.30; H, 3.80; N, 11.70; S, 5.45.
- 2-((3-Allyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)-N-(4-(5-cyano-6-oxo-4-phenyl-1,6-dihydropyridin-2-yl)phenyl)acetamide (17). Yellow powder; yield (0.409 g, 75%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.25 (s, 2H, SCH2), 4.75 (d, J = 5.2 Hz, 2H, CH2CH=CH2), 5.16–5.27 (m, 2H, CH2CH=CH2), 5.92–5.98 (m, 1H, CH2CH=CH2), 6.82 (s, 1H, pyridine-C5–H), 7.41–7.48 (m, 4H, Ar-H), 7.57–7.60 (m, 3H, Ar-H), 7.77 (d, J = 8.2 Hz, 3H, Ar-H), 7.92 (d, J = 8.5 Hz, 2H, Ar-H), 8.08 (d, J = 7.9 Hz, 1H, Ar-H), 10.73 (s, 1H, CONH), 12.67 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.20, 46.50, 91.09, 105.51, 114.69, 116.99, 118.17, 118.43, 119.05, 119.63, 125.67, 126.30, 126.79, 127.03, 128.85, 129.14, 130.38, 131.64, 132.49, 135.49, 141.59, 147.16, 156.74, 158.94, 161.01, 161.29, 167.05, 174.30. Anal. Calcd. For C31H23N5O3S (545.62): C, 68.24; H, 4.25; N, 12.84; S, 5.88. Found: C, 68.34; H, 4.40; N, 12.90; S, 5.93.
- 2-((3-Allyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)-N-(4-(5-cyano-4-(4-methoxy phenyl)-6-oxo-1,6-dihydropyridin-2-yl)phenyl)acetamide (18). Yellow powder; yield (0.303 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 3.83 (s, 3H, OCH3), 4.25 (s, 2H, SCH2), 4.73–4.76 (m, 2H, CH2CH=CH2), 5.15–5.26 (m, 2H, CH2CH=CH2), 5.91–6.00 (m, 1H, CH2CH=CH2), 6.71 (s, 1H, pyridine-C5–H), 7.05–7.10 (m, 2H, Ar-H), 7.40–7.48 (m, 2H, Ar-H), 7.62–7.67 (m, 2H, Ar-H), 7.71–7.79 (m, 3H, Ar-H), 7.90–7.95 (m, 2H, Ar-H), 8.06–8.10 (m, 1H, Ar-H), 10.70 (s, 1H, CONH), 12.56 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.20, 46.50, 55.90, 91.09, 105.51, 114.69, 118.17, 118.43, 119.05, 119.63, 125.67, 126.30, 126.79, 127.03, 128.85, 129.14, 130.38, 131.64, 132.49, 135.49, 141.59, 147.16, 156.74, 158.94, 161.01, 161.29, 167.05, 174.30. Anal. Calcd. For C32H25N5O4S (575.64): C, 66.77; H, 4.38; N, 12.17; S, 5.57. Found: C, 66.80; H, 4.58; N, 12.29; S, 5.70.
- 2-((3-Allyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)-N-(4-(4-(4-chlorophenyl)-5-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)acetamide (19). Yellow powder; yield (0.293 g, 78%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.26 (s, 2H, SCH2), 4.74–4.80 (m, 2H, CH2CH=CH2), 5.16–5.29 (m, 2H, CH2CH=CH2), 5.91–6.02 (m, 1H, CH2CH=CH2), 6.83 (s, 1H, pyridine-C5–H), 7.41–7.51 (m, 3H, Ar-H), 7.63–7.67 (m, 2H, Ar-H), 7.75–7.77 (m, 2H, Ar-H), 7.77–7.79 (m, 2H, Ar-H), 7.89–7.96 (m, 2H, Ar-H), 8.05–8.10 (m, 1H, Ar-H), 10.75 (s, 1H, CONH), 12.73 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.20, 46.50, 91.54, 105.51, 114.69, 118.17, 118.43, 119.05, 119.63, 126.30, 126.79, 127.03, 128.85, 129.14, 130.38, 131.64, 132.49, 135.49, 141.59, 147.16, 156.74, 158.94, 161.01, 161.29, 167.05, 170.13, 174.30. Anal. Calcd. For C31H22ClN5O3S (580.06): C, 64.19; H, 3.82; N, 12.07; S, 5.53. Found: C, 64.22; H, 3.90; N, 12.17; S, 5.64.
