The Essential Oil from Oliveria decumbens Vent. (Apiaceae) as Inhibitor of Breast Cancer Cell (MCF-7) Growth
Abstract
1. Introduction
2. Results
2.1. Chemical Identification by GC/MS (Gas Chromatography–Mass Spectrometry) Analysis
2.2. Assessment of Antioxidant Activity
2.3. Cytotoxic Activity
2.4. Alteration in the Morphology of MCF-7 Cells by Acridine Orange/Ethidium Bromide (AO/EB) Double-Staining
2.5. DNA Fragmentation Assay
2.6. Expression of BIM, Bcl-2, PTEN, and AURKA Genes in OEO-Treated MCF-7 Cells
2.7. Relationship between Gene Expressions in OEO-Treated MCF-7 Cells
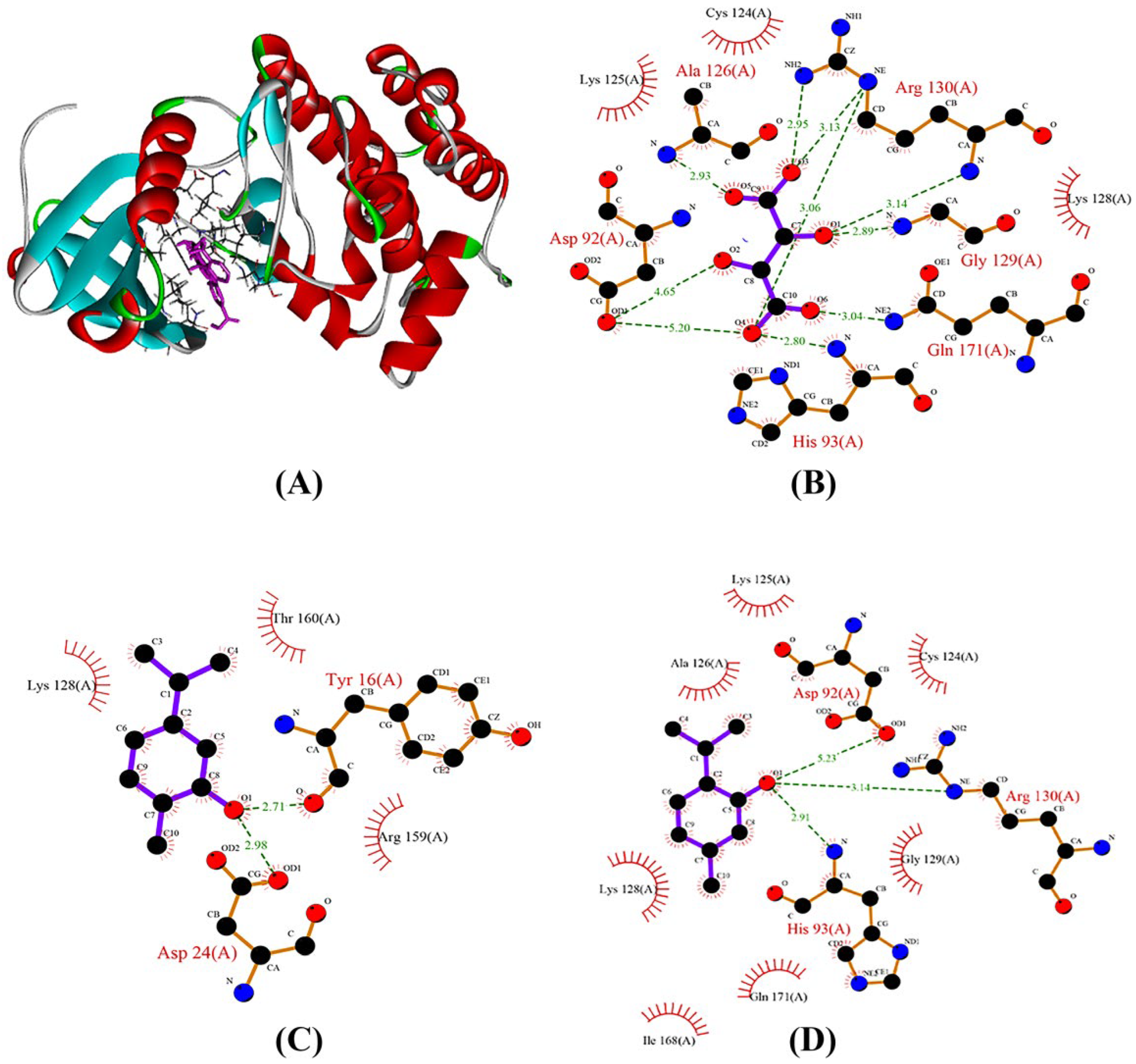
2.8. Analysis of Protein–Ligand Interaction of PTEN with Main Compounds of OEO
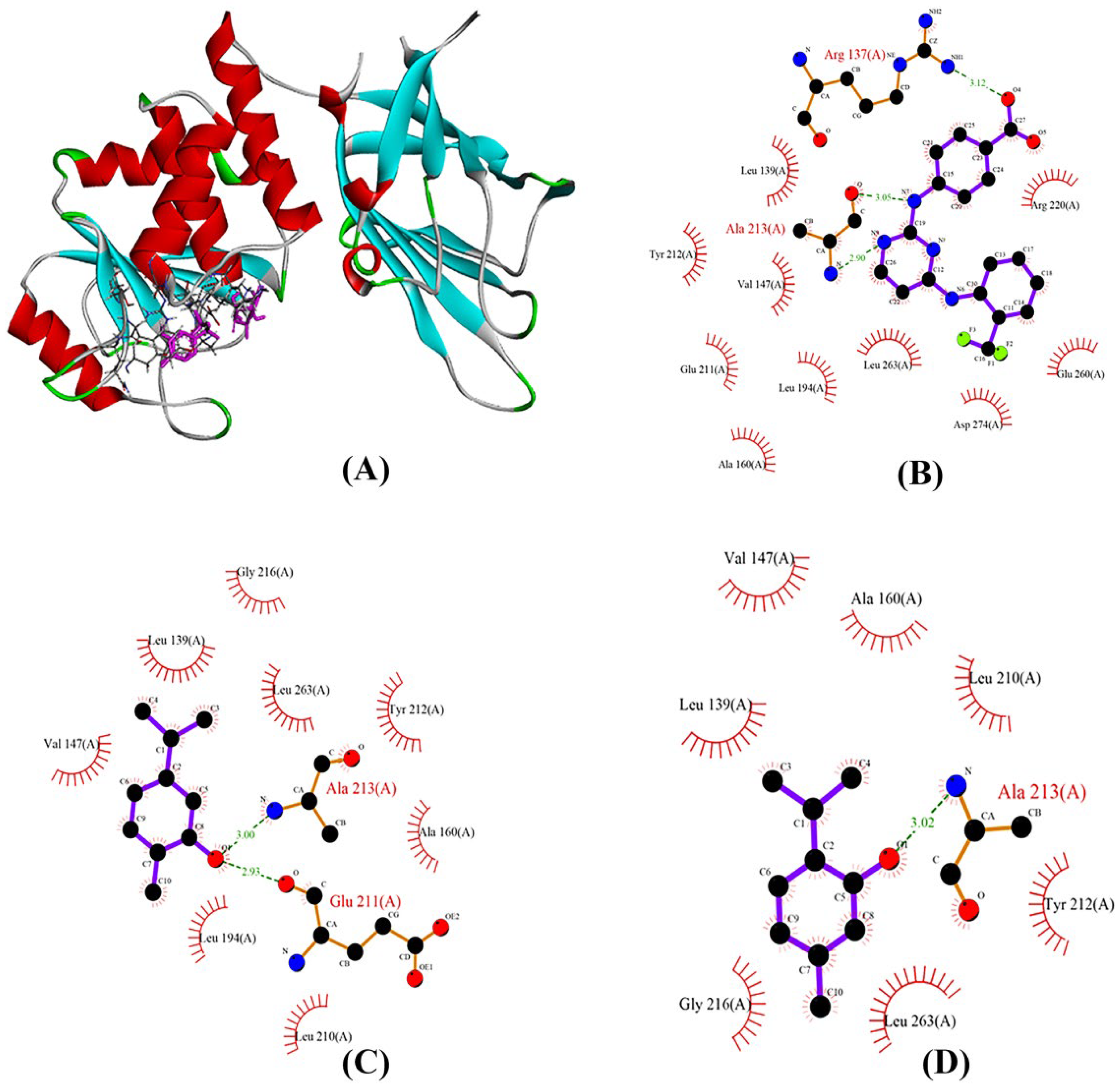
2.9. AURKA Interaction with Ligands during Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Preparation of O. decumbens Vent Essential Oil
4.2. Identification of Oliveria decumbens Vent Essential Oil Components
4.3. DPPH Assay
4.4. Cell Culture
4.5. Cell Viability Test
4.6. Acridine Orange/Ethidium Bromide Staining
4.7. DNA Extraction
4.8. PPI Network Analysis and Hub Gene Identification
4.9. RNA Extraction and cDNA Synthesis Genomics
4.10. Primer Design and Quantitative Expression Examination
4.11. Binding Site and Docking
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKT | serine/threonine kinase |
| AURKA | Aurora kinase A |
| Bcl-2 | b-lymphocytoma-2 gene |
| BIM | BCL2 like 11 |
| CTAB | Cetrimonium bromide |
| DPPH | di(phenyl)-(2,4,6-trinitrophenyl) iminoazanium |
| GC/MS | Gas chromatography–mass spectrometry |
| MTT OEO | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide O. decumbens essential oil |
| TBHQ | Tertiary butylhydroquinone |
| PiP3 Pi3K PPi PTEN | phosphatidylinositol (3,4,5)-triphoshpate phosphatidylinositol-3 kinase Protein Protein Interaction Phosphatase and tensin homolog |
References
- He, Z.; Chen, Z.; Tan, M.; Elingarami, S.; Liu, Y.; Li, T.; Deng, Y.; He, N.; Li, S.; Fu, J.; et al. A Review on Methods for Diagnosis of Breast Cancer Cells and Tissues. Cell Prolif. 2020, 53, e12822. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.; Huynh-Do, U. The Role of PTEN in Tumor Angiogenesis. J. Oncol. 2012, 2012, 141236. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Garcia, V.; Tawil, Y.; Wise, H.M.; Leslie, N.R. Mechanisms of PTEN Loss in Cancer: It’s All about Diversity. Semin. Cancer Biol. 2019, 59, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, D.; Longy, M. Mutations of the Human PTEN Gene. Hum. Mutat. 2000, 16, 109–122. [Google Scholar] [CrossRef]
