Abstract
Osteogenesis imperfecta (OI) is a group of connective tissue disorders leading to abnormal bone formation, mainly due to mutations in genes encoding collagen type I (Col I). Osteogenesis is regulated by a number of molecules, including microRNAs (miRNAs), indicating their potential as targets for OI therapy. The goal of this study was to identify and analyze the expression profiles of miRNAs involved in bone extracellular matrix (ECM) regulation in patients diagnosed with OI type I caused by mutations in COL1A1 or COL1A2. Primary skin fibroblast cultures were used for DNA purification and sequence analysis, followed by analysis of miRNA expression. Sequencing analysis revealed mutations of the COL1A1 or COL1A2 genes in all OI patients, including four previously unreported. Amongst the 40 miRNAs analyzed, 9 were identified exclusively in OI cells and 26 in both OI patients and the controls. In the latter case, the expression of six miRNAs (hsa-miR-10b-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, has-miR-204-5p, has-miR-216a-5p, and hsa-miR-449a) increased, while four (hsa-miR-129-5p, hsa-miR-199b-5p, hsa-miR-664a-5p, and hsa-miR-30a-5p) decreased significantly in OI cells in comparison to their expression in the control cells. The identified mutations and miRNA expression profiles shed light on the intricate processes governing bone formation and ECM regulation, paving the way for further research and potential therapeutic advancements in OI and other genetic diseases related to bone abnormality management.
1. Introduction
Osteogenesis imperfecta (OI) is a group of genetically related connective tissue disorders, diverse in terms of both genotypes and phenotypes, characterized by increased bone fragility. Clinically, five types of OI ranging from mild (type I) to moderate (types III and IV) and severe or lethal (type II) are distinguished [1,2,3]. So far, numerous mutations in genes encoding collagen type I (Col I) or involved in collagen modification, e.g., cartilage-associated protein (CRTAP) and leucine proline-enriched proteoglycan 1 (LEPRE1) [4,5]; folding, e.g., serpin peptidase inhibitor (SERPINH1) [6]; and processing, e.g., bone morphogenetic protein 1 (BMP1) [7], have been shown to cause various types of OI. Nevertheless, a majority of OI cases inherited in an autosomal-dominant manner are caused by mutations in the collagen type I α1 or α2 encoding genes (COL1A1 and COL1A2, respectively), resulting in a reduced amount or abnormal structure of collagen [8,9,10].
The earliest stage when an OI diagnosis can be made is the prenatal period. It depends on the severity of the OI and is based on family history; genetic tests, i.e., sequencing of genomic DNA and specific mutation identification; and prenatal tests, i.e., a decrease in the femoral length (FL) [11]. In the post-natal period, positive family history and pivotal clinical symptom (bone fragility, typical features of blue sclera) notification with bone density measurement (BMD) implementation are routinely used in OI diagnosis [12,13]. Additional diagnostic methods include magnetic resonance imaging (MRI), radiography of the uterus, and other genetic tests, such as multiplex ligation-dependent probe amplification (MLPA) or chorionic villus sampling (CVS) [12,14]. If the typical clinical symptoms and positive family history are noted by the clinician, routine diagnostic methods are sufficient to diagnose OI; however, about 10–15% of patients suffer from less common variants of the disease. Moreover, any diagnostic method used may be clear-cut and have drawbacks, e.g., employment of the FL carries the risk of other skeletal dysplasia’s recognition instead of OI [13,15]. In such circumstances, further procedures, mostly based on molecular diagnostics, are needed [11].
MicroRNA (miRNA) constitutes short, about 20-nucleotide, single-stranded, and endogenous RNA molecules that have been observed to negatively regulate gene expression in two manners: i) by attaching to their target messenger RNA (mRNA) at different mRNA sites, most often 100% complementary, and cutting it; or ii) by binding to the 3′-untranslated region (3′UTR) of the target mRNA and inhibiting translation, an action that does not require 100% complementarity [16]. The principal role of miRNAs is the regulation of a number of biological processes, such as embryogenesis, apoptosis, hosting immune responses during infections, or cancerogenesis [16]. miRNAs fulfill a crucial role in the regulation of gene expression and are involved in bone formation and bone diseases [17,18,19]. For instance, miR-217 has been reported to promote cell proliferation and osteogenic differentiation during the development of steroid-associated osteonecrosis in bone marrow mesenchymal stem cells [20,21], whereas miR-200a-3p has been linked with the synthesis of ECM components in fibrotic diseases [22].
Over the past two decades, the mechanisms of human extracellular matrix (ECM) regulation by miRNAs have been extensively studied, including the genes involved in collagen synthesis. For example, the negative influence of hsa-miR-133a-3p, which has been assigned a possible antifibrotic role [23,24]; hsa-miR-27b-3p, which increases collagen expression [25]; and hsa-miR-382-5p, which promotes epidural fibrosis [26,27], has been demonstrated. Furthermore, many miRNAs that can regulate genes involved in the enzymatic processing or degradation of natively synthesized collagen have been recently discovered. For example, hsa-miR-377-3p has been shown to inhibit the synthesis of matrix metalloproteinase-2 (MMP-2) and MMP-16, whereas hsa-miR-200b-3p has been proven to regulate tissue inhibition of metalloproteinase-2 (TIMP2) [26]. Moreover, miRNAs can affect signaling pathways involved in ECM synthesis and degradation; e.g., hsa-miR-216a-5p has been shown to regulate Janus kinase 2 (JAK2) [28,29], while hsa-miR-25-3p has been shown to control the transforming growth factor β receptor I (TGFβR1) and neurogenic locus notch homolog protein 1 (NOTCH1) signaling pathways [30,31]. To date, however, only a few miRNAs, such as let-7a-5p, miR-92a, and miR-29b, have been studied in OI [17,18].
The aim of this study was to identify and analyze the expression profiles of miRNAs in osteogenesis imperfecta (OI) type I caused by mutations in the COL1A1 and COL1A2 genes.
2. Results
2.1. Mutation Analysis in the COL1A1 and COL1A2 Genes from OI Type I
Osteogenesis imperfecta type I in the majority of cases (~95%) is caused by mutations in the COL1A1 and COL1A2 genes; hence, in this study, we focused on sequencing analyses of those genes [1,2]. Mutations in COL1A1 were found in 8 study subjects and in COL1A2 in 2 out of 10 study subjects diagnosed with OI type I. The missense mutations were detected in both COL1A1 (donor nos. 1–4) and COL1A2 (donor nos. 9 and 10). In COL1A1, other mutation types were also found, including a single-nucleotide deletion resulting in a frameshift in three donors (nos. 5–7) and an exon skipping in one patient (no. 8). The missense mutations detected in the COL1A1 gene resulted in three different substitutions, namely glycine by alanine in exon 17 (patient no. 1), glycine by valine in exon 9 (patient no. 2), and a third one also found in exon 17 that resulted in valine-to-phenylalanine alteration (patient no. 3). Additionally, in two other patients (nos. 6 and 7), two deletions of T located in exon 30 and exon 17 of the COL1A1 gene resulted in the production of an altered protein. In contrast, both missense mutations in the COL1A2 gene resulted in the substitution of glycine by serine in exon 31 (patient no. 9) or exon 38 (patient no. 10).