- 2-((3-Allyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)-N-(4-(4-(4-bromophenyl)-4-cyano-6-oxo-1,6-dihydropyridin-2-yl)phenyl)acetamide (20). Yellow powder; yield (0.279 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.26 (s, 2H, SCH2), 4.74–4.80 (m, 2H, CH2CH=CH2), 5.16–5.29 (m, 2H, CH2CH=CH2), 5.91–6.02 (m, 1H, CH2CH=CH2), 6.83 (s, 1H, pyridine-C5–H), 7.41–7.51 (m, 3H, Ar-H), 7.63–7.67 (m, 2H, Ar-H), 7.75–7.77 (m, 2H, Ar-H), 7.77–7.79 (m, 2H, Ar-H), 7.89–7.96 (m, 2H, Ar-H), 8.05–8.10 (m, 1H, Ar-H), 10.75 (s, 1H, CONH), 12.73 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.20, 46.50, 91.54, 105.51, 114.69, 118.17, 118.43, 119.05, 119.63, 126.30, 126.79, 127.03, 128.85, 129.14, 130.38, 131.64, 132.49, 135.49, 141.59, 147.16, 156.74, 158.94, 161.01, 161.29, 167.05, 170.13, 174.30. Anal. Calcd. For C31H22BrN5O3S (624.51): C, 59.62; H, 3.55; N, 11.21; S, 5.13. Found: C, 59.70; H, 3.60; N, 11.41; S, 5.26.
- 2-((3-Allyl-4-oxo-3,4-dihydroquinazolin-2-yl)thio)-N-(4-(5-cyano-4-(4-fluorophenyl) -6-oxo-1,6-dihydropyridin-2-yl)phenyl)acetamide (21). Yellow powder; yield (0.309 g, 80%); m.p.: > 300 °C; 1H NMR (500 MHz, DMSO-d6) δ (ppm): 4.26 (s, 2H, SCH2), 4.74–4.80 (m, 2H, CH2CH=CH2), 5.16–5.29 (m, 2H, CH2CH=CH2), 5.91–6.02 (m, 1H, CH2CH=CH2), 6.83 (s, 1H, pyridine-C5–H), 7.41–7.51 (m, 3H, Ar-H), 7.63–7.67 (m, 2H, Ar-H), 7.75–7.77 (m, 2H, Ar-H), 7.77–7.79 (m, 2H, Ar-H), 7.89–7.96 (m, 2H, Ar-H), 8.05–8.10 (m, 1H, Ar-H), 10.75 (s, 1H, CONH), 12.73 (br s, 1H, pyridine-NH). 13C NMR (125 MHz, DMSO-d6) δ (ppm): 37.20, 46.50, 91.54, 105.51, 114.69, 118.17, 118.43, 119.05, 119.63, 126.30, 126.79, 127.03, 128.85, 129.14, 130.38, 131.64, 132.49, 135.49, 141.59, 147.16, 156.74, 158.94, 161.01, 161.29, 167.05, 170.13, 174.30. Anal. Calcd. For C31H22FN5O3S (563.61): C, 66.06; H, 3.93; N, 12.43; S, 5.69. Found: C, 66.20; H, 3.95; N, 12.46; S, 5.69.
3.2. Biology
3.2.1. Cell Viability Assay
The cell viability test utilized the human mammary gland epithelial cell line (MCF-10A) [62]. Compounds 7–21 were incubated with MCF-10A cells for a period of 4 days, followed by an assessment of cell survival through the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [9]. For more details, see Section A (Supplementary File).
3.2.2. Antiproliferative Assay
Compounds 7–21 were examined to determine their ability to inhibit cell proliferation in four types of human cancer cells: pancreatic cancer (Panc-1), breast cancer (MCF-7), colon cancer (HT-29), and epithelial cancer (A-549). The MTT assay [63,64] was employed for this assessment, with Doxorubicin as the reference drug. See Section A (Supplementary File).
3.2.3. EGFR Inhibitory Activity
The most effective antiproliferative hybrids, 8, 9, 18, and 19, were tested for EGFR-TK inhibitory activity as a possible target for their antiproliferative activity. See Section A (Supplementary File).