- Ton, A.-T.; Singh, K.; Morin, H.; Ban, F.; Leblanc, E.; Lee, J.; Lallous, N.; Cherkasov, A. Dual-Inhibitors of N-Myc and AURKA as Potential Therapy for Neuroendocrine Prostate Cancer. Int. J. Mol. Sci. 2020, 21, 8277. [Google Scholar] [CrossRef]
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora Kinases: Novel Therapy Targets in Cancers. Oncotarget 2017, 8, 23937–23954. [Google Scholar] [CrossRef]
- Curčić, M.G.; Stanković, M.S.; Mrkalić, E.M.; Matović, Z.D.; Banković, D.D.; Cvetković, D.M.; Dačić, D.S.; Marković, S.D. Antiproliferative and Proapoptotic Activities of Methanolic Extracts from Ligustrum vulgare L. as an Individual Treatment and in Combination with Palladium Complex. Int. J. Mol. Sci. 2012, 13, 2521–2534. [Google Scholar] [CrossRef]
- Abdullah, A.-S.H.; Mohammed, A.S.; Abdullah, R.; Mirghani, M.E.S.; Al-Qubaisi, M. Cytotoxic Effects of Mangifera Indica L. Kernel Extract on Human Breast Cancer (MCF-7 and MDA-MB-231 Cell Lines) and Bioactive Constituents in the Crude Extract. BMC Complement. Altern. Med. 2014, 14, 199. [Google Scholar] [CrossRef]
- Greenwell, M.; Rahman, P.K.S.M. Medicinal Plants: Their Use in Anticancer Treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103–4112. [Google Scholar] [CrossRef]
- Talib, W.H.; Daoud, S.; Mahmod, A.I.; Hamed, R.A.; Awajan, D.; Abuarab, S.F.; Odeh, L.H.; Khater, S.; Al Kury, L.T. Plants as a source of anticancer agents: From bench to bedside. Molecules 2022, 27, 4818. [Google Scholar] [CrossRef]
- Fitsiou, E.; Pappa, A. Anticancer Activity of Essential Oils and Other Extracts from Aromatic Plants Grown in Greece. Antioxidants 2019, 8, 290. [Google Scholar] [CrossRef]
- Samadi, N.; Masoum, S.; Mehrara, B.; Hosseini, H. Application of Linear Multivariate Calibration Techniques to Identify the Peaks Responsible for the Antioxidant Activity of Satureja hortensis L. and Oliveria decumbens Vent. Essential Oils by Gas Chromatography–Mass Spectrometry. J. Chromatogr. B 2015, 1001, 75–81. [Google Scholar] [CrossRef]
