Synthesis and Cytotoxicity Evaluation of Novel Coumarin–Palladium(II) Complexes against Human Cancer Cell Lines
Abstract
1. Introduction
2. Results and Discussion
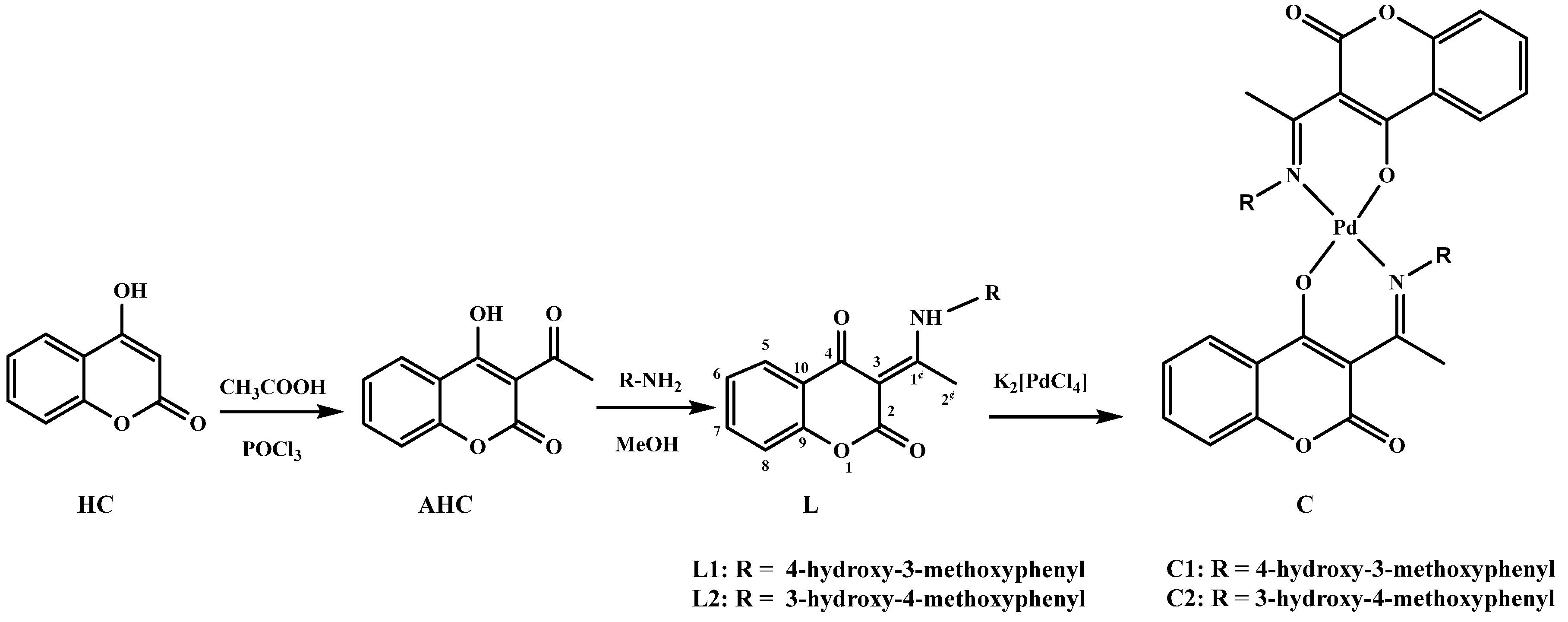
2.1. Chemistry
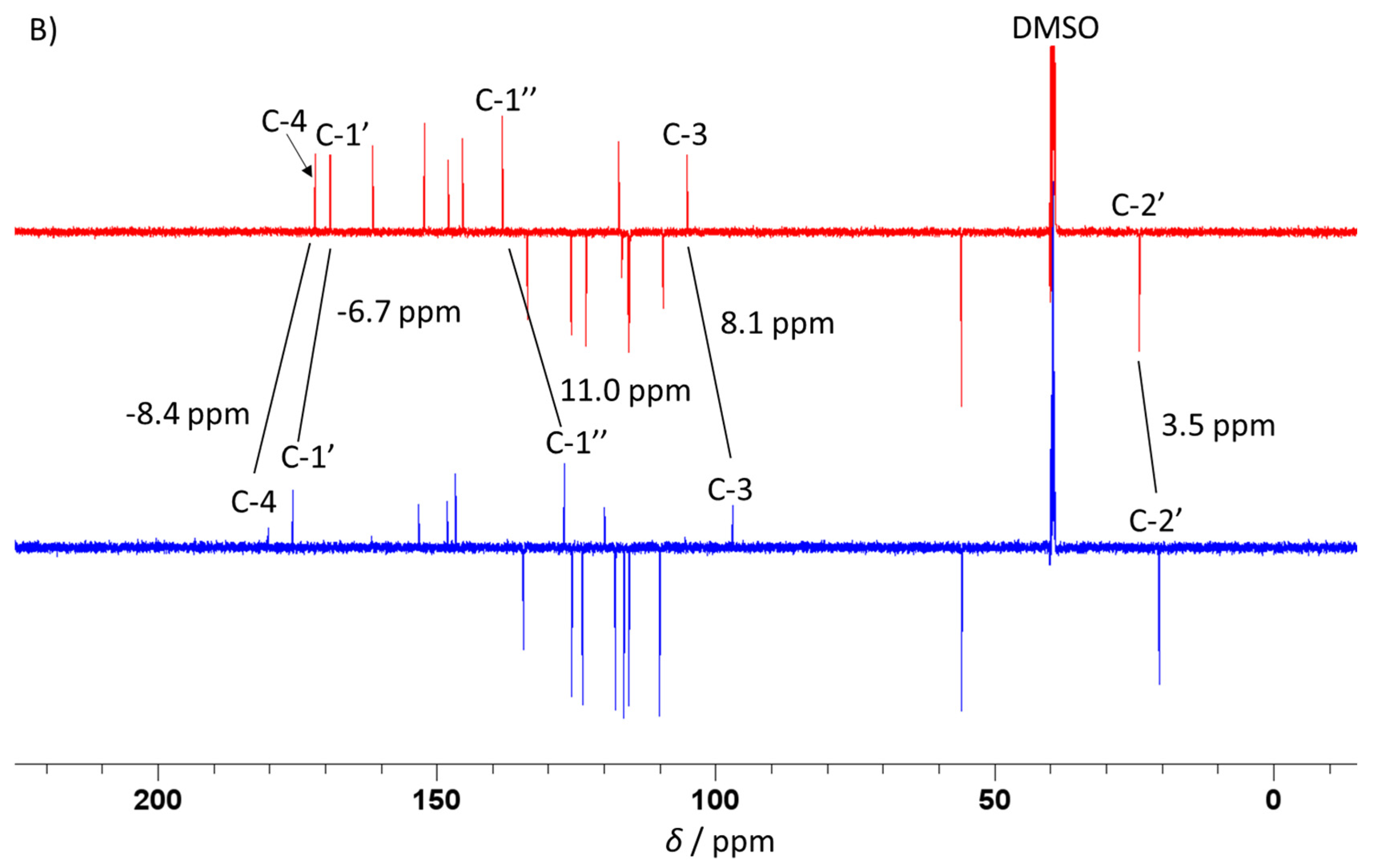
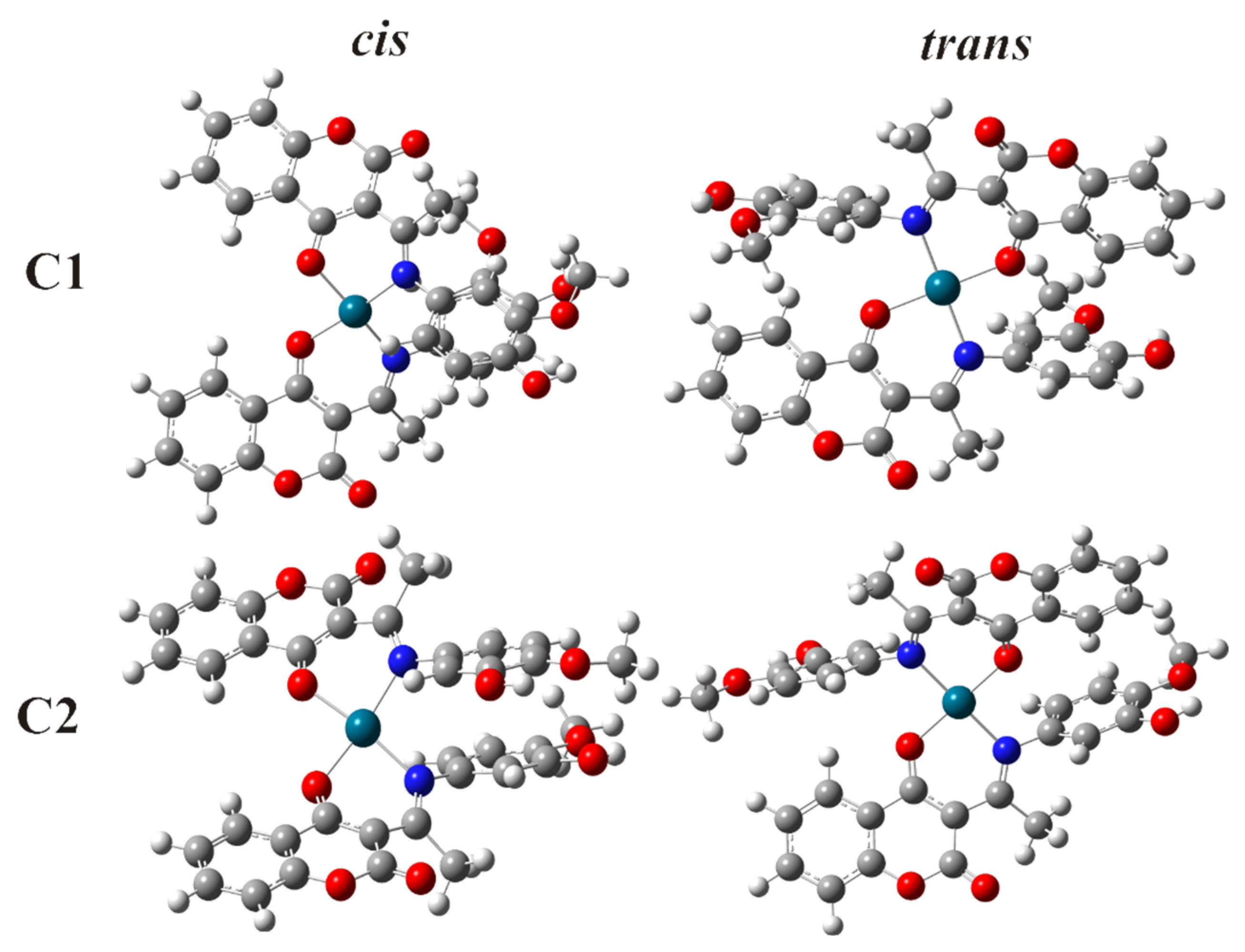
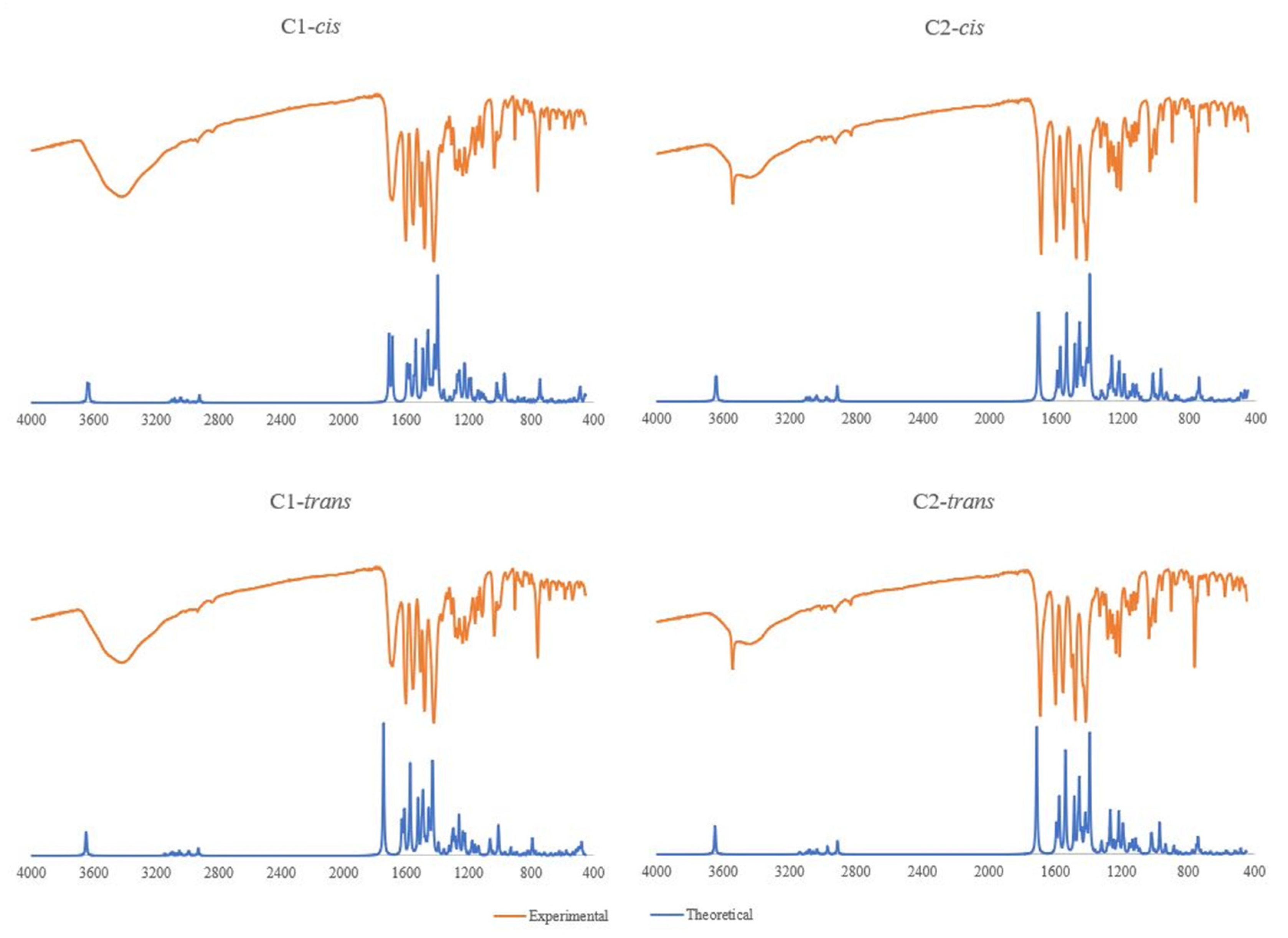
2.2. Spectroscopic and DFT Characterization
2.3. In Vitro Cytotoxicity
2.4. Cell Cycle Analysis
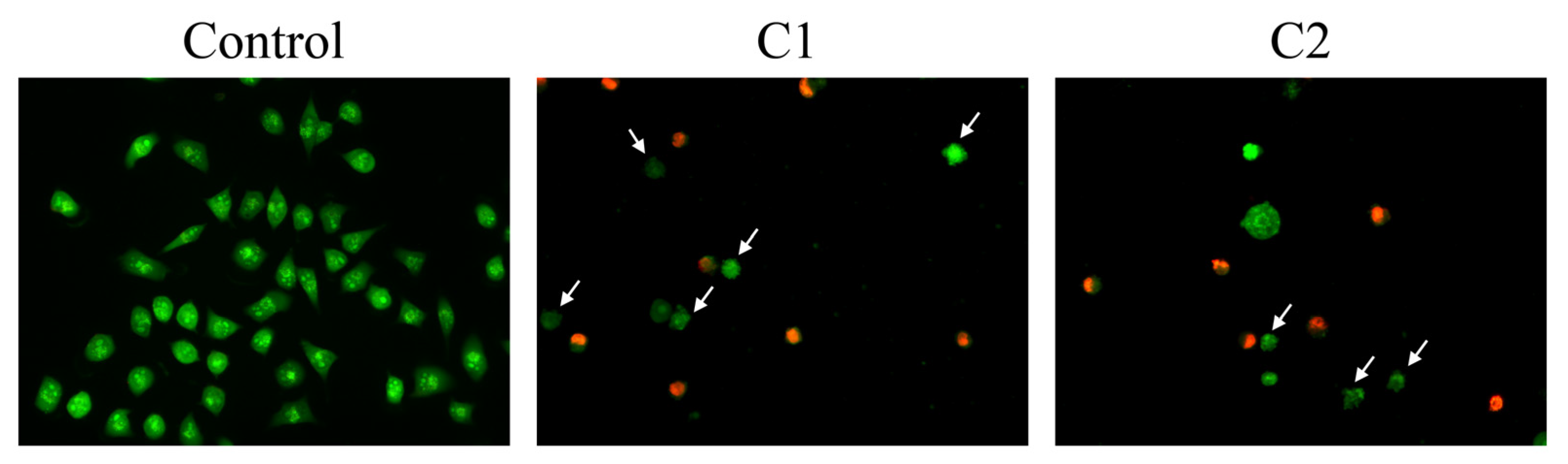
2.5. Fluorescence Microscopy
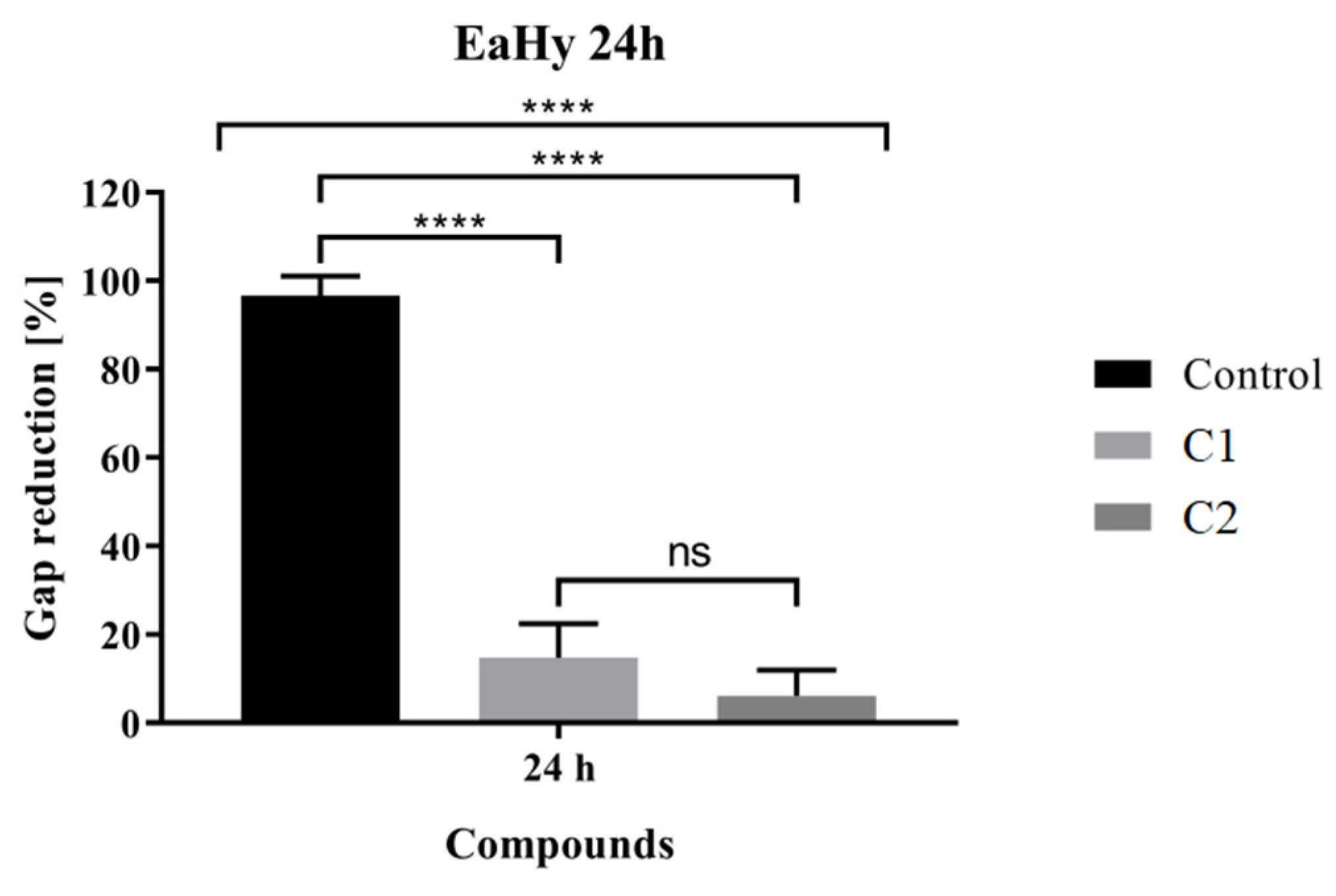
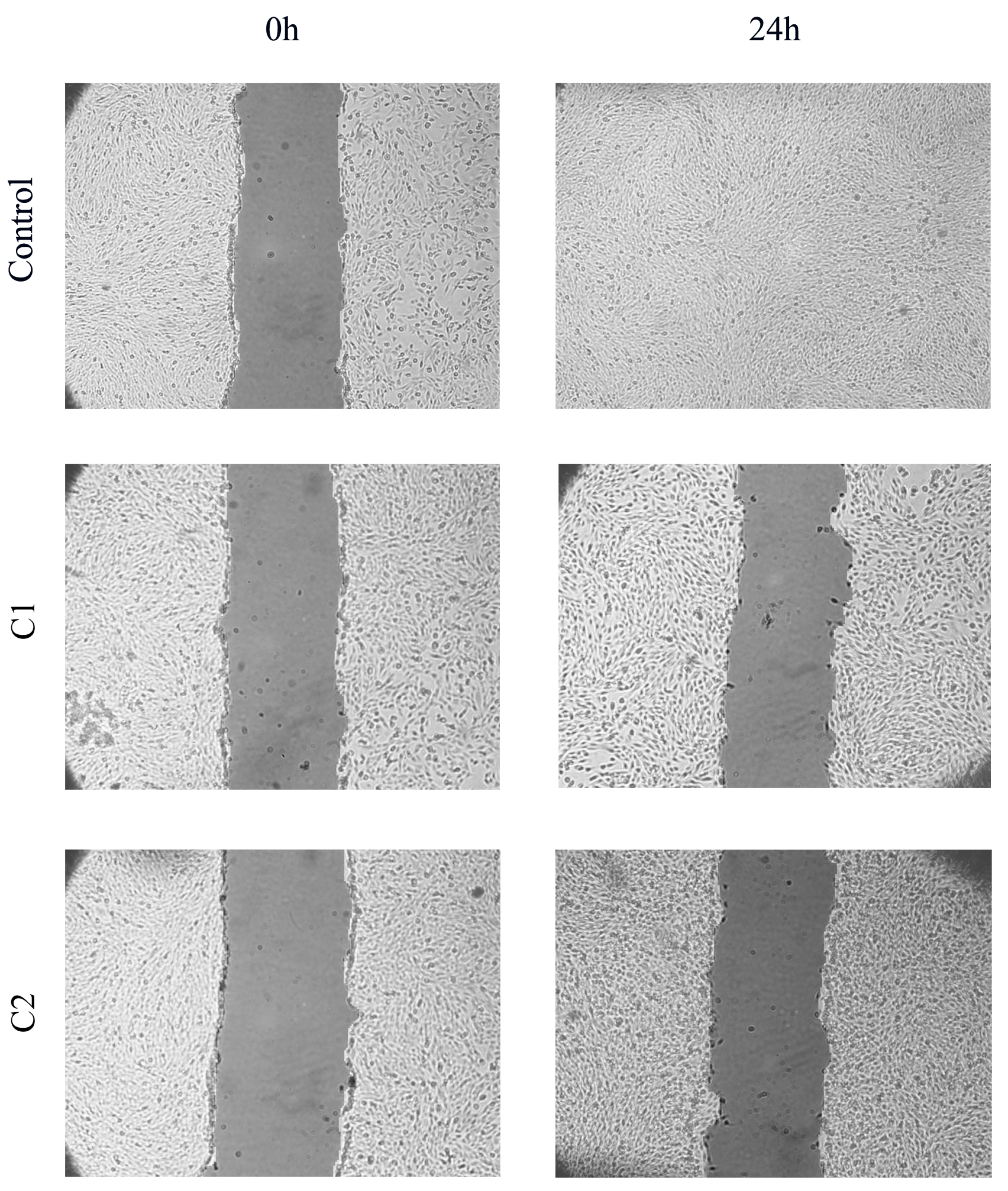
2.6. In Vitro Scratch Assay
2.7. Tube Formation Assay
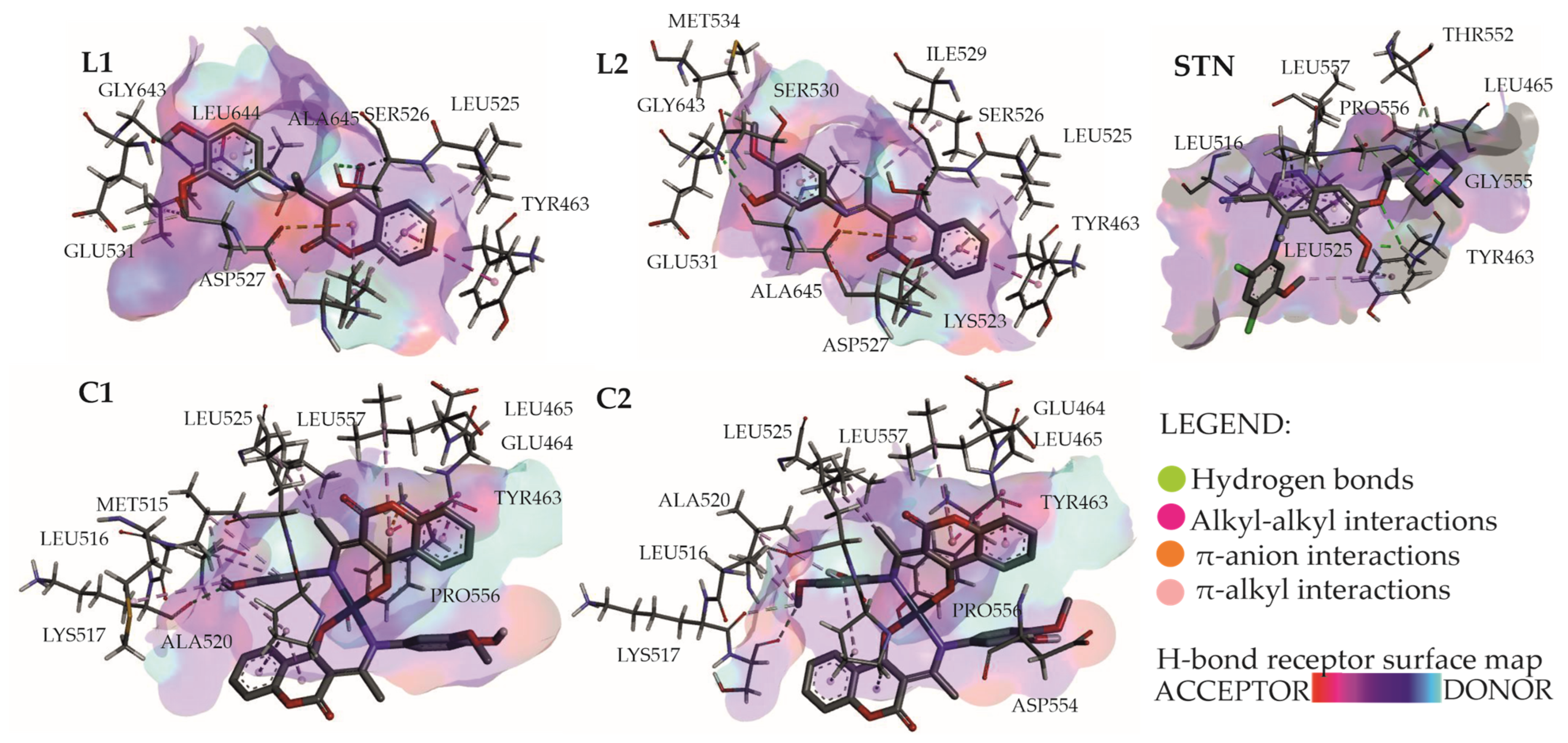
2.8. Molecular Docking Study
3. Materials and Methods
3.1. Chemical Studies
3.1.1. Rationale for Choosing Ligand
3.1.2. General Procedure for Synthesis and Spectral Data of Palladium(II) Complexes
3.1.3. DFT Calculations
3.2. Biological Studies
3.2.1. In Vitro Cytotoxicity
3.2.2. Cell Cycle Analysis
3.2.3. Fluorescence Microscopy
3.2.4. In Vitro Scratch Assay
3.2.5. Tube Formation Assay
3.2.6. Antimicrobial Activity
3.2.7. Statistical Processing of the Information
3.2.8. Protocol of Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weinberg, R.A. How cancer arises. SciAm 1996, 275, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, C.K. Classification of multiple cancer types by multicategory support vector machines using gene expression data. Bioinformatics 2003, 19, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Abu-Surrah, A.S.; Safieh, K.A.A.; Ahmad, I.M.; Abdalla, M.Y.; Ayoub, M.T.; Qaroush, A.K.; Abu-Mahtheieh, A.M. New palladium (II) complexes bearing pyrazole-based Schiff base ligands: Synthesis, characterization and cytotoxicity. Eur. J. Med. Chem. 2010, 45, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Abu-Surrah, A.S.; Al-Allaf, T.A.; Rashan, L.J.; Klinga, M.; Leskelä, M. Synthesis, crystal structure and initial biological evaluation of the new enantiomerically pure chiral palladium (II) complex trans-bis {endo-(1R)-1,7,7-trimethylbicyclo [2.2.1]-heptan-2-amino} palladium (II) dichloride. Eur. J. Med. Chem. 2002, 37, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Tušek-Božić, L.; Furlani, A.; Scarcia, V.; De Clercq, E.; Balzarini, J. Spectroscopic and biological properties of palladium (II) complexes of ethyl 2-quinolylmethylphosphonate. J. Inorg. Biochem. 1998, 72, 201–210. [Google Scholar] [CrossRef]