3.2.4. BRAFV600E Inhibitory Activity
An in vitro study was carried out to evaluate the activity of the newly synthesized compounds 8, 9, 18, and 19 against BRAFV600E [59]. See Section A (Supplementary File).
3.3. In Silico Studies
3.3.1. Docking Study
For the molecular docking study, we used BIOVIA Discovery Studio 2021 software v21.1.0.20.298. The selected proteins underwent preparation for docking analysis through the Protein Preparation Wizard. Subsequently, ligands were mapped onto a three-dimensional model and subjected to energy minimization using LigPrep. To potentially improve binding, a receptor grid was created for the selected binding site using the Receptor Grid Generation Tool. Finally, the Glide tool was employed to evaluate both docking scores and various binding modes for the ligands.
3.3.2. In Silico ADMET Analysis
BIOVIA I Discovery Studio 2016 was used for ADMET investigations. The chemical structures of all substances were imported, and ADMET descriptors were predicted using integrated models that included Lipinski’s Rule of Five and assessments of absorption, distribution, metabolism, excretion, and toxicity. The obtained results were examined to determine the drug-likeness and safety profiles of the compounds.
4. Conclusions
A novel series of hybrids, combining quinazolin-4-one and 3-cyanopyrid-2-one in a single compact structure, was synthesized and developed as dual-targeting antiproliferative agents. Using the MTT assay, these newly synthesized compounds were tested for antiproliferative activities against four cancer cell lines. Compounds 18 and 19 were the most potent antiproliferative agents, with GI50 values of 1.20 µM and 1.40 µM, respectively, compared to Doxorubicin’s GI50 of 1.10 µM. Compounds 18 and 19 suppressed EGFR efficiently, with IC50 values of 110 nM and 140 nM, respectively. Furthermore, with IC50 values of 140 nM and 190 nM, these hybrid compounds showed promising inhibition of BRAFV600E. As a result of their combined inhibition of EGFR and BRAFV600E, they have the potential to operate as effective antiproliferative agents. Molecular docking studies indicated that compound 18 is a potent inhibitor for EGFR and BRAFV600E kinase domains. Moreover, an ADMET study suggested that most compounds have low toxicity and limited adverse effects in the tested models. Additional in vitro and in vivo studies and potential chemical modifications may be required to advance the development of highly effective antiproliferative medicines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph16111522/s1.
Author Contributions
H.A.H., A.M.H. and M.A.-A.: Supervision, conceptualization, editing and revision. B.G.M.Y., M.H.: methodology, formal analysis, writing, editing and revision. S.B.: formal analysis, writing and editing. H.A.A.-Z.: Docking Study, L.H.A.-W.: Funding Acquisition, editing and revision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R3), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Data is contained within the article and supplementary material.
Acknowledgments
The authors acknowledge the support by Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R3), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors also acknowledge support from the KIT-Publication Fund of the Karlsruhe Institute of Technology.
Conflicts of Interest
The authors reported no potential conflict of interest.
References