- Li, Y.; Yeung, C.; Chiu, L.C.M.; Cen, Y.; Ooi, V.E.C. Chemical Composition and Antiproliferative Activity of Essential Oil from the Leaves of a Medicinal Herb, Schefflera heptaphylla. Phytother. Res. 2009, 23, 140–142. [Google Scholar] [CrossRef]
- Sibanda, S.; Chigwada, G.; Poole, M.; Gwebu, E.T.; Noletto, J.A.; Schmidt, J.M.; Rea, A.I.; Setzer, W.N. Composition and Bioactivity of the Leaf Essential Oil of Heteropyxis dehniae from Zimbabwe. J. Ethnopharmacol. 2004, 92, 107–111. [Google Scholar] [CrossRef]
- Yousefzadi, M.; Riahi-Madvar, A.; Hadian, J.; Rezaee, F.; Rafiee, R.; Biniaz, M. Toxicity of Essential Oil of Satureja khuzistanica: In Vitro Cytotoxicity and Anti-Microbial Activity. J. Immunotoxicol. 2014, 11, 50–55. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Sharifi-Rad, M.; Hoseini-Alfatemi, S.M.; Iriti, M.; Sharifi-Rad, M.; Sharifi-Rad, M. Composition, Cytotoxic and Antimicrobial Activities of Satureja intermedia CA Mey Essential Oil. Int. J. Mol. Sci. 2015, 16, 17812–17825. [Google Scholar] [CrossRef]
- Karami, A.; Khoshbakht, T.; Esmaeili, H.; Maggi, F. Essential Oil Chemical Variability in Oliveria decumbens (Apiaceae) from Different Regions of Iran and Its Relationship with Environmental Factors. Plants 2020, 9, 680. [Google Scholar] [CrossRef]
- Saidi, M. Antioxidant Activities and Chemical Composition of Essential Oils from Satureja khuzestanica, Oliveria decumbens and Thymus daenensis. J. Essent. Oil Bear. Plants 2014, 17, 513–521. [Google Scholar] [CrossRef]
- Khajehie, N.; Golmakani, M.-T.; Eblaghi, M.; Eskandari, M.H. Evaluating the Effects of Microwave-Assisted Hydrodistillation on Antifungal and Radical Scavenging Activities of Oliveria decumbens and Chaerophyllum macropodum Essential Oils. J. Food Prot. 2017, 80, 783–791. [Google Scholar] [CrossRef]
- Khosravinezhad, M.; Talebi, E.; Shivakumar; Nemati, Z.; Nasrollahi, I. Essential Oil Composition and Antimicrobial, Antioxidant Activities of Oliveria decumbens Vent. Int. J. Herb. Med. 2017, 5, 102–106. [Google Scholar]
- Ribble, D.; Goldstein, N.B.; Norris, D.A.; Shellman, Y.G. A Simple Technique for Quantifying Apoptosis in 96-Well Plates. BMC Biotechnol. 2005, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Karami-Tehrani, F.; Ghavami, S.; Maddika, S.; Los, M. Adenosine and Deoxyadenosine Induces Apoptosis in Oestrogen Receptor-Positive and -Negative Human Breast Cancer Cells via the Intrinsic Pathway. Cell Prolif. 