Moreover, we identified a series of new mutations in both genes: substitution of guanine for thymine in exon 9 in patient no. 2 and deletion of 10 nucleotides in exon 50 (frameshift) adjacent to a missense mutation (substitution of glutamine by valine) in OI patient no. 4, both in the COL1A1 gene. Another alteration comprised a guanine deletion in exon 2 of the COL1A1 gene and was identified in patient no. 5. A new mutation found in the COL1A2 gene was represented by a substitution of guanine for adenine in exon 31 of patient no. 9.
A brief clinical description of OI patients harboring previously reported mutations as well as the new mutations in COL1A1 and COL1A2 identified in this study are presented in Table 1.

Table 1.
Brief clinical summary and characterization of mutations detected in the COL1A1 and COL1A2 genes of OI donors enrolled in this study.
2.2. Identification of miRNAs in OI Type I
First, we performed identification analyses of the miRNAs in the OI-derived cells and control cells. As shown in Table 2, the results were categorized into the three following parts: (A) miRNAs detected in OI but not in the control fibroblasts; (B) miRNAs detected in both OI and the control cells; (C) miRNAs not detected in OI nor the control cells.

Table 2.
Characteristics of analyzed miRNAs.
As presented in Table 2, nine specific miRNAs, i.e., hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-133a-3p, hsa-miR-200b-3p, hsa-miR-204-5p, 216a-5p, hsa-miR-377-3p, hsa-miR-449a, and hsa-miR-590-5p, were identified in the fibroblasts of 30% to 70% of OI donors, but not in the control fibroblasts. Other 26 miRNA molecules were detected in both the OI and the control cells, with the percentage of OI donors with a specific miRNA detection ranging from 70% to 100%. Finally, five miRNAs selected for analysis, i.e., hsa-miR-1-3p, hsa-miR-141-3p, hsa-miR-142-3p, hsa-miR-200a-3p, and hsa-miR-217, were not detected in the OI or the control cells.
2.3. Expression Analysis of miRNAs in OI
Based on the miRNA identification, an expression analysis was performed for miRNAs detected in >50% of OI patients and compared with the controls. As depicted in Figure 1, increased expression was observed in the case of six miRNAs (hsa-miR-10b-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, has-miR-204-5p, has-miR-216a-5p, and hsa-miR-449a), while decreased expression was observed in the case of four miRNAs (hsa-miR-129-5p, hsa-miR-199b-5p, hsa-miR-664a-5p, and hsa-miR-30a-5p) in OI as compared to the controls. Among the miRNAs analyzed, miR-449a’s expression increased about 20-fold, followed by an almost 5-fold increase in miR-19b-3p and a ~4-fold increase in miR-19a-3p. Meanwhile, an almost 6-fold decrease in expression in OI was found in the case of miR-664a-5p, whereas a slightly less decrease was observed for miR-129-5p (3.6-fold), miR-199b-5p (2.8-fold), miR-30a-5p (2.2-fold), and miR-25-3p (1.9-fold) (Table 3).
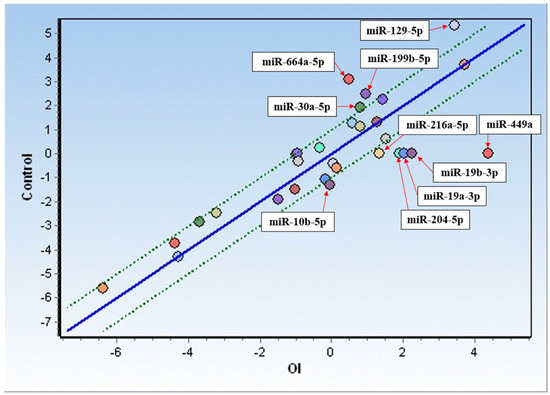
Figure 1.
Comparison of miRNA expression in fibroblasts from OI donors (OI) and control cells. Only upregulated (below the green line) and downregulated (above the green line) miRNAs have been presented.

Table 3.
Fold change in miRNA expression in OI vs. controls.
2.4. miRNA Correlation Analysis in OI
Among the miRNAs that were overexpressed in the OI cells, two sets displaying moderate-to-strong positive correlations within each group were observed (Table 4). The first group consisted of miR-10a-5p, miR-10b-5p, miR-199a-5p, and miR-382-5p, and the second consisted of miR-19a-3p, miR-19b-3p, miR-204-5p, miR-216a-5p, and miR-449a. miR-17-5p, on the other hand, did not show strong correlations with any of the overexpressed miRNAs.

Table 4.
Spearman coefficient for miRNAs overexpressed in OI. Moderate correlation (0.4–0.6), light-green background; strong correlation (0.6–0.9), green background; complete correlation (1), yellow background; p-values are depicted in Supplementary Materials—Table S2.
A significantly decreased expression in the OI group was noted for 13 miRNAs (Table 5). In this group, an almost complete correlation was found for miR-21-5p, miR-25-3p, miR-26a-5p, miR-26b-5p, miR-27a-3p, miR-27b-3p, miR-29a-3p, and miR-29c-3p, which have been previously reported to regulate the expression of genes associated with fibrosis (cystic fibrosis in children or liver fibrosis) and cancerogenesis [24,25,48,49,56]. Further, miR-30a-5p showed a strong correlation with almost all miRNAs presented in Table 5.

Table 5.
Spearman coefficient for miRNAs downregulated in OI. Moderate correlation (0.4–0.6), light-green background; strong correlation (0.6–0.9), green background; complete correlation (1), yellow background; moderate negative correlation ((−0.6)–(−0.4)), light-blue background; p-values are depicted in Supplementary Materials—Table S2.
In contrast, a moderately negative correlation of miR-129-5p with miR-21-5p, miR-26b-5p, miR-29a-3p, and miR-29c-3p was observed. This may indicate an antagonistic function of miR-129-5p in the regulation of collagen synthesis. Finally, the weakest correlations with other tested downregulated miRNAs were observed for miR-199b-5p and miR-29b-3p (Table 5).
2.5. Association of Identified miRNAs with Mutations in COL1A1 and COL1A2
In the group of OI patients harboring mutations in COL1A1, upregulated expression of miR-17-5p, miR-21-5p, miR-26b-3p, miR27a-3p, miR27b-3p, miR29a-3p, mir29c-3p, miR-30a-5p, miR-34a-5p, miR-92a-3p, and miR143-3p was found. In contrast, in the OI caused by mutations in COL1A2, only miR-25-3p was found to be upregulated (Table 6).

Table 6.
List of downregulated and overexpressed miRNAs analyzed individually for each OI donor. The table depicts the expression trends of miRNAs vs. controls.
Interestingly, the expression of miR-132-3p was comparable in the controls and the COL1A1-mutated fibroblasts but increased in the group of OI caused by mutations in COL1A2 (Table 6). These observations may suggest that miR-25-3p and miR-132-3p could play a particularly significant role in the pathogenetic processes of OI induced by mutations in the COL1A2 gene.