- DeVita, V.T., Jr.; Rosenberg, S.A. Two hundred years of cancer research. N. Engl. J. Med. 2012, 366, 2207–2214. [Google Scholar] [CrossRef]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hyman, D.M.; Davids, M.S.; Siu, L.L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet 2020, 395, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, N.M.; Pingili, D.; Kadasi, S.; Mettu, A.; Prasad, S. Dual or multi-targeting inhibitors: The next generation anticancer agents. Eur. J. Med. Chem. 2018, 143, 1277–1300. [Google Scholar] [CrossRef] [PubMed]
- Elwaie, T.A.; Abbas, S.E.; Aly, E.I.; George, R.F.; Ali, H.; Kraiouchkine, N.; Abdelwahed, K.S.; Fandy, T.E.; El Sayed, K.A.; Ali, H.I.; et al. HER2 Kinase-Targeted Breast Cancer Therapy: Design, Synthesis, and In Vitro and In Vivo Evaluation of Novel Lapatinib Congeners as Selective and Potent HER2 Inhibitors with Favorable Metabolic Stability. J. Med. Chem. 2020, 63, 15906–15945. [Google Scholar] [CrossRef] [PubMed]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef]
- Youssif, B.G.; Abdelrahman, M.H.; Abdelazeem, A.H.; Ibrahim, H.M.; Salem, O.I.; Mohamed, M.F.; Bukhari, S.N.A. Design, synthesis, mechanistic and histopathological studies of small-molecules of novel indole-2-carboxamides and pyrazino [1, 2-a] indol-1 (2H)-ones as potential anticancer agents effecting the reactive oxygen species production. Eur. J. Med. Chem. 2018, 146, 260–273. [Google Scholar] [CrossRef]
- Youssif, B.G.; Gouda, A.M.; Moustafa, A.H.; Abdelhamid, A.A.; Gomaa, H.A.; Kamal, I.; Marzouk, A.A. Design and synthesis of new triarylimidazole derivatives as dual inhibitors of BRAFV600E/p38α with potential antiproliferative activity. J. Mol. Struct. 2022, 1253, 132218. [Google Scholar] [CrossRef]
- Seton-Rogers, S. Cytokine cues. Nat. Rev. Cancer 2011, 11, 690. [Google Scholar] [CrossRef]
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging new therapeutic antibody derivatives for cancer treatment. Signal Transduct. Target. Ther. 2022, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Cross, D.; Janne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Yahya, E.B.; Alqadhi, A.M. Recent trends in cancer therapy: A review on the current state of gene delivery. Life Sci. 2021, 269, 119087. [Google Scholar] [CrossRef]
- Avendaño, C.; Menendez, J.C. Medicinal Chemistry of Anticancer Drugs; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, T.E.; Glazer, P.M.; Slack, F.J.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015, 518, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Troike, K.; Lathia, J.D. Optimising gene editing for cancer therapy. Nat Cell Biol. 2020, 22, 259–261. [Google Scholar] [CrossRef]
- Tebbutt, N.; Pedersen, M.W.; Johns, T.G. Targeting the ERBB family in cancer: Couples therapy. Nat. Rev. Cancer 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Yarden, Y. Biology of HER2 and its importance in breast cancer. Oncology 2001, 61 (Suppl. 2), 1–13. [Google Scholar] [CrossRef]
- Pennock, S.; Wang, Z. Stimulation of cell proliferation by endosomal epidermal growth factor receptor as revealed through two distinct phases of signaling. Mol. Cell. Biol. 2003, 23, 5803–5815. [Google Scholar] [CrossRef]
- Barberan, S.; Cebria, F. The role of the EGFR signaling pathway in stem cell differentiation during planarian regeneration and homeostasis. Semin. Cell Dev. Biol. 2019, 87, 45–57. [Google Scholar] [CrossRef]