2005, 38, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Depowski, P.L.; Rosenthal, S.I.; Ross, J.S. Loss of Expression of the PTEN Gene Protein Product Is Associated with Poor Outcome in Breast Cancer. Mod. Pathol. 2001, 14, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Grigorieva, I.; Steele, R.; Hoover, R.G.; Ray, R.B. PTEN Transcriptionally Modulates C-Myc Gene Expression in Human Breast Carcinoma Cells and Is Involved in Cell Growth Regulation. Gene 1999, 235, 85–91. [Google Scholar] [CrossRef]
- Katsha, A.; Belkhiri, A.; Goff, L.; El-Rifai, W. Aurora Kinase A in Gastrointestinal Cancers: Time to Target. Mol. Cancer 2015, 14, 106. [Google Scholar] [CrossRef]
- Lee, J.-O.; Yang, H.; Georgescu, M.-M.; Di Cristofano, A.; Maehama, T.; Shi, Y.; Dixon, J.E.; Pandolfi, P.; Pavletich, N.P. Crystal Structure of the PTEN Tumor Suppressor: Implications for Its Phosphoinositide Phosphatase Activity and Membrane Association. Cell 1999, 99, 323–334. [Google Scholar] [CrossRef]
- Chinnasamy, S.; Selvaraj, G.; Kaushik, A.C.; Kaliamurthi, S.; Chandrabose, S.; Singh, S.K.; Thirugnanasambandam, R.; Gu, K.; Wei, D.-Q. Molecular Docking and Molecular Dynamics Simulation Studies to Identify Potent AURKA Inhibitors: Assessing the Performance of Density Functional Theory, MM-GBSA and Mass Action Kinetics Calculations. J. Biomol. Struct. Dyn. 2020, 38, 4325–4335. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef]
- Tan, M.-H.; Mester, J.L.; Ngeow, J.; Rybicki, L.A.; Orloff, M.S.; Eng, C. Lifetime Cancer Risks in Individuals with Germline PTEN Mutations. Clin. Cancer Res. 2012, 18, 400–407. [Google Scholar] [CrossRef]
- Kim, D.; Suh, J.; Surh, Y.; Na, H. Regulation of the Tumor Suppressor PTEN by Natural Anticancer Compounds. Ann. N. Y. Acad. Sci. 2017, 1401, 136–149. [Google Scholar] [CrossRef]
- Li, L.-L.; Wei, L.; Zhang, N.; Wei, W.-Y.; Hu, C.; Deng, W.; Tang, Q.-Z. Levosimendan Protects against Doxorubicin-Induced Cardiotoxicity by Regulating the PTEN/Akt Pathway. BioMed Res. Int. 2020, 2020, 8593617. [Google Scholar] [CrossRef]
- Esmaeili, H.; Karami, A.; Maggi, F. Essential Oil Composition, Total Phenolic and Flavonoids Contents, and Antioxidant Activity of Oliveria Decumbens Vent. (Apiaceae) at Different Phenological Stages. J. Clean. Prod. 2018, 198, 91–95. [Google Scholar] [CrossRef]