Considering the type of mutation, it was noted that patients no. 1, 2, and 3 with missense mutations in the COL1A1 gene revealed similar overexpression patterns for several miRNAs, indicating that these mutations may have similar effects on miRNA regulation. Patients no. 4, 6, and 7, with frameshift mutations in the COL1A1 gene, displayed similar overexpression patterns for some miRNAs but also individually dependent differences among particular types of miRNAs, and the profiles of expression also varied from those found in OI with missense mutations. In turn, patient no. 5, with a frameshift mutation in the COL1A1 gene, had a unique expression pattern of overexpressed and downregulated miRNAs. In summary, the miRNA expression patterns observed in the individually analyzed OI patients indicate that particular mutations may influence the regulation of specific miRNAs, potentially leading to downstream effects on genes involved in bone development and maintenance (Table 6).
3. Discussion
Despite similar and uniform phenotypic symptoms, the molecular mechanisms of pathogenetic processes contributing to OI remain unclear. This disorder is difficult to treat as it can be caused not only by direct defects in the amount of type I collagen produced but also by the influence of various proteins and regulatory RNA molecules, like miRNAs, on the processing of synthesized collagen, including the pool of aberrant proteins.
Around 5% of OI cases have an unknown genetic background. DNA sequencing analyses revealed the presence of mutations in both genes of all OI donors enrolled in this study, including four novel and previously unreported, of which three were identified in the COL1A1 gene and one in the COL1A2 gene. The missense mutations detected in the COL1A1 and COL1A2 genes resulted in the substitution of glycine and other amino acids, leading to disruptions in the collagen chain interactions and destabilization of the collagen triple helix [1,76]. Frameshift mutations, including single-nucleotide deletion, ten-nucleotide deletion, and exon skipping in the COL1A1 gene, altered the reading frame, resulting in truncated or absent Col I chains. Additionally, a single-nucleotide deletion in exon 2 of COL1A1 disrupted the formation of the collagen triple helix. These mutations likely contribute to the development of OI by affecting the stability and packing of the collagen molecule [77]. The mutations identified in this study align with previous reports on OI, reaffirming that mutations in the COL1A1 and COL1A2 genes are responsible for the majority of OI cases [78,79].
This research focused on the investigation of the miRNAs that regulate the expression of the ECM, including Col I processing and trafficking in the context of OI pathogenesis. Nine miRNAs that were identified in OI but not in the controls, namely hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-133a-3p, hsa-miR-200b-3p, hsa-miR-204-5p, 216a-5p, hsa-miR-377-3p, hsa-miR-449a, and hsa-miR-590-5p, have been previously reported to regulate the TGF-β and Wnt signaling pathways, indicating their essential role in the regulation of genes involved in bone formation and turnover [28,29,32,33,34].
Five out of forty analyzed miRNAs, i.e., hsa-miR-1-3p, hsa-miR-141-3p, hsa-miR-142-3p, hsa-miR-200a-3p, and hsa-miR-217, were absent in the OI group and the controls. These molecules affected suppressor genes, such as the phosphatase and tensin homolog (PTEN) and the dickkopf WNT signaling pathway inhibitor 1 (DKK1), as well as genes involved in ECM turnover, i.e., Rho-associated coiled-coil-containing protein kinase 2 (ROCK2) [20,21,22,71,72,73,74,75]. These miRNAs have not been directly linked to genes related to collagen synthesis and secretion or connective tissue diseases; however, they participate in the inhibition of the epithelial–mesenchymal transition in renal tubular epithelial cells (e.g., miR-141-3p) and the promotion of cell proliferation and osteogenic differentiation during the development of steroid-associated osteonecrosis in bone marrow mesenchymal stem cells (e.g., miR-217) [21,73] (Table 2). Moreover, hsa-miR-200a-3p has been found to participate in the overproduction of ECM proteins in fibrotic diseases.
The involvement of miRNAs in ECM regulation has recently become an area of intense research, as their ability to modulate gene expression post-transcriptionally makes them potent regulators of crucial cellular processes [17,18,19]. Understanding how these miRNAs affect ECM components will be of great importance in the search for new therapeutic targets for therapies for diseases associated with ECM dysregulation.
The expression analysis performed herein showed altered expression of 10 miRNAs in the fibroblasts of OI patients compared to the controls. The expression of six, i.e., hsa-miR-10b-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, has-miR-204-5p, has-miR-216a-5p, and hsa-miR-449a, was substantially increased, while the expression of four (hsa-miR-129-5p, hsa-miR-199b-5p, hsa-miR-664a-5p, and hsa-miR-30a-5p) decreased.
Further, the fold change analysis provided robust validation of the observed miRNA expression alterations in OI compared to the controls, displaying a significant multiplication in up- or downregulation. Remarkably, miR-449a’s expression increased about 20-fold, while miR-449a was exclusively detected in OI patients, which may suggest the potential relevance of both miRNAs in the pathogenesis of OI.
In contrast, we noted a six-fold expression decrease in miR-664a-5p. Though this miRNA has been implicated in the modulation of tumor suppressor p53 and the expression of p53 target genes and has recently been associated with osteosarcoma [70], this finding adds an intriguing dimension to this study, hinting at potential links between OI and p53-related pathways that warrant further investigation.
The intricate relationships between miRNAs may offer a unique opportunity to establish connections between molecular processes, providing insights into the pathogenesis of diverse diseases. For better clarification of the impact of miRNAs on bone ECM changes in OI, we explored their potential correlations, whether positive or negative. Correlation analyses were carried out within the groups with upregulated and downregulated expressions of miRNAs. The miRNAs overexpressed in OI that strongly correlate with each other are involved in the regulation of osteoblast differentiation and osteogenesis [26,27,44,45,46,66,67], while the miRNAs presented in the latter group have been previously associated with various biological processes and diseases, including cancer, inflammation, and neuronal differentiation [28,29,32,33,34,36,37,38]. With the exception of miR-129-5p, which showed a moderate negative correlation with four other miRNAs associated with various cancers, both groups of analyzed miRNAs revealed positive correlations with miR-29 family members [49,52,56]. The miR-29 family negatively regulates collagen expression by directly targeting COL1A1’s and COL1A2’s mRNA transcripts, while the other miRNAs are linked to fibrosis and cancers, suggesting their effect on the synthesis of the ECM [30,31,48,49,50,51].
In the present study, 40 miRNAs in total were analyzed in 10 OI donors in whom mutations, including four unreported ones, were identified in COL1A1/COL1A2. Overexpression of miR-10b-5p and miR-382-5p in some patients suggests their involvement in the pathogenesis of OI. On the other hand, decreased expression of miR-129-5p, miR-199b-5p, miR-29b-3p, and miR-664a-5p in most patients suggests their importance in normal bone development and function (Table 6). Kaneto et al. reported decreased expression of miR-29b during the osteoblastic differentiation of mesenchymal stem cells (MSCs). Likewise, in our research, we observed a modest decrease in the expression of miR-29b in skin fibroblasts from a group of patients diagnosed with OI type I. Upon conducting separate analyses for each OI patient, we observed an upregulation of miR-29b expression in certain individuals, which contrasts with the overall trend observed in the entire OI group. This variability may be attributed to the diverse range of patient demographics, including age and gender. miR-29 family members, including miR-29a, miR-29b, and miR-29c, have been implicated in the regulation of ECM proteins, such as collagen, which are important for bone development and homeostasis [52,53,54,55,56]. The aberrant regulation of those miRNAs in patients with OI type I induced by mutations in COL1A1 may contribute to reduced collagen synthesis and impaired bone formation. Regarding OI patients harboring mutations in the COL1A2 gene, we observed overexpression of miRNA-25-3p. Noticeably, this miRNA has been implicated in the regulation of bone mineralization and may play a role in osteoblast differentiation [30,31]. We also observed some discrepancies in the expression of certain miRNAs, such as let-7a, which was found to be overexpressed in some patients with missense mutations, but its expression was downregulated in patients with an exon-skipping mutation. Interestingly, one patient (no. 5) with a novel identified mutation that resulted in a frameshift in exon 2 of the COL1A1 gene displayed a distinct miRNA expression pattern compared to other examined patients, which may suggest a unique background of OI in this individual.