- Chiasson-MacKenzie, C.; McClatchey, A.I. EGFR-induced cytoskeletal changes drive complex cell behaviors: The tip of the iceberg. Sci. Signal. 2018, 11, eaas9473. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Schmidt, M.H.H. EGFR and EGFRvIII Promote Angiogenesis and Cell Invasion in Glioblastoma: Combination Therapies for an Effective Treatment. Int. J. Mol. Sci. 2017, 18, 1295. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.A.; Rakus, D.; Gizak, A.; Steelman, L.S.; Abrams, S.L.; Lertpiriyapong, K.; Fitzgerald, T.L.; Yang, L.V.; Fitzgerald, T.L.; Martelli, A.M.; et al. Effects of mutations in Wnt/β-catenin, hedgehog, Notch and PI3K pathways on GSK-3 activity—Diverse effects on cell growth, metabolism and cancer. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2016, 1863, 2942–2976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Castanaro, C.; Luan, B.; Yang, K.; Fan, L.; Fairhurst, J.L.; Rafique, A.; Potocky, T.B.; Shan, J.; Daly, C.; et al. ERBB3/HER2 signaling promotes resistance to EGFR blockade in head and neck and colorectal cancer models. Mol. Cancer Ther. 2014, 13, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Chmielecki, J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer 2010, 10, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Jadhav, H.R. Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov. Today 2018, 23, 745–753. [Google Scholar] [CrossRef]
- Yan, N.; Guo, S.; Zhang, H.; Zhang, Z.; Shen, S.; Li, X. BRAF-Mutated Non-Small Cell Lung Cancer: Current Treatment Status and Future Perspective. Front. Oncol. 2022, 12, 863043. [Google Scholar] [CrossRef]
- Murugesan, S.; Murugesan, J.; Palaniappan, S.; Palaniappan, S.; Murugan, T.; Siddiqui, S.S.; Loganathan, S. Tyrosine Kinase Inhibitors (TKIs) in Lung Cancer Treatment: A Comprehensive Analysis. Curr. Cancer Drug Targets 2021, 21, 55–69. [Google Scholar] [CrossRef]
- Cicenas, J.; Tamosaitis, L.; Kvederaviciute, K.; Tarvydas, R.; Staniute, G.; Kalyan, K.; Meskinyte-Kausiliene, E.; Stankevicius, V.; Valius, M. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med. Oncol. 2017, 34, 26. [Google Scholar] [CrossRef]
- Pakneshan, S.; Salajegheh, A.; Smith, R.A.; Lam, A.K.-Y. Clinicopathological relevance of BRAF mutations in human cancer. Pathology 2013, 45, 346–356. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A Review on Recent Advances in Nitrogen-Containing Molecules and Their Biological Applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Hisham, M.; Hassan, H.A.; Gomaa, H.A.M.; Youssif, B.G.M.; Hayallah, A.M.; Abdel-Aziz, M. Structure-based design, synthesis and antiproliferative action of new quinazoline-4-one/chalcone hybrids as EGFR inhibitors. J. Mol. Struct. 2022, 1254, 132422. [Google Scholar] [CrossRef]
- Dawood, D.H.; Nossier, E.S.; Abdelhameed, M.F.; Asaad, G.F.; Abd El-Rahman, S.S. Design, synthesis, anti-inflammatory evaluation and molecular docking of novel thiophen-2-ylmethylene-based derivatives as potential TNF-α production inhibitors. Bioorganic Chem. 2022, 122, 105726. [Google Scholar] [CrossRef] [PubMed]
- Hisham, M.; Hassan, H.A.; Gomaa, H.A.M.; Youssif, B.G.M.; Hayalah, A.M.; Abdel-Aziz, M. Design, Synthesis, and Antiproliferative Activity of Quinazolin-4-One/Chalcone Hybrids via the Egfr Inhibition Pathway. Anti-Cancer Agents Med. Chem. 2023, 23, 1932–1943. [Google Scholar] [CrossRef]
- Dutta, A.; Trivedi, P.; Gehlot, P.S.; Gogoi, D.; Hazarika, R.; Chetia, P.; Kumar, A.; Chaliha, A.K.; Chaturvedi, V.; Sarma, D. Design and Synthesis of Quinazolinone-Triazole Hybrids as Potent Anti-Tubercular Agents. ACS Appl. Bio Mater. 2022, 5, 4413–4424. [Google Scholar] [CrossRef]
- Zayed, M.F. Medicinal Chemistry of Quinazolines as Analgesic and Anti-Inflammatory Agents. Chemengineering 2022, 6, 94. [Google Scholar] [CrossRef]