- Sampaio, L.A.; Pina, L.T.S.; Serafini, M.R.; dos Santos Tavares, D.; Guimaraes, A.G. Antitumor Effects of Carvacrol and Thymol: A Systematic Review. Front. Pharmacol. 2021, 12, 702487. [Google Scholar] [CrossRef]
- Arunasree, K.M. Anti-Proliferative Effects of Carvacrol on a Human Metastatic Breast Cancer Cell Line, MDA-MB 231. Phytomedicine 2010, 17, 581–588. [Google Scholar] [CrossRef]
- Kowalczyk, A.; Przychodna, M.; Sopata, S.; Bodalska, A.; Fecka, I. Thymol and Thyme Essential Oil—New Insights into Selected Therapeutic Applications. Molecules 2020, 25, 4125. [Google Scholar] [CrossRef]
- Jamali, T.; Kavoosi, G.; Jamali, Y.; Mortezazadeh, S.; Ardestani, S.K. In-Vitro, in-Vivo, and in-Silico Assessment of Radical Scavenging and Cytotoxic Activities of Oliveria decumbens Essential Oil and Its Main Components. Sci. Rep. 2021, 11, 14281. [Google Scholar] [CrossRef]
- Kang, S.-H.; Kim, Y.-S.; Kim, E.-K.; Hwang, J.-W.; Jeong, J.-H.; Dong, X.; Lee, J.-W.; Moon, S.-H.; Jeon, B.-T.; Park, P.-J. Anticancer Effect of Thymol on AGS Human Gastric Carcinoma Cells. J. Microbiol. Biotechnol. 2016, 26, 28–37. [Google Scholar] [CrossRef]
- Mondal, S.K.; Sen, M.K. Loss of Phosphatase Activity in PTEN (Phosphatase and Tensin Homolog Deleted on Chromosome Ten) Results in Endometrial Carcinoma in Humans: An in-Silico Study. Heliyon 2020, 6, e03106. [Google Scholar] [CrossRef]
- Georgescu, M.-M. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer 2010, 1, 1170–1177. [Google Scholar] [CrossRef]
- Boi, D.; Souvalidou, F.; Capelli, D.; Polverino, F.; Marini, G.; Montanari, R.; Pochetti, G.; Tramonti, A.; Contestabile, R.; Trisciuoglio, D.; et al. PHA-680626 Is an Effective Inhibitor of the Interaction between Aurora-A and N-Myc. Int. J. Mol. Sci. 2021, 22, 13122. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Park, J.; Ahn, S.; Lee, Y.; Lee, J.; Shin, S.Y.; Koh, D.; Lim, Y. Design, Synthesis, and Biological Evaluation of Polyphenols with 4, 6-Diphenylpyrimidin-2-Amine Derivatives for Inhibition of Aurora Kinase A. DARU J. Pharm. Sci. 2019, 27, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Chu, P.; Chen, G.; Wang, K.; Hsu, W.; Ahmed, A.; Ma, W.; Cheng, W.; Wu, Y.; Yang, J. Natural Anthraquinone Compound Emodin as a Novel Inhibitor of Aurora A Kinase: A Pilot Study. Chem. Biol. Drug Des. 2022, 99, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.P.; Zhu, J.