- Keating, G.J.; O’Kennedy, R. The chemistry and occurrence of coumarins. In Coumarins: Biology, Applications and Mode of Action; Wiley: Hoboken, NJ, USA, 1997; pp. 23–66. [Google Scholar]
- Miranda Martínez, M.; Cuéllar-Cuéllar, A. Farmacognosia Y Productos Naturales; Editorial Félix Varela: Habana, Cuba, 2001; pp. 147–170. [Google Scholar]
- Ojala, T.; Remes, S.; Haansuu, P.; Vuorela, H.; Hiltunen, R.; Haahtela, K.; Vuorela, P. Antimicrobial activity of some coumarin containing herbal plants growing in Finland. J. Ethnopharmacol. 2000, 73, 299–305. [Google Scholar] [CrossRef]
- Cottigli, F.; Loy, G.; Garau, D.; Floris, C.; Caus, M.; Pompei, R.A.F.F.; Bonsignore, L. Antimicrobial evaluation of coumarins and flavonoids from the stems of Daphne gnidium L. Phytomedicine 2001, 8, 302–305. [Google Scholar] [CrossRef]
- Basile, A.; Sorbo, S.; Spadaro, V.; Bruno, M.; Maggio, A.; Faraone, N.; Rosselli, S. Antimicrobial and antioxidant activities of coumarins from the roots of Ferulago campestris (Apiaceae). Molecules 2009, 14, 939–952. [Google Scholar] [CrossRef]
- Bhavsar, D.; Trivedi, J.; Parekh, S.; Savant, M.; Thakrar, S.; Bavishi, A.; Radadiya, A.; Vala, H.; Lunagariya, J.; Parmar, M.; et al. Synthesis and in vitro anti-HIV activity of N-1, 3-benzo [d] thiazol-2-yl-2-(2-oxo-2H-chromen-4-yl) acetamide derivatives using MTT method. Bioorg. Med. Chem. 2011, 21, 3443–3446. [Google Scholar] [CrossRef]
- Kostova, I. Coumarins as inhibitors of HIV reverse transcriptase. Curr. HIV Res. 2006, 4, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, H.R.; Liu, H.S.; Cheng, M.; Xia, P.; Qian, K.; Wu, P.C.; Lai, C.Y.; Xia, Y.; Yang, Z.Y.; et al. Antitumor agents 292. Design, synthesis and pharmacological study of S-and O-substituted 7-mercapto-or hydroxy-coumarins and chromones as potent cytotoxic agents. Eur. J. Med. Chem. 2012, 49, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Singla, R.; Jaitak, V. Coumarins as anticancer agents: A review on synthetic strategies, mechanism of action and SAR studies. Eur. J. Med. Chem. 2015, 101, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.O.; Bari, S.B.; Firke, S.D.; Deshmukh, P.K.; Donda, S.T.; Patil, D.A. A comprehensive review on synthesis and designing aspects of coumarin derivatives as monoamine oxidase inhibitors for depression and Alzheimer’s disease. Bioorg. Med. Chem. 2013, 21, 2434–2450. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, X.B.; Wang, T.; Kong, L.Y. Design, synthesis, and acetylcholinesterase inhibitory activity of novel coumarin analogues. Bioorg. Med. Chem. 2008, 16, 8011–8021. [Google Scholar] [CrossRef]
- Kontogiorgis, C.; Hadjipavlou-Litina, D. Biological evaluation of several coumarin derivatives designed as possible anti-inflammatory/antioxidant agents. J. Enzyme Inhib. Med. Chem. 2003, 18, 63–69. [Google Scholar] [CrossRef]
- Kontogiorgis, C.A.; Hadjipavlou-Litina, D.J. Synthesis and antiinflammatory activity of coumarin derivatives. J. Med. Chem. 2005, 48, 6400–6408. [Google Scholar] [CrossRef]
- Dimić, D.S.; Marković, Z.S.; Saso, L.; Avdović, E.H.; Đorović, J.R.; Petrović, I.P.; Stanisavljević, D.D.; Stevanović, M.J.; Potočňák, I.; Samoľová, E.; et al. Synthesis and Characterization of 3-(1-((3, 4-Dihydroxyphenethyl) amino) ethylidene)-chroman-2, 4-dione as a Potential Antitumor Agent. Oxid. Med. Cell. Longev. 2019, 2019, 12–25. [Google Scholar] [CrossRef]
- Avdović, E.H.; Petrović, I.P.; Stevanović, M.J.; Saso, L.; Dimitrić Marković, J.M.; Filipović, N.D.; Živić, M.Ž.; Cvetić Antić, T.N.; Žižić, M.V.; Todorović, N.V.; et al. Synthesis and Biological Screening of New 4-Hydroxycoumarin Derivatives and Their Palladium (II) Complexes. Oxid. Med. Cell. Longev. 2021, 2021, 8849568. [Google Scholar] [CrossRef]
- Avdović, E.H.; Milenković, D.; Marković, J.M.D.; Đorović, J.; Vuković, N.; Vukić, M.D.; Jevtić, V.V.; Trifunović, S.R.; Potočňák, I.; Marković, Z. Synthesis, spectroscopic characterization (FT-IR, FT-Raman, and NMR), quantum chemical studies and molecular docking of 3-(1-(phenylamino) ethylidene)-chroman-2, 4-dione. Spectrochim. Acta A Mol. 2018, 195, 31–40. [Google Scholar] [CrossRef]
- Avdović, E.H.; Milenković, D.; Dimitrić-Marković, J.M.; Vuković, N.; Trifunović, S.R.; Marković, Z. Structural, spectral and NBO analysis of 3-(1-(3-hydroxypropylamino) ethylidene) chroman-2, 4-dione. J. Mol. Struct. 2017, 1147, 69–75. [Google Scholar] [CrossRef]