- Łysakowska, M.; Głowacka, I.E.; Andrei, G.; Schols, D.; Snoeck, R.; Lisiecki, P.; Szemraj, M.; Piotrowska, D.G. Design, Synthesis, Anti-Varicella-Zoster and Antimicrobial Activity of (Isoxazolidin-3-yl)Phosphonate Conjugates of N1-Functionalised Quinazoline-2,4-Diones. Molecules 2022, 27, 6526. [Google Scholar] [CrossRef]
- Cohen, M.H.; Johnson, J.R.; Chen, Y.F.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Erlotinib (Tarceva) tablets. Oncologist 2005, 10, 461–466. [Google Scholar] [CrossRef]
- Das, D.; Hong, J. Recent advancements of 4-aminoquinazoline derivatives as kinase inhibitors and their applications in medicinal chemistry. Eur. J. Med. Chem. 2019, 170, 55–72. [Google Scholar] [CrossRef]
- Abdelgalil, A.A.; Al-Kahtani, H.M.; Al-Jenoobi, F.I. Erlotinib. Profiles Drug Subst. Excip. Relat. Methodol. 2020, 45, 93–117. [Google Scholar]
- Faidallah, H.M.; Rostom, S.A.F.; Badr, M.H.; Ismail, A.E.; Almohammadi, A.M. Synthesis of Some 1,4,6-Trisubstituted-2-oxo-1,2-dihydropyridine-3-carbonitriles and Their Biological Evaluation as Cytotoxic and Antimicrobial Agents. Arch. Pharm. 2015, 348, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Al-Etaibi, A.M.; El-Apasery, M.A. A Comprehensive Review on the Synthesis and Versatile Applications of Biologically Active Pyridone-Based Disperse Dyes. Int. J. Environ. Res. Public Health 2020, 17, 4714. [Google Scholar] [CrossRef] [PubMed]
- Mamedov, I.; Naghiyev, F.; Maharramov, A.; Uwangue, O.; Farewell, A.; Sunnerhagen, P.; Erdelyi, M. Antibacterial activity of 2-amino-3-cyanopyridine derivatives. Mendeleev Commun. 2020, 30, 498–499. [Google Scholar] [CrossRef]
- Abo-Ghalia, M.H.; Amr, A.E.-G.E.; Abdalah, M.M. Synthesis of Some New (Nα-Dipicolinoyl)-bis-L-leucyl-DL-norvalyl Linear tetra and Cyclic octa Bridged Peptides as New Antiinflammatory Agents. Z. Naturforschung B 2003, 58, 903–910. [Google Scholar] [CrossRef]
- Amr Ael, G.; Sayed, H.H.; Abdulla, M.M. Synthesis and reactions of some new substituted pyridine and pyrimidine derivatives as analgesic, anticonvulsant and antiparkinsonian agents. Arch. Pharm. 2005, 338, 433–440. [Google Scholar] [CrossRef]
- Abou-Zied, H.A.; Beshr, E.A.; Gomaa, H.A.; Mostafa, Y.A.; Youssif, B.G.; Hayallah, A.M.; Abdel-Aziz, M. Discovery of new cyanopyridine/chalcone hybrids as dual inhibitors of EGFR/BRAF(V600E) with promising antiproliferative properties. Arch. Pharm. 2023, 356, e2200464. [Google Scholar] [CrossRef]
- Ismail, M.M.F.; Farrag, A.M.; Harras, M.F.; Ibrahim, M.H.; Mehany, A.B.M. Apoptosis: A target for anticancer therapy with novel cyanopyridines. Bioorganic Chem. 2020, 94, 103481. [Google Scholar] [CrossRef]
- Abd Elhameid, M.K.; Ryad, N.; Al-Shorbagy, M.Y.; Mohammed, M.R.; Ismail, M.M.; El Meligie, S. Design, Synthesis and Screening of 4,6-Diaryl Pyridine and Pyrimidine Derivatives as Potential Cytotoxic Molecules. Chem. Pharm. Bull. 2018, 66, 939–952. [Google Scholar] [CrossRef]
- Abadi, A.H.; Abouel-Ella, D.A.; Lehmann, J.; Tinsley, H.N.; Gary, B.D.; Piazza, G.A.; Abdel-Fattah, M.A. Discovery of colon tumor cell growth inhibitory agents through a combinatorial approach. Eur. J. Med. Chem. 2010, 45, 90–97. [Google Scholar] [CrossRef][Green Version]
- Sabour, R.; Harras, M.F.; Mehany, A.B.M. Design, synthesis, cytotoxicity screening and molecular docking of new 3-cyanopyridines as survivin inhibitors and apoptosis inducers. Bioorganic Chem. 2020, 94, 103358. [Google Scholar] [CrossRef]
- Abdelaziz, M.E.; El-Miligy, M.M.M.; Fahmy, S.M.; Mahran, M.A.; Hazzaa, A.A. Design, synthesis and docking study of pyridine and thieno [2,3-b] pyridine derivatives as anticancer PIM-1 kinase inhibitors. Bioorganic Chem. 2018, 80, 674–692. [Google Scholar] [CrossRef] [PubMed]