-Y.; Lawrence, H.R.; Pireddu, R.; Luo, Y.; Alam, R.; Ozcan, S.; Sebti, S.M.; Lawrence, N.J.; Schönbrunn, E. A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A. ACS Chem. Biol. 2012, 7, 698–706. [Google Scholar] [CrossRef]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatograpy/Mass Spectrometry, 4th ed.; Allured Publishing Corpporation: Carol Stream, IL, USA, 2007; pp. 804–806. [Google Scholar]
- Molyneux, P. The Use of the Stable Free Radical Diphenylpicrylhydrazyl (DPPH) for Estimating Antioxidant Activity. Songklanakarin J. Sci. Technol. 2004, 26, 211–219. [Google Scholar]
- Saadat, Y.R.; Saeidi, N.; Vahed, S.Z.; Barzegari, A.; Barar, J. An Update to DNA Ladder Assay for Apoptosis Detection. BioImpacts BI 2015, 5, 25. [Google Scholar] [CrossRef]


| No | Compounds | Class a | RI b | RI c | Relative Percentage (%) |
|---|---|---|---|---|---|
| 1 | α-Thujene | MH | 927 | 930 | 0.14 ± 0.05 |
| 2 | α-Pinene | MH | 934 | 939 | 0.18 ± 0.02 |
| 3 | β-Pinene | MH | 973 | 979 | 1.53 ± 0.08 |
| 4 | Limonene | MH | 1023 | 1029 | 1.00 ± 0.12 |
| 5 | β–Phellandrene | MH | 1021 | 1030 | 0.88 ± 0.10 |
| 6 | p-Cymene | MH | 1024 | 1024 | 19.95 ± 0.63 |
| 7 | γ-Terpinene | MH | 1050 | 1059 | 19.65 ± 0.30 |
| 8 | cis-Limonene oxide | MO | 1117 | 1132 | 1.09 ± 0.04 |
| 9 | trans-Carveol | MO | 1200 | 1215 | 0.46 ± 0.12 |
| 10 | Thymol | MO | 1271 | 1290 | 26.63 ± 1.42 |
| 11 | Carvacrol | MO | 1283 | 1298 | 24.12 ± 1.23 |
| 12 | Eugenol | PP | 1339 | 1356 | 0.14 ± 0.07 |
| 13 | Spathulenol | SO | 1566 | 1577 | 0.17 ± 0.11 |
| 14 | Myristicin | PP | 1494 | 1522 | 1.88 ± 0.24 |
| Total | - | - | 97.84 |
| Time (h) | OEO (µg/mL) | Doxorubicin (µg/mL) |
|---|---|---|
| 24 | 84.07 * ± 6.66 | - |
| 48 | 39.54 * ± 2.35 | 0.215 * ± 0.028 |
| 72 | 33.32 * ± 1.14 | 0.111 * ± 0.036 |
| Aurka | Bim | Bcl-2 | ||
|---|---|---|---|---|
| PTEN | Pearson’s r | −0.947 ** | 0.886 * | −0.967 ** |
| p-value | 0.004 | 0.019 | 0.002 | |
| Aurka | Pearson’s r | −0.793 | 0.962 ** | |
| p-value | 0.060 | 0.002 | ||
| Bim | Pearson’s r | −0.879 * | ||
| p-value | 0.021 | |||
| Compound IDs | Docking Score | H-Bond Interactions | Hydrophobic Interaction |
|---|---|---|---|