One of the potential therapies for OI based on targeting the miRNAs involved in the regulation of bone formation and remodeling can be applied by either increasing the expression of beneficial miRNAs or decreasing the expression of harmful ones [80,81,82]. However, there are several challenges that need to be addressed before its implementation into clinical practice, including: (i) the identification of disease-specific and most effective targeted miRNAs (as discussed in the current work); (ii) the validation of relevant delivery methods that would not only effectively “reach” the targeted micromolecule but also efficiently deliver miRNAs to the bone; and (iii) the overcoming of the immune response to exogenous miRNAs [83].
4. Conclusions
Despite similar and uniform phenotypic symptoms in individuals with OI, the molecular basis of this disease seems to be varied and still unclear. Moreover, it is difficult to treat, as it can be caused not only by direct defects in the amount of type I collagen produced but also by the influence of various proteins and regulatory RNA molecules, including miRNAs, on the processing of synthesized collagen and the pool of aberrant proteins. Therefore, understanding the function of miRNAs that can affect collagen or signaling pathways that indirectly regulate the amount or quality of collagen is extremely important and necessary. Overall, the current study underscores the importance of the ECM in the development of OI and other genetic diseases of connective tissue. The identified mutations and miRNA expression profiles shed light on the intricate processes governing bone formation and ECM regulation, paving the way for further research and potential therapeutic advancements in OI and other genetic diseases related to bone abnormality management.
5. Materials and Methods
5.1. Study Participants and Control Material
This study was carried out in accordance with the Declaration of Helsinki and was approved by the Bioethics Committee of the Jagiellonian University in Krakow, Poland (protocol no. DK/KB/CM/0031/689/2010). Clinical examination and medical histories of the subjects were taken by a clinician from the Chair of Pediatrics, Department of Medical Genetics, UJCM, Krakow, Poland. All subjects (or representing individuals) read and signed a written informed consent form prior to inclusion in this study.
Ten subjects (ranging in age from 0.5 to 29 years, of both sexes, and diagnosed with OI type I) were enrolled in this study. The diagnosis was confirmed on the basis of multiple (from ~9 to 20) bone fractures in the post-natal period and/or blue sclera or osteoporosis. The inclusion criterion was a clinically confirmed diagnosis of OI type I. Exclusion criteria included the clinical diagnosis of other types of OI and absence of mutations in the COL1A1 and/or COL1A2 genes. A clinical and molecular description of the OI type I subjects enrolled in this study is presented in Table 1. As a control, the human fibroblast cell line BJ (CRL-2522, ATCC, Manassas, VA, USA) was used.
5.2. Isolation Procedure of Skin Fibroblasts from OI Donors
Skin biopsy samples were collected from 10 patients diagnosed with OI type I. To establish a fibroblast culture, skin biopsies of a size of about 3–5 mm2 were cut into smaller pieces and placed in a Petri dish with 10 mL of Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 4.5 g/L glucose, 10% fetal calf serum (FCS), 50 μg/mL streptomycin, 50 U/mL penicillin, and 25 µg/mL amphotericin B (PAA, Austria). After one week, the remaining parts of the skin explants were removed from the dish, and the fibroblast culture was kept until 90% confluence. Subsequently, the cells were harvested using trypsin/EDTA, plated in fresh DMEM with 4.5 g/L glucose, 10% FCS, streptomycin, penicillin, and amphotericin B in plastic flasks, and kept until homogeneous cultures were obtained. The homogeneity of the fibroblast culture was confirmed after first passage by immunofluorescent staining against vimentin. Cell viability amounted to 98% using Trypan blue.
5.3. Fibroblast Cultures
For the experiments, 2 × 105 cells per well were plated in 6-well culture plates and maintained for 7 days in DMEM with 4% FBS supplemented every second day with a fresh L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich, St. Louis, MO, USA) at a concentration of 40 μg/mL. The experiments were carried out between the third and sixth passages. Control human fibroblast cell line BJ (CRL-2522, ATCC, Manassas, VA, USA) was cultured and treated identically as the primary skin fibroblasts recovered from the study group.
5.4. Fibroblast DNA Isolation and Amplification
Lysis and DNA isolation from primary skin fibroblasts and the control cell line were conducted according to the manufacturer’s protocol using DNA purification kit Blood Mini (A&A Biotechnology, Gdansk, Poland). The DNA amplification reactions were conducted using FastStartTaq DNA polymerase kit (Roche, Basel, Switzerland). The composition of the reaction mixture and conditions used are described elsewhere [84,85]. Briefly, for each gene, 10 amplification reactions were conducted, and the DNA fragments were visualized in 1% agarose gel (Agarose, LE Analytical Grade, Promega, Madison, WI, USA) in TAE buffer.
5.5. Sequence Analyses of COL1A1 and COL1A2 Genes from OI Donors
Sequencing PCR for the modified Sanger’s enzymatic method was conducted according to the recommendation provided by the manufacturer of the ABI Prism 3130xl DNA Analyzer. In brief, 20 ng of each purified amplified PCR product was mixed with 5 pM of the appropriate sequencing primer and Big Dye Terminator v.3.1, as previously described [84,85]. Before sequencing analysis, DNA fragments were purified employing the BigDye XTerminator Purification Kit (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s manual. Bioinformatic analysis of the DNA sequences obtained was conducted by the Chromas Lite 2.01 and NCBI Nucleotide Blast software against the COL1A1 (NG_007400.1) and COL1A2 (NG_007405.1) genes as the reference sequences.
5.6. Fibroblast RNA Isolation
Primary skin fibroblasts and control cell line cultures were harvested with trypsin/EDTA (PAA Laboratories, Colbe, Germany) and washed twice with sterile phosphate buffered saline (PBS) (PAA Laboratories, Colbe, Germany). The cell amount was counted using Trypan Blue (BIO-RAD, Hercules, CA, USA) in the TC-20 Automated Cell Counter (BIO-RAD, Hercules, CA, USA). Cell lysis and purification of total RNA and miRNA were conducted using 2.5 × 106 cells in total. The isolation procedure was performed with the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The purity and concentration of the collected RNA samples were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
5.7. Real-Time Quantitative Polymerase Chain Reaction
The miScript II RT Kit (Qiagen, Hilden, Germany) and an RNA concentration of 250 ng were used to synthesize cDNA. The obtained cDNA was used to examine the defined miRNA expression (Table 2). For this purpose, expression analysis of Custom miScript miRNA PCR Array, conf. no. CMIHS02722F (Qiagen, Hilden, Germany), was performed in a thermal cycler, the LightCycler 480 II (Roche, Basel, Switzerland). The quantitative polymerase chain reaction analysis was carried out in quadruplicate with miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) according to the manufacturer’s manual.