- Abouzid, K.A.M.; Al-Ansary, G.H.; El-Naggar, A.M. Eco-friendly synthesis of novel cyanopyridine derivatives and their anticancer and PIM-1 kinase inhibitory activities. Eur. J. Med. Chem. 2017, 134, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Al-Wahaibi, L.H.; Abou-Zied, H.A.; Hisham, M.; Beshr, E.A.M.; Youssif, B.G.M.; Bräse, S.; Hayallah, A.M.; Abdel-Aziz, M. Design, Synthesis, and Biological Evaluation of Novel 3-Cyanopyridone/Pyrazoline Hybrids as Potential Apoptotic Antiproliferative Agents Targeting EGFR/BRAF(V600E) Inhibitory Pathways. Molecules 2023, 28, 6586. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.C.; Liao, W.Y.; Lin, C.A.; Shih, J.Y.; Yu, C.J.; Yang, J.C. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J. Thorac. Oncol. 2017, 12, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.; Markman, B.; Ananda, S.; Tebbutt, N.C.; Michael, M.; Solomon, B.J.; McArthur, G.A.; Tie, J.; Gibbs, P.; Ritchie, D.; et al. A phase I/II trial of combined BRAF and EGFR inhibition in patients (pts) with BRAF V600E mutated (BRAFm) metastatic colorectal (mCRC): The EViCT (Erlotinib and Vemurafenib in Combination Trial) study. J. Clin. Oncol. 2017, 35, 3557. [Google Scholar] [CrossRef]
- Remon, J.; Morán, T.; Majem, M.; Reguart, N.; Dalmau, E.; Márquez-Medina, D.; Lianes, P. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: A new era begins. Cancer Treat. Rev. 2014, 40, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Turner, E.; Chen, L.; Foulke, J.G.; Gu, Z.; Tian, F. CRISPR/Cas9 Edited RAS & MEK Mutant Cells Acquire BRAF and MEK Inhibitor Resistance with MEK1 Q56P Restoring Sensitivity to MEK/BRAF Inhibitor Combo and KRAS G13D Gaining Sensitivity to Immunotherapy. Cancers 2022, 14, 5449. [Google Scholar]
- Al-Wahaibi, L.H.; Gouda, A.M.; Abou-Ghadir, O.F.; Salem, O.I.; Ali, A.T.; Farghaly, H.S.; Abdelrahman, M.H.; Trembleau, L.; Abdu-Allah, H.H.; Youssif, B.G. Design and synthesis of novel 2, 3-dihydropyrazino [1, 2-a] indole-1, 4-dione derivatives as antiproliferative EGFR and BRAFV600E dual inhibitors. Bioorganic Chem. 2020, 104, 104260. [Google Scholar] [CrossRef]
- Gomaa, H.A.; Shaker, M.E.; Alzarea, S.I.; Hendawy, O.; Mohamed, F.A.; Gouda, A.M.; Ali, A.T.; Morcoss, M.M.; Abdelrahman, M.H.; Trembleau, L.; et al. Optimization and SAR investigation of novel 2, 3-dihydropyrazino [1, 2-a] indole-1, 4-dione derivatives as EGFR and BRAFV600E dual inhibitors with potent antiproliferative and antioxidant activities. Bioorganic Chem. 2022, 120, 105616. [Google Scholar] [CrossRef]
- Hisham, M.; Youssif, B.G.M.; Osman, E.E.A.; Hayallah, A.M.; Abdel-Aziz, M. Synthesis and biological evaluation of novel xanthine derivatives as potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019, 176, 117–128. [Google Scholar] [CrossRef]
- Youssif, B.G.; Mohamed, A.M.; Osman, E.E.A.; Abou-Ghadir, O.F.; Elnaggar, D.H.; Abdelrahman, M.H.; Treamblu, L.; Gomaa, H.A. 5-Chlorobenzofuran-2-carboxamides: From allosteric CB1 modulators to potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019, 177, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, H.A.M.; El-Sherief, H.A.M.; Hussein, S.; Gouda, A.M.; Salem, O.I.A.; Alharbi, K.S.; Hayallah, A.M.; Youssif, B.G.M. Novel 1, 2, 4-triazole derivatives as apoptotic inducers targeting p53: Synthesis and antiproliferative activity. Bioorganic Chem. 2020, 105, 104369. [Google Scholar] [CrossRef] [PubMed]