- Ilić, D.R.; Jevtić, V.V.; Radić, G.P.; Arsikin, K.; Ristić, B.; Harhaji-Trajković, L.; Trifunović, S.R. Synthesis, characterization and cytotoxicity of a new palladium (II) complex with a coumarine-derived ligand. Eur. J. Med. Chem. 2014, 74, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Budzisz, E.; Keppler, B.K.; Giester, G.; Wozniczka, M.; Kufelnicki, A.; Nawrot, B. Synthesis, Crystal Structure and Biological Characterization of a Novel Palladium(II) Complex with a Coumarin-Derived Ligand. EurJIC 2004, 2004, 4412–4419. [Google Scholar] [CrossRef]
- Krstic, A.; Pavic, A.; Avdovic, E.; Markovic, Z.; Stevanovic, M.; Petrovic, I. Coumarin-Palladium (II) Complex Acts as a Potent and Non-Toxic Anticancer Agent against Pancreatic Carcinoma Cells. Molecules 2022, 27, 2115. [Google Scholar] [CrossRef] [PubMed]
- Avdović, E.H.; Stojković, D.L.; Jevtić, V.V.; Kosić, M.; Ristić, B.; Harhaji-Trajković, L.; Vukić, M.; Vuković, N.; Marković, Z.S.; Potočňák, I.; et al. Synthesis, characterization and cytotoxicity of a new palladium (II) complex with a coumarin-derived ligand 3-(1-(3-hydroxypropylamino) ethylidene) chroman-2, 4-dione. Crystal structure of the 3-(1-(3-hydroxypropylamino) ethylidene)-chroman-2, 4-dione. Inorg. Chim. Acta 2017, 466, 188–196. [Google Scholar] [CrossRef]
- Avdović, E.H.; Milanović, Ž.B.; Živanović, M.N.; Šeklić, D.S.; Radojević, I.D.; Čomić, L.R.; Trifunović, S.R.; Amić, A.; Marković, Z.S. Synthesis, spectroscopic characterization, biological activity, DFT and molecular docking study of novel 4-hydroxycoumarine derivatives and corresponding palladium (II) complexes. Inorg. Chim. Acta 2020, 504, 119465. [Google Scholar] [CrossRef]
- Milenković, D.A.; Dimić, D.S.; Avdović, E.H.; Marković, Z.S. Several coumarin derivatives and their Pd (II) complexes as potential inhibitors of the main protease of SARS-CoV-2, an in silico approach. RSC Adv. 2020, 10, 35099–35108. [Google Scholar] [CrossRef]
- Dimić, D.S.; Kaluđerović, G.N.; Avdović, E.H.; Milenković, D.A.; Živanović, M.N.; Potočňák, I.; Samoľová, E.; Dimitrijević, M.S.; Saso, L.; Marković, Z.S.; et al. Synthesis, Crystallographic, quantum chemical, antitumor, and molecular docking/dynamic studies of 4-hydroxycoumarin-neurotransmitter derivatives. Int. J. Mol. Sci. 2022, 23, 1001. [Google Scholar] [CrossRef]