5.8. Statistical Analyses
The expression analysis of miRNAs was performed by GenEx ver6 software (MultiD Analyses AB). Raw data were normalized to the reference genes (SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A, and RNU6-2) according to the manufacturer’s protocol. The Kolmogorov–Smirnov test was employed to determine the distribution of the data (Supplementary Table S1). For miRNA data that presented normal distribution, a parametric, unpaired, one-tailed t-test was conducted. In the case of miRNAs that showed non-normal distribution of data, a one-tailed Mann–Whitney test was used. The results were considered statistically significant at p < 0.05. The correlation analyses of miRNAs were performed using the Spearman method. Comparative expression of miRNAs in the study group and the control cell line was determined by scatter plot analysis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ph16101414/s1: Figure S1: The relative expression of the tested miRNAs in subjects with mutations in the COL1A1 and COL1A2 genes and the control cells; Table S1: Normality test; Table S2: p-Value for Spearman coefficient.
Author Contributions
M.B. designed the experiments and contributed to data acquisition, manuscript writing, and preparation; A.A.-D. contributed to data acquisition, manuscript writing, and preparation; M.L., Ł.S., A.D.-K., J.W., M.A., and A.M.-T. contributed to data acquisition and interpretation; M.B.-M. and A.G. critically revised the manuscript; A.L.S. designed the experiments and drafted and revised the manuscript; K.G. designed and supervised the experiments and wrote and prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Medical University of Silesia, Katowice, Poland—“Młody Naukowiec” Project (KWN-2-O09/N/8/N awarded to M.B.).
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Bioethics Committee of the Jagiellonian University in Krakow, Poland (protocol no. DK/KB/CM/0031/689/2010).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sillence, D.O.; Senn, A.; Danks, D.M. Genetic heterogeneity in osteogenesis imperfecta. J. Med. Genet. 1979, 16, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Byers, P.H.; Cole, W.G. Osteogenesis Imperfecta. In Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects; Wiley: Hoboken, NJ, USA; pp. 385–430. [CrossRef]
- Van Dijk, F.S.; Pals, G.; Van Rijn, R.R.; Nikkels, P.G.J.; Cobben, J.M. Classification of Osteogenesis Imperfecta revisited. Eur. J. Med. Genet. 2010, 53, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Barnes, A.M.; Chang, W.; Morello, R.; Cabral, W.A.; Weis, M.; Eyre, D.R.; Leikin, S.; Makareeva, E.; Kuznetsova, N.; Uveges, T.E.; et al. Deficiency of Cartilage-Associated Protein in Recessive Lethal Osteogenesis Imperfecta. N. Engl. J. Med. 2006, 355, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Cabral, W.A.; Chang, W.; Barnes, A.M.; Weis, M.; Scott, M.A.; Leikin, S.; Makareeva, E.; Kuznetsova, N.V.; Rosenbaum, K.N.; Tifft, C.J.; et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007, 39, 359–365. [Google Scholar] [CrossRef]
- Christiansen, H.E.; Schwarze, U.; Pyott, S.M.; AlSwaid, A.; Al Balwi, M.; Alrasheed, S.; Pepin, M.G.; Weis, M.A.; Eyre, D.R.; Byers, P.H. Homozygosity for a Missense Mutation in SERPINH1, which Encodes the Collagen Chaperone Protein HSP47, Results in Severe Recessive Osteogenesis Imperfecta. Am. J. Hum. Genet. 2010, 86, 389. [Google Scholar] [CrossRef] [PubMed]
- Asharani, P.V.; Keupp, K.; Semler, O.; Wang, W.; Li, Y.; Thiele, H.; Yigit, G.; Pohl, E.; Becker, J.; Frommolt, P.; et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am. J. Hum. Genet. 2012, 90, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Roughley, P.J.; Rauch, F.; Glorieux, F.H. Osteogenesis imperfecta—clinical and molecular diversity. Eur. Cell. Mater. 2003, 5, 41–47; discussion 47. [Google Scholar] [CrossRef]
- Augusciak-Duma, A.; Witecka, J.; Sieron, A.L.; Janeczko, M.; Pietrzyk, J.J.; Ochman, K.; Galicka, A.; Borszewska-Kornacka, M.K.; Pilch, J.; Jakubowska-Pietkiewicz, E.; et al. Mutations in the COL1A1 and COL1A2 genes associated with osteogenesis imperfecta (OI) types I or III. Acta Biochim. Pol. 2018, 65, 79–86. [Google Scholar] [CrossRef]
- Botor, M.; Fus-Kujawa, A.; Uroczynska, M.; Stepien, K.L.; Galicka, A.; Gawron, K.; Sieron, A.L. Osteogenesis Imperfecta: Current and Prospective Therapies. Biomolecules 2021, 11, 1493. [Google Scholar] [CrossRef]
- Trejo, P.; Rauch, F. Osteogenesis imperfecta in children and adolescents—New developments in diagnosis and treatment. Osteoporos. Int. 2016, 27, 3427–3437. [Google Scholar] [CrossRef]
- Alharbi, S.A. A Systematic Overview of Osteogenesis Imperfecta. Mol. Biol. 2015, 5, 150. [Google Scholar] [CrossRef]
- Deguchi, M.; Tsuji, S.; Katsura, D.; Kasahara, K.; Kimura, F.; Murakami, T. Current Overview of Osteogenesis Imperfecta. Medicina 2021, 57, 464. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, F.S.; Huizer, M.; Kariminejad, A.; Marcelis, C.L.; Plomp, A.S.; Terhal, P.A.; Meijers-Heijboer, H.; Weiss, M.M.; Van Rijn, R.R.; Cobben, J.M.; et al. Complete COL1A1 allele deletions in osteogenesis imperfecta. Genet. Med. 2010, 12, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Mortier, G.R.; Cohn, D.H.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Mundlos, S.; Nishimura, G.; Robertson, S.; Sangiorgi, L.; Savarirayan, R.; et al. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. A 2019, 179, 2393–2419. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation, Genomics. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Kaneto, C.M.; Lima, P.S.P.; Zanette, D.L.; Prata, K.L.; Neto, J.M.P.; de Paula, F.J.A.; Silva, W.A. COL1A1 and miR-29b show lower expression levels during osteoblast differentiation of bone marrow stromal cells from Osteogenesis Imperfecta patients. BMC Med. Genet. 2014, 15, 45. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, Y.; Zhang, X.; Xu, C.; Han, J.; Ren, X.; Wang, Y.; Li, Z. Serum microRNA is a promising biomarker for osteogenesis imperfecta. Intractable Rare Dis. Res. 2012, 1, 81. [Google Scholar] [CrossRef]
- Kocijan, R.; Muschitz, C.; Geiger, E.; Skalicky, S.; Baierl, A.; Dormann, R.; Plachel, F.; Feichtinger, X.; Heimel, P.; Fahrleitner-Pammer, A.; et al. Circulating microRNA Signatures in Patients With Idiopathic and Postmenopausal Osteoporosis and Fragility Fractures. J. Clin. Endocrinol. Metab. 2016, 101, 4125–4134. [Google Scholar] [CrossRef]
- Yang, G.; Wang, J.; Huang, B. Loss of miR-217 promotes osteosarcoma cell proliferation through targeting SETD8. Pharmazie 2018, 73, 711–714. [Google Scholar] [CrossRef]
- Dai, Z.; Jin, Y.; Zheng, J.; Liu, K.; Zhao, J.; Zhang, S.; Wu, F.; Sun, Z. MiR-217 promotes cell proliferation and osteogenic differentiation of BMSCs by targeting DKK1 in steroid-associated osteonecrosis. Biomed. Pharmacother. 2019, 109, 1112–1119. [Google Scholar] [CrossRef]
- Chen, R.; Wu, J.C.; Liu, T.; Qu, Y.; Lu, L.G.; Xu, M.Y. MicroRNA profile analysis in the liver fibrotic tissues of chronic hepatitis B patients. J. Dig. Dis. 2017, 18, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, G.; di Gioia, C.R.T.; Bombardi, C.; Catalucci, D.; Corradi, B.; Gualazzi, M.G.; Leopizzi, M.; Mancini, M.; Zerbini, G.; Condorelli, G.; et al. MiR-133a regulates collagen 1A1: Potential role of miR-133a in myocardial fibrosis in angiotensin II-dependent hypertension. J. Cell. Physiol. 2012, 227, 850–856. [Google Scholar] [CrossRef]
- Roderburg, C.; Luedde, M.; Cardenas, D.V.; Vucur, M.; Mollnow, T.; Zimmermann, H.W.; Koch, A.; Hellerbrand, C.; Weiskirchen, R.; Frey, N.; et al. miR-133a mediates TGF-β-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J. Hepatol. 2013, 58, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Tavallaee, G. Role of microRNA-27b-3p in the Regulation of Key Extracellular Matrix Markers in the Synovium During Osteoarthritis. Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2020; p. 172. Available online: http://proxy.library.vcu.edu/login?url=https://www.proquest.com/dissertations-theses/role-microrna-27b-3p-regulation-key-extracellular/docview/2425107599/se-2?accountid=14780%0Ahttps://libkey.io/libraries/468/openurl?genre=dissertations&au=Tavallaee%2C+Gh (accessed on 31 August 2023).