- Marzouk, A.A.; Abdel-Aziz, S.A.; Abdelrahman, K.S.; Wanas, A.S.; Gouda, A.M.; Youssif, B.G.; Abdel-Aziz, M. Design and synthesis of new 1, 6-dihydropyrimidin-2-thio derivatives targeting VEGFR-2: Molecular docking and antiproliferative evaluation. Bioorganic Chem. 2020, 102, 104090. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.A.; Mohammed, A.F.; Salem, O.I.; Gomaa, H.A.; Youssif, B.G. New 1, 3, 4-oxadiazoles linked with the 1, 2, 3-triazole moiety as antiproliferative agents targeting the EGFR tyrosine kinase. Arch. Pharm. 2022, 355, 2200009. [Google Scholar] [CrossRef] [PubMed]
- Jejurikar, B.L.; Rohane, S.H. Drug designing in discovery studio. Indian J. 2021, 14, 135–138. [Google Scholar]
- Ibrahim, T.S.; Bokhtia, R.M.; Al-Mahmoudy, A.M.; Taher, E.S.; AlAwadh, M.A.; Elagawany, M.; Abdel-Aal, E.H.; Panda, S.; Gouda, A.M.; Asfour, H.Z.; et al. Design, synthesis and biological evaluation of novel 5-((substituted quinolin-3-yl/1-naphthyl) methylene)-3-substituted imidazolidin-2, 4-dione as HIV-1 fusion inhibitors. Bioorganic Chem. 2020, 99, 103782. [Google Scholar] [CrossRef] [PubMed]
- Shaykoon, M.S.; Marzouk, A.A.; Soltan, O.M.; Wanas, A.S.; Radwan, M.M.; Gouda, A.M.; Youssif, B.G.; Abdel-Aziz, M. Design, synthesis and antitrypanosomal activity of heteroaryl-based 1, 2, 4-triazole and 1, 3, 4-oxadiazole derivatives. Bioorganic Chem. 2020, 100, 103933. [Google Scholar] [CrossRef]
- Bhat, M.A.; Tüzün, B.; Alsaif, N.A.; Khan, A.A.; Naglah, A.M. Synthesis, characterization, molecular modeling against EGFR target and ADME/T analysis of novel purine derivatives of sulfonamides. J. Mol. Struct. 2022, 1257, 132600. [Google Scholar] [CrossRef]
- Umar, A.B.; Uzairu, A.; Shallangwa, G.A.; Uba, S. QSAR modelling and molecular docking studies for anti-cancer compounds against melanoma cell line SK-MEL-2. Heliyon 2020, 6, e03640. [Google Scholar] [CrossRef]
- Wu, Z.; Lei, T.; Shen, C.; Wang, Z.; Cao, D.; Hou, T. ADMET evaluation in drug discovery. 19. Reliable prediction of human cytochrome P450 inhibition using artificial intelligence approaches. J. Chem. Inf. Model. 2019, 59, 4587–4601. [Google Scholar] [CrossRef]
- Abd El-Sattar, N.E.; Badawy, E.H.; AbdEl-Hady, W.H.; Abo-Alkasem, M.I.; Mandour, A.A.; Ismail, N.S. Design and synthesis of new CDK2 inhibitors containing thiazolone and thiazolthione scafold with apoptotic activity. Chem. Pharm. Bull. 2021, 69, 106–117. [Google Scholar] [CrossRef]
- Rai, H.; Barik, A.; Singh, Y.P.; Suresh, A.; Singh, L.; Singh, G.; Nayak, U.Y.; Dubey, V.K.; Modi, G. Molecular docking, binding mode analysis, molecular dynamics, and prediction of ADMET/toxicity properties of selective potential antiviral agents against SARS-CoV-2 main protease: An effort toward drug repurposing to combat COVID-19. Mol. Divers. 2021, 25, 1905–1927. [Google Scholar] [CrossRef]
- Roy, P.P.; Roy, K. QSAR Studies of CYP2D6 Inhibitor Aryloxypropanolamines Using 2D and 3D Descriptors. Chem. Biol. Drug Des. 2009, 73, 442–455. [Google Scholar] [CrossRef]
- Xia, X.; Maliski, E.G.; Gallant, P.; Rogers, D. Classification of Kinase Inhibitors Using a Bayesian Model. J. Med. Chem. 2004, 47, 4463–4470. [Google Scholar] [CrossRef] [PubMed]
- Goodrnan, G.; Wilson, R. Comparison of the Dependence of the TD50 on Maximum Tolerated Dose for Mutagens and Nonmutagens. Risk Anal. 1992, 12, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Gonella Diaza, R.; Manganelli, S.; Esposito, A.; Roncaglioni, A.; Manganaro, A.; Benfenati, E. Comparison of in silico tools for evaluating rat oral acute toxicity. SAR QSAR Environ. Res. 2015, 26, 1–27. [Google Scholar] [CrossRef]
- Wilhelmus, K.R. The Draize Eye Test. Surv. Ophthalmol. 2001, 45, 493–515. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).