- Milanović, Ž.B.; Dimić, D.S.; Avdović, E.H.; Milenković, D.A.; Marković, J.D. Synthesis and comprehensive spectroscopic (X-ray, NMR, FTIR, UV–Vis), quantum chemical and molecular docking investigation of 3-acetyl-4-hydroxy-2-oxo-2H-chromen-7-yl acetate. J. Mol. Struct. 2021, 1225, 129256. [Google Scholar] [CrossRef]
- Avdović, E.H.; Dimić, D.S.; Fronc, M.; Kožišek, J.; Klein, E.; Milanović, B.; Kesić, A.; Marković, Z.S. Structural and theoretical analysis, molecular docking/dynamics investigation of 3-(1-m-chloridoethylidene)-chromane-2, 4-dione: The role of chlorine atom. J. Mol. Struct. 2021, 1231, 129962. [Google Scholar] [CrossRef]
- Savić, A.; Gligorijević, N.; Aranđelović, S.; Dojčinović, B.; Kaczmarek, A.M.; Radulović, S.; Van Hecke, K. Antitumor activity of organoruthenium complexes with chelate aromatic ligands, derived from 1, 10-phenantroline: Synthesis and biological activity. J. Inorg. Biochem. 2020, 202, 110869. [Google Scholar] [CrossRef] [PubMed]
- Milanović, Ž.B.; Marković, Z.S.; Dimić, D.S.; Klisurić, O.R.; Radojević, I.D.; Šeklić, D.S.; Živanović, M.N.; Dimitrić Marković, J.; Radulović, M.; Avdović, E.H. Synthesis, structural characterization, biological activity and molecular docking study of 4, 7-dihydroxycoumarin modified by aminophenol derivatives. Comptes Rendus Chim. 2021, 24, 215–232. [Google Scholar] [CrossRef]
- Kargar, H.; Behjatmanesh-Ardakani, R.; Torabi, V.; Sarvian, A.; Kazemi, Z.; Chavoshpour-Natanzi, Z.; Ashfaq, M. Novel copper (II) and zinc (II) complexes of halogenated bidentate N, O-donor Schiff base ligands: Synthesis, characterization, crystal structures, DNA binding, molecular docking, DFT and TD-DFT computational studies. Inorg. Chim. Acta 2021, 514, 120004. [Google Scholar] [CrossRef]
- Raymond, E.; Faivre, S.; Armand, J.P. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs 2000, 60, 15–23. [Google Scholar] [CrossRef]
- Purnama, A.; Rizki, D.R.; Qanita, I.; Iqhrammullah, M.; Ahmad, K.; Mardina, V.; Puspita, K.; Hasballah, K. Molecular docking investigation of calotropone as a potential natural therapeutic agent against pancreatic cancer. JAPTR 2022, 13, 44. [Google Scholar]
- Oliveira-Cunha, M.; Newman, W.G.; Siriwardena, A.K. Epidermal growth factor receptor in pancreatic cancer. Cancers 2011, 3, 1513–1526. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J.; Ferguson, K.M. The EGFR family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2014, 6, a020768. [Google Scholar] [CrossRef]