- Rak, B.; Mehlich, D.; Garbicz, F.; Domosud, Z.; Paskal, W.; Marczewska, J.M.; Włodarski, P.K. Post-transcriptional Regulation of MMP16 and TIMP2 Expression via miR-382, miR-410 and miR-200b in Endometrial Cancer. Cancer Genom. Proteom. 2017, 14, 389–401. [Google Scholar] [CrossRef]
- Lin, C.A.; Duan, K.Y.; Wang, X.W.; Zhang, Z.S. Study on the role of Hsa-miR-382-5p in epidural fibrosis. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3663–3668. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhang, L.; Yu, Q.; Zhang, Y.; Yan, L.; Chen, Z.J. The estrogen-regulated lncRNA H19/miR-216a-5p axis alters stromal cell invasion and migration via ACTA2 in endometriosis. Mol. Hum. Reprod. 2019, 25, 550–561. [Google Scholar] [CrossRef]
- Rong, Y.; Zhang, J.; Jiang, D.; Ji, C.; Wang, J.; Ge, X.; Tang, P.; Yu, S.; Cui, W.; Cai, W. Hypoxic pretreatment of small extracellular vesicles mediates cartilage repair in osteoarthritis by delivering miR-216a-5p. Acta Biomater. 2021, 122, 325–342. [Google Scholar] [CrossRef]
- Genz, B.; Coleman, M.A.; Irvine, K.M.; Kutasovic, J.R.; Miranda, M.; Gratte, F.D.; Tirnitz-Parker, J.E.E.; Olynyk, J.K.; Calvopina, D.A.; Weis, A.; et al. Overexpression of miRNA-25-3p inhibits Notch1 signaling and TGF-β-induced collagen expression in hepatic stellate cells. Sci. Rep. 2019, 9, 8541. [Google Scholar] [CrossRef]
- He, X.; Deng, L. Potential of miR-25-3p in protection of chondrocytes: Emphasis on osteoarthritis. Folia Histochem. Cytobiol. 2021, 59, 30–39. [Google Scholar] [CrossRef]
- Zou, M.; Wang, F.; Gao, R.; Wu, J.; Ou, Y.; Chen, X.; Wang, T.; Zhou, X.; Zhu, W.; Li, P.; et al. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci. Rep. 2016, 6, 24747. [Google Scholar] [CrossRef]
- Duca, R.B.; Massillo, C.; Dalton, G.N.; Farré, P.L.; Graña, K.D.; Gardner, K.; De Siervi, A. MiR-19b-3p and miR-101-3p as potential biomarkers for prostate cancer diagnosis and prognosis. Am. J. Cancer Res. 2021, 11, 2802. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8263646/ (accessed on 27 April 2023). [PubMed]
- Nikulin, S.V.; Knyazev, E.N.; Gerasimenko, T.N.; Shilin, S.A.; Gazizov, I.N.; Zakharova, G.S.; Poloznikov, A.A.; Shkurnikov, M.Y. Non-Invasive Evaluation of Extracellular Matrix Formation in the Intestinal Epithelium. Bull. Exp. Biol. Med. 2018, 166, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.P.; Zhang, C.L.; Ma, X.L.; Hu, J.P.; Cai, T.; Zhang, L. Analyzing the Interactions of mRNAs and ncRNAs to Predict Competing Endogenous RNA Networks in Osteosarcoma Chemo-Resistance. Mol. Ther. 2019, 27, 518. [Google Scholar] [CrossRef]
- Luan, W.; Qian, Y.; Ni, X.; Bu, X.; Xia, Y.; Wang, J.; Ruan, H.; Ma, S.; Xu, B. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. Onco. Targets. Ther. 2017, 10, 1237. [Google Scholar] [CrossRef]
- Paik, S.; Jung, H.S.; Lee, S.; Yoon, D.S.; Park, M.S.; Lee, J.W. miR-449a Regulates the Chondrogenesis of Human Mesenchymal Stem Cells Through Direct Targeting of Lymphoid Enhancer-Binding Factor-1. Stem Cells Dev. 2012, 21, 3298. [Google Scholar] [CrossRef]
- Martinez-Anton, A.; Sokolowska, M.; Kern, S.; Davis, A.S.; Alsaaty, S.; Taubenberger, J.K.; Sun, J.; Cai, R.; Danner, R.L.; Eberlein, M.; et al. Changes in microRNA and mRNA expression with differentiation of human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 384–395. [Google Scholar] [CrossRef]
- Masè, M.; Grasso, M.; Avogaro, L.; Giacomaz, M.N.; D’Amato, E.; Tessarolo, F.; Graffigna, A.; Denti, M.A.; Ravelli, F. Upregulation of mir-133b and mir-328 in patients with atrial dilatation: Implications for stretch-induced atrial fibrillation. Front. Physiol. 2019, 10, 1133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Dou, X.; Li, S.; Jia, Q.; Ling, P.; Liu, H.; Han, Q.; Sun, S. miR-590-5p affects chondrocyte proliferation, apoptosis, and inflammation by targeting FGF18 in osteoarthritis. Am. J. Transl. Res. 2021, 13, 8728. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8430182/ (accessed on 27 April 2023).