| 10364 | −5.3 | Tyr16, Asp24 | Lys128, Arg159, Thr160 |
| 6989 | −5.4 | Asp92, His93, Arg130 | Cys124, Lys125, Ala126, Lys128, Gly129, Ile168, Gln171 |
| 7461 | −4.6 | - | Tyr16, Asp24, Lys28, Gly127, Arg159, Thr160 |
| 7463 | −4.7 | - | Tyr16, Asp24, Lys28, Gly127, Arg159, Thr160 |
| Compound IDs | Docking Score | H-Bond Interactions | Hydrophobic Interaction |
|---|---|---|---|
| 10364 | −7.0 | Glu211, Ala213 | Leu139, Val147, Ala160, Leu194, Leu210, Tyr212, Gly216, Leu263 |
| 6989 | −6.2 | Ala213 | Leu139, Val147, Ala160, Leu210, Tyr212, Gly216, Leu263 |
| 7461 | −6.7 | - | Leu139, Val147, Ala160, Leu194, Leu210, Tyr212, Ala213, Gly216, Leu263 |
| 7463 | −6.8 | - | Leu139, Val147, Ala160, Leu194, Leu210, Tyr212, Ala213, Gly216, Leu263 |
| Gene | Primer Sequence 5′→3′ | Ta (°C) | Product Length (bp) |
|---|---|---|---|
| Bim | F: CCACCAGCACCATAGAAGAAT | 63 | 135 |
| R: TAAGGAGCAGGCACAGAGA | |||
| PTEN | F: CAGTAGAGGAGCCGTCAA | 58.5 | 108 |
| R: CAGAGTCAGTGGTGTCAGA | |||
| Aurka | F: CATAGAGACATTAAGCCAGAGA | 59 | 157 |
| R: GCATCCGACCTTCAATCA | |||
| Bcl-2 | F: AGTGATAATCAAGTCCTTT | 60 | 155 |
| R: GGCAGTCCAGATGAACCG | |||
| β-actin | F: GCCTTTGCCGATCCGC | 65 | 160 |
| R: GCCGTAGCCGTTGTCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shariatzadeh, M.; Karami, A.; Moghadam, A.; Lotfi, M.; Maggi, F.; Ebrahimie, E. The Essential Oil from Oliveria decumbens Vent. (Apiaceae) as Inhibitor of Breast Cancer Cell (MCF-7) Growth. Pharmaceuticals 2023, 16, 59. https://doi.org/10.3390/ph16010059
Shariatzadeh M, Karami A, Moghadam A, Lotfi M, Maggi F, Ebrahimie E. The Essential Oil from Oliveria decumbens Vent. (Apiaceae) as Inhibitor of Breast Cancer Cell (MCF-7) Growth. Pharmaceuticals. 2023; 16(1):59. https://doi.org/10.3390/ph16010059
Chicago/Turabian StyleShariatzadeh, Mandana, Akbar Karami, Ali Moghadam, Mahbobeh Lotfi, Filippo Maggi, and Esmaeil Ebrahimie. 2023. "The Essential Oil from Oliveria decumbens Vent. (Apiaceae) as Inhibitor of Breast Cancer Cell (MCF-7) Growth" Pharmaceuticals 16, no. 1: 59. https://doi.org/10.3390/ph16010059
APA StyleShariatzadeh, M., Karami, A., Moghadam, A., Lotfi, M., Maggi, F., & Ebrahimie, E. (2023). The Essential Oil from Oliveria decumbens Vent. (Apiaceae) as Inhibitor of Breast Cancer Cell (MCF-7) Growth. Pharmaceuticals, 16(1), 59. https://doi.org/10.3390/ph16010059