- Bold, R.J.; Virudachalam, S.; McConkey, D.J. BCL2 expression correlates with metastatic potential in pancreatic cancer cell lines. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2001, 92, 1122–1129. [Google Scholar] [CrossRef]
- Cohen, M.H.; Johnson, J.R.; Chen, Y.F.; Sridhara, R.; Pazdur, R. FDA drug approval summary: Erlotinib (Tarceva®) tablets. Oncologist 2005, 10, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Isfort, S.; Amsberg, G.K.V.; Schafhausen, P.; Koschmieder, S.; Brümmendorf, T.H. Bosutinib: A novel second-generation tyrosine kinase inhibitor. Small Mol. Oncol. 2014, 201, 81–97. [Google Scholar] [CrossRef]
- Wilson, W.H.; O’Connor, O.A.; Czuczman, M.S.; LaCasce, A.S.; Gerecitano, J.F.; Leonard, J.P.; Humerickhouse, R.A. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: A phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010, 11, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- Vargesson, N. Thalidomide-induced limb defects: Resolving a 50-year-old puzzle. Bioessays 2009, 31, 1327–1336. [Google Scholar] [CrossRef]
- Kraskouskaya, D.; Duodu, E.; Arpin, C.C.; Gunning, P.T. Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 2013, 42, 3337–3370. [Google Scholar] [CrossRef]
- Morlacchi, P.; Robertson, F.M.; Klostergaard, J.; McMurray, J.S. Targeting SH2 domains in breast cancer. Future Med. Chem. 2014, 6, 1909–1926. [Google Scholar] [CrossRef]
- Bae, J.H.; Lew, E.D.; Yuzawa, S.; Tomé, F.; Lax, I.; Schlessinger, J. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell 2009, 138, 514–524. [Google Scholar] [CrossRef]
- Sanches, K.; Dias, R.V.R.; da Silva, P.H.; Fossey, M.A.; Caruso, Í.P.; de Souza, F.P.; de Melo, F.A. Grb2 dimer interacts with Coumarin through SH2 domains: A combined experimental and molecular modeling study. Heliyon 2019, 5, e02869. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, L.; Jiang, Y.; Li, Z.; Feng, L.; Zhuang, X.; Xu, B. Preclinical Evaluation of the Multiple Tyrosine Kinases Inhibitor Anlotinib in Leukemia Stem Cells. Pharmaceuticals 2022, 15, 1313. [Google Scholar] [CrossRef]
- Chin, Y.P.; Huang, W.J.; Hsu, F.L.; Lin, Y.L.; Lin, M.H. Synthesis and evaluation of antibacterial activities of 5,7-dihydroxycoumarin derivatives. Arch. Pharm. 2011, 344, 386–393. [Google Scholar] [CrossRef]