- Martínez-González, E.; Brochado-Kith, Ó.; Gómez-Sanz, A.; Martín-Carbonero, L.; Jimenez-Sousa, M.Á.; Martínez-Román, P.; Resino, S.; Briz, V.; Fernández-Rodríguez, A. Comparison of methods and characterization of small RNAs from plasma extracellular vesicles of HIV/HCV coinfected patients. Sci. Rep. 2020, 10, 11140. [Google Scholar] [CrossRef]
- Chen, Y.N.; Ren, C.C.; Yang, L.; Nai, M.M.; Xu, Y.M.; Zhang, F.; Liu, Y. MicroRNA let-7d-5p rescues ovarian cancer cell apoptosis and restores chemosensitivity by regulating the p53 signaling pathway via HMGA1. Int. J. Oncol. 2019, 54, 1771–1784. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Shao, Z.; Li, C.; Li, Y.; Liu, Q.; Zhang, Y.; Tan, B.; Liu, Y. Identifying multiple collagen gene family members as potential gastric cancer biomarkers using integrated bioinformatics analysis. PeerJ 2020, 8, e9123. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, H.-Y.; Bai, W.-D.; Su, L.-L.; Liu, J.-Q.; Cai, W.-X.; Zhao, B.; Gao, J.-X.; Han, S.-C.; Li, J.; et al. MiR-10a and miR-181c regulate collagen type I generation in hypertrophic scars by targeting PAI-1 and uPA. FEBS Lett. 2015, 589, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Z.; Zhao, Z.; Li, L. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum. Pathol. 2013, 44, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Cai, Y.; Rong, X.; Chen, J.; Zheng, D.; Chen, L.; Zhang, J. MiR-661 promotes tumor invasion and metastasis by directly inhibiting RB1 in non small cell lung cancer. Mol. Cancer 2017, 16, 122. [Google Scholar] [CrossRef]
- Wang, W.; Dong, R.; Guo, Y.; He, J.; Shao, C.; Yi, P.; Yu, F.; Gu, D.; Zheng, J. CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell. Mol. Med. 2019, 23, 5486. [Google Scholar] [CrossRef]
- Liu, E.; Lv, L.; Zhan, Y.; Ma, Y.; Feng, J.; He, Y.; Wen, Y.; Zhang, Y.; Pu, Q.; Ji, F.; et al. METTL3/N6-methyladenosine/ miR-21-5p promotes obstructive renal fibrosis by regulating inflammation through SPRY1/ERK/NF-κB pathway activation. J. Cell. Mol. Med. 2021, 25, 7660–7674. [Google Scholar] [CrossRef]
- Fukumoto, I.; Kikkawa, N.; Matsushita, R.; Kato, M.; Kurozumi, A.; Nishikawa, R.; Goto, Y.; Koshizuka, K.; Hanazawa, T.; Enokida, H.; et al. Tumor-suppressive microRNAs (miR-26a/b, miR-29a/b/c and miR-218) concertedly suppressed metastasis-promoting LOXL2 in head and neck squamous cell carcinoma. J. Hum. Genet. 2016, 61, 109–118. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Q.; Guo, F.; Ma, X.; Wang, J.; Zhao, Y.; Yan, Y.; Qin, G. Involvement of miR-27a-3p in diabetic nephropathy via affecting renal fibrosis, mitochondrial dysfunction, and endoplasmic reticulum stress. J. Cell. Physiol. 2021, 236, 1454–1468. [Google Scholar] [CrossRef]
- Xu, Y.; Li, D.; Zhu, Z.; Li, L.; Jin, Y.; Ma, C.; Zhang, W. miR-27a-3p negatively regulates osteogenic differentiation of MC3T3-E1 preosteoblasts by targeting osterix. Mol. Med. Rep. 2020, 22, 1717–1726. [Google Scholar] [CrossRef]
- Yan, Y.; Du, C.; Duan, X.; Yao, X.; Wan, J.; Jiang, Z.; Qin, Z.; Li, W.; Pan, L.; Gu, Z.; et al. Inhibiting collagen I production and tumor cell colonization in the lung via miR-29a-3p loading of exosome-/liposome-based nanovesicles. Acta Pharm. Sin. B 2022, 12, 939. [Google Scholar] [CrossRef]
- Shen, H.; Wang, L.; Chen, Q.; Xu, J.; Zhang, J.; Fang, L.; Wang, J.; Fan, W. The prognostic value of COL3A1/FBN1/COL5A2/SPARC-mir-29a-3p-H19 associated ceRNA network in Gastric Cancer through bioinformatic exploration. J. Cancer 2020, 11, 4933. [Google Scholar] [CrossRef]
- Cao, B.; Dai, X. Platelet lysate induces chondrogenic differentiation of umbilical cord-derived mesenchymal stem cells by regulating the lncRNA H19/miR-29b-3p/SOX9 axis. FEBS Open Bio. 2020, 10, 2656. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Wang, X.; Zhou, F. Liver microRNA-29b-3p positively correlates with relative enhancement values of magnetic resonance imaging and represses liver fibrosis. J. Biochem. 2020, 168, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jin, J.; Tian, X.; Wu, L. hsa-miR-29c-3p regulates biological function of colorectal cancer by targeting SPARC. Oncotarget 2017, 8, 104508. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jiang, T.; Hu, X.; Liu, Z.; Zhao, L.; Liu, H.; Liu, Z.; Ma, L. Downregulation of microRNA-30a in bronchoalveolar lavage fluid from idiopathic pulmonary fibrosis patients. Mol. Med. Rep. 2018, 18, 5799–5806. [Google Scholar] [CrossRef] [PubMed]
- Haghi, M.; Taha, M.F.; Javeri, A. Suppressive effect of exogenous miR-16 and miR-34a on tumorigenesis of breast cancer cells. J. Cell. Biochem. 2019, 120, 13342–13353. [Google Scholar] [CrossRef]
- Chou, K.Y.U.; Chang, A.N.C.; Tsai, T.E.F.U.; Lin, Y.I.C.; Chen, H.E.N.; Ho, C.Y.E.N.; Chen, P.O.C.; Hwang, T.I.S. MicroRNA-34a-5p serves as a tumor suppressor by regulating the cell motility of bladder cancer cells through matrix metalloproteinase-2 silencing. Oncol. Rep. 2021, 45, 911–920. [Google Scholar] [CrossRef]
- Mao, G.; Zhang, Z.; Huang, Z.; Chen, W.; Huang, G.; Meng, F.; Zhang, Z.; Kang, Y. MicroRNA-92a-3p regulates the expression of cartilage-speci fi c genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr. Cartil. 2017, 25, 521–532. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, J.; Zhang, D.; Wang, L.; Zhao, W.; Lin, D.Y.T.; Chen, R.; Xie, H.; Hu, X.; Fang, X.; et al. MicroRNA expression profiles of scar and normal tissue from patients with posterior urethral stricture caused by pelvic fracture urethral distraction defects. Int. J. Mol. Med. 2018, 41, 2733–2743. [Google Scholar] [CrossRef]
- Zhou, X.; Luo, D.; Sun, H.; Qi, Y.; Xu, W.; Jin, X.; Li, C.; Lin, Z.; Li, G. MiR-132-3p regulates ADAMTS-5 expression and promotes chondrogenic differentiation of rat mesenchymal stem cells. J. Cell. Biochem. 2018, 119, 2579–2587. [Google Scholar] [CrossRef]
- Zhou, L.L.; Zhu, Y.M.; Qian, F.Y.; Yuan, C.C.; Yuan, D.P.; Zhou, X.P. MicroRNA-143-3p contributes to the regulation of pain responses in collagen-induced arthritis. Mol. Med. Rep. 2018, 18, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, X.; Tang, Y.; Chen, C.; Jing, R.; Liu, T. miR-155-5p Implicates in the Pathogenesis of Renal Fibrosis via Targeting SOCS1 and SOCS6. Oxidative Med. Cell. Longev. 2020, 2020, 6263921. [Google Scholar] [CrossRef]
- Chen, G.; Wang, D.; Zhao, X.; Cao, J.; Zhao, Y.; Wang, F.; Bai, J. miR-155-5p modulates malignant behaviors of hepatocellular carcinoma by directly targeting CTHRC1 and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling. Cancer Cell Int. 2017, 17, 118. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lu, L.; Liang, J.; Guo, X.; Zhang, P.H.; Luo, S. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet. Mol. Res. 2014, 13, 2727–2738. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gu, S.; Chen, B.F.; Shen, W.L.; Yin, Z.; Xu, G.W.; Hu, J.J.; Zhu, T.; Li, G.; Wan, C.; et al. Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials 2015, 53, 239–250. [Google Scholar] [CrossRef]
- Wu, A.; Chen, Y.; Liu, Y.; Lai, Y.; Liu, D. miR-199b-5p inhibits triple negative breast cancer cell proliferation, migration and invasion by targeting DDR1. Oncol. Lett. 2018, 16, 4889–4896. [Google Scholar] [CrossRef]
- Winship, A.; Ton, A.; Van Sinderen, M.; Menkhorst, E.; Rainczuk, K.; Griffiths, M.; Cuman, C.; Dimitriadis, E. Mouse double minute homologue 2 (MDM2) downregulation by miR-661 impairs human endometrial epithelial cell adhesive capacity. Reprod. Fertil. Dev. 2018, 30, 477–486. [Google Scholar] [CrossRef]
- Sahin, Y.; Altan, Z.; Arman, K.; Bozgeyik, E.; Ozer, M.K.; Arslan, A. Inhibition of miR-664a interferes with the migration of osteosarcoma cells via modulation of MEG3. Biochem. Biophys. Res. Commun. 2017, 490, 1100–1105. [Google Scholar] [CrossRef]
- Ding, R.; Liu, X.; Zhang, J.; Yuan, J.; Zheng, S.; Cheng, X.; Jia, J. Downregulation of miR-1-3p expression inhibits the hypertrophy and mineralization of chondrocytes in DDH. J. Orthop. Surg. Res. 2021, 16, 512. [Google Scholar] [CrossRef]
- Guan, G.; Niu, X.; Qiao, X.; Wang, X.; Liu, J.; Zhong, M. Upregulation of Neural Cell Adhesion Molecule 1 (NCAM1) by hsa-miR-141-3p Suppresses Ameloblastoma Cell Migration. Med. Sci. Monit. 2020, 26, e923491-1–e923491-8. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, C.; Hou, L.; Wu, Y. Silencing of the lncRNA TUG1 attenuates the epithelial-mesenchymal transition of renal tubular epithelial cells by sponging miR-141-3p via regulating β-catenin. Am. J. Physiol. Renal Physiol. 2020, 319, F1125–F1134. [Google Scholar] [CrossRef] [PubMed]
- Makino, K.; Jinnin, M.; Kajihara, I.; Honda, N.; Sakai, K.; Masuguchi, S.; Fukushima, S.; Inoue, Y.; Ihn, H. Circulating miR-142-3p levels in patients with systemic sclerosis. Clin. Exp. Dermatology Exp. Dermatol. 2011, 37, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Börschel, C.S.; Stejskalova, A.; Schäfer, S.D.; Kiesel, L.; Götte, M. miR-142-3p Reduces the Size, Migration, and Contractility of Endometrial and Endometriotic Stromal Cells by Targeting Integrin- and Rho GTPase-Related Pathways That Regulate Cytoskeletal Function. Biomedicines 2020, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Sillence, D.O.; Rimoin, D.L. Classification of osteogenesis imperfect. Lancet 1978, 1, 1041–1042. [Google Scholar] [CrossRef] [PubMed]
- Kuivaniemi, H.; Tromp, G.; Prockop, D.J. Mutations in collagen genes: Causes of rare and some common diseases in humans. FASEB J. 1991, 5, 2052–2060. [Google Scholar] [CrossRef]
- Forlino, A.; Cabral, W.A.; Barnes, A.M.; Marini, J.C. New perspectives on osteogenesis imperfecta. Nat. Rev. Endocrinol. 2011, 7, 540–557. [Google Scholar] [CrossRef]
- Prockop, D.J.; Colige, A.; Helminen, H.; Khillan, J.S.; Pereira, R.; Vandenberg, P. Mutations in Type 1 Procollagen That Cause Osteogenesis Imperfecta: Effects of the Mutations on the Assembly of Collagen into Fibrils, the Basis of Phenotypic Variations, and Potential Antisense Therapies. J. Bone Miner. Res. 1993, 8, S489–S492. [Google Scholar] [CrossRef]
- Sun, K.; Wang, J.; Liu, F.; Ji, Z.; Guo, Z.; Zhang, C.; Yao, M. Ossotide promotes cell differentiation of human osteoblasts from osteogenesis imperfecta patients by up-regulating miR-145. Biomed. Pharmacother. 2016, 83, 1105–1110. [Google Scholar] [CrossRef]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Invest. 2017, 127, 761–771. [Google Scholar] [CrossRef]
- Leimena, C.; Qiu, H. Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension. Int. J. Mol. Sci. 2018, 19, 927. [Google Scholar] [CrossRef]
- Fus-Kujawa, A.; Mendrek, B.; Bajdak-Rusinek, K.; Diak, N.; Strzelec, K.; Gutmajster, E.; Janelt, K.; Kowalczuk, A.; Trybus, A.; Rozwadowska, P.; et al. Gene-repaired iPS cells as novel approach for patient with Osteogenesis imperfecta. Front. Bioeng. Biotechnol. 2023, 11, 1205122. [Google Scholar] [CrossRef] [PubMed]
- Witecka, J.; Auguściak-Duma, A.M.; Kruczek, A.; Szydło, A.; Lesiak, M.; Krzak, M.; Pietrzyk, J.J.; Männikkö, M.; Sieroń, A.L. Two novel COL1A1 mutations in patients with osteogenesis imperfecta (OI) affect the stability of the collagen type I triple-helix. J. Appl. Genet. 2008, 49, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Majka, M.; Janeczko, M.; Goździk, J.; Jarocha, D.; Auguściak-Duma, A.; Witecka, J.; Lesiak, M.; Koryciak-Komarska, H.; Sieroń, A.L.; Józef, J.J. Cell therapy of a patient with type III osteogenesis imperfecta caused by mutation in COL1A2 gene and unstable collagen type I*. Open J. Genet. 2013, 3, 49–60. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).