- Avdović, E.H.; Milanović, Ž.B.; Molčanov, K.; Roca, S.; Vikić-Topić, D.; Mrkalić, E.M.; Jelić, R.M.; Marković, Z.S. Synthesis, characterization and investigating the binding mechanism of novel coumarin derivatives with human serum albumin: Spectroscopic and computational approach. J. Mol. Struct. 2022, 1254, 132366. [Google Scholar] [CrossRef]
- Frisch, M.E.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Fox, D.J. Gaussian 16; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Check, C.E.; Faust, T.O.; Bailey, J.M.; Wright, B.J.; Gilbert, T.M.; Sunderlin, L.S. Addition of polarization and diffuse functions to the LANL2DZ basis set for p-block elements. J. Phys. Chem. A. 2001, 105, 8111–8116. [Google Scholar] [CrossRef]
- Andersson, M.P.; Uvdal, P. New scale factors for harmonic vibrational frequencies using the B3LYP density functional method with the triple-ζ basis set 6-311+ G. (d, p). J. Phys. Chem. A. 2005, 109, 2937–2941. [Google Scholar] [CrossRef]
- Becke, A.D.; Johnson, E.R. A density-functional model of the dispersion interaction. J. Chem. Phys. 2005, 123, 154101. [Google Scholar] [CrossRef]
- Wolinski, K.; Hinton, J.F.; Pulay, P. Efficient implementation of the gauge-independent atomic orbital method for NMR chemical shift calculations. J. Am. Chem. Soc. 1990, 112, 8251–8260. [Google Scholar] [CrossRef]
- Kargar, H.; Torabi, V.; Akbari, A.; Behjatmanesh-Ardakani, R.; Sahraei, A.; Tahir, M.N. Pd (II) and Ni (II) complexes containing an asymmetric Schiff base ligand: Synthesis, X-ray crystal structure, spectroscopic investigations and computational studies. J. Mol. Struct. 1205, 2020, 127642. [Google Scholar] [CrossRef]
- Takano, Y.; Houk, K.N. Benchmarking the conductor-like polarizable continuum model (CPCM) for aqueous solvation free energies of neutral and ionic organic molecules. J. Chem. Theory Comput. 2005, 1, 70–77. [Google Scholar] [CrossRef]
- Grozdanic, N.; Zdunić, G.; Ŝavikin, K.; Duričić, I.; Kosanic, M.; Mačic, V.; Matic, I.; Stanojkovic, T. Seasonal variation in biopharmaceutical activity and fatty acid content of endemic Fucus virsoides algae from Adriatic Sea. Acta Pol. Pharm. 2019, 76, 833–844. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 24 September 2022).
- Bailey, N.T. Statistical Methods in Biology; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.W.; Beran, B.; Bi, C.; Bluhm, W.F.; Dimitropoulos, D.; Goodsell, D.S.; Prlic, A.; Quesada, M.; Quinn, G.B.; Westbrook, J.D.; et al. The RCSB Protein Data Bank: Redesigned web site and web services. Nucleic Acids Res. 2010, 39, D392–D401. [Google Scholar] [CrossRef] [PubMed]
- Jejurikar, B.L.; Rohane, S.H. Drug designing in discovery studio. Asian J. Res. Chem. 2021, 14, 135–138. [Google Scholar]
- Rizvi, S.M.D.; Shakil, S.; Haneef, M. A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 2013, 12, 831. [Google Scholar]
- Avdović, E.H.; Stojković, D.L.; Jevtić, V.V.; Milenković, D.; Marković, Z.S.; Vuković, N.; Potočňák, I.; Radojević, I.D.; Čomić, L.R.; Trifunović, S.R. Preparation and antimicrobial activity of a new palladium (II) complexes with a coumarin-derived ligands. Crystal structures of the 3-(1-(o-toluidino) ethylidene)-chroman-2,4-dione and 3-(1-(m-toluidino) ethylidene)-chroman-2,4-dione. Inorg. Chim. Acta 2019, 484, 52–59. [Google Scholar]
- Ardakani, A.A.; Kargar, H.; Feizi, N.; Tahir, M.N. Synthesis, characterization, crystal structures and antibacterial activities of some Schiff bases with N2O2 donor sets. J. Iran. Chem. Soc. 2018, 15, 1495–1504. [Google Scholar]
- Arulmurugan, S.; Kavitha, H.P.; Venkatraman, B.R. Biological activities of Schiff base and its complexes: A review. Rasayan J. Chem. 2010, 3, 385–410. [Google Scholar]




| IC50, µM * | FemX | A549 | Panc-1 | HeLa |
|---|---|---|---|---|
| L1 | 21.43 ± 1.59 | 78.83 ± 11.48 | 28.17 ± 7.40 | >500 |
| L2 | 27.56 ± 5.54 | 404.90 ± 6.94 | 72.71 ± 11.80 | >500 |
| C1 | 6.97 ± 0.36 | 10.7 ± 0.71 | 7.67 ± 0.74 | 5.68 ± 0.69 |
| C2 | 7.67 ± 0.09 | 11.06 ± 0.41 | 10.43 ± 0.34 | 4.25 ± 0.85 |
| Cis-Pt | 6.16 ± 0.31 | 12.74 ± 1.26 | 16.44 ± 1.56 | 4.00 ± 0.47 |
| Protein | EGFR | RTK | BCL-2 | ||||
|---|---|---|---|---|---|---|---|
| Ligand/Complex | ΔGbind | ki (µM) | ΔGbind | ki (µM) | ΔGbind | ki (µM) | |
| L1 | −5.80 | 56.05 | −8.80 | 0.35 | −7.94 | 1.51 | |
| L2 | −6.26 | 25.79 | −8.39 | 0.70 | −8.39 | 0.71 | |
| C1-trans | −6.47 | 18.09 | −10.22 | 0.03 | −8.56 | 0.53 | |
| C2-trans | −6.61 | 14.28 | −9.40 | 0.13 | −6.87 | 9.21 | |
| Standards | −4.93 | 240.00 | −8.20 | 0.98 | −8.69 | 0.43 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avdović, E.H.; Antonijević, M.; Simijonović, D.; Roca, S.; Topić, D.V.; Grozdanić, N.; Stanojković, T.; Radojević, I.; Vojinović, R.; Marković, Z. Synthesis and Cytotoxicity Evaluation of Novel Coumarin–Palladium(II) Complexes against Human Cancer Cell Lines. Pharmaceuticals 2023, 16, 49. https://doi.org/10.3390/ph16010049
Avdović EH, Antonijević M, Simijonović D, Roca S, Topić DV, Grozdanić N, Stanojković T, Radojević I, Vojinović R, Marković Z. Synthesis and Cytotoxicity Evaluation of Novel Coumarin–Palladium(II) Complexes against Human Cancer Cell Lines. Pharmaceuticals. 2023; 16(1):49. https://doi.org/10.3390/ph16010049
Chicago/Turabian StyleAvdović, Edina H., Marko Antonijević, Dušica Simijonović, Sunčica Roca, Dražen Vikić Topić, Nađa Grozdanić, Tatjana Stanojković, Ivana Radojević, Radiša Vojinović, and Zoran Marković. 2023. "Synthesis and Cytotoxicity Evaluation of Novel Coumarin–Palladium(II) Complexes against Human Cancer Cell Lines" Pharmaceuticals 16, no. 1: 49. https://doi.org/10.3390/ph16010049
APA StyleAvdović, E. H., Antonijević, M., Simijonović, D., Roca, S., Topić, D. V., Grozdanić, N., Stanojković, T., Radojević, I., Vojinović, R., & Marković, Z. (2023). Synthesis and Cytotoxicity Evaluation of Novel Coumarin–Palladium(II) Complexes against Human Cancer Cell Lines. Pharmaceuticals, 16(1), 49. https://doi.org/10.3390/ph16010049










