Abstract
The p53 protein has appropriately been named the “guardian of the genome”. In almost all human cancers, the powerful tumor suppressor function of p53 is compromised by a variety of mechanisms, including mutations with either loss of function or gain of function and inhibition by its negative regulators MDM2 and/or MDMX. We review herein the progress made on different therapeutic strategies for targeting p53.
Keywords:
wild-type p53; mutant p53; activators; MDM2; MDMX; Y220C; inhibitors; PROTACS; degraders; molecular glue 1. Introduction
Appropriately named the guardian of the genome, p53 is the hub in which cell fate is determined [1]. p53 is a transcription factor that functions as a tumor suppressor, which is induced in response to various stresses, such as DNA damage, oncogene expression, oxidative stress, nutrient deprivation, hypoxia, telomere attrition, or ribosomal dysfunction through both transcription-dependent and -independent mechanisms [2]. In response to these stresses, p53 can activate or repress the expression of many target genes that induce responses, such as cell cycle arrest, DNA repair, senescence, apoptosis, metabolism, autophagy, or ferroptosis [3].
The p53 protein has 393 amino acids, which are encoded by the TP53 gene. Structurally, p53 comprises several domains with different functions, including: an N-terminal transactivation domain (1–62) and proline rich region (63–94); a DNA binding domain (95–292); an oligomerization domain (325–356); and a C-terminal regulatory domain (357–393) (Figure 1) [2,4,5]. In unstressed cells, low levels of p53 are maintained by direct interactions with its negative regulators MDM2 and MDMX [6,7,8]. However, upon stress stimuli, such as DNA damage or oncogene activation, p53 is phosphorylated (P) by upstream kinases [9,10] or acetylated (Ac) [11,12]. Both of these post-translational modifications (PTM) stabilize and activate p53 by dislocating it from and decreasing the binding to its negative regulator MDM2 [9,10,11,12]. p53 is highly disordered and consequently has intrinsically low thermodynamic and kinetic stability and a body temperature half-life of about 9 min during which it switches rapidly between its unfolded and folded states [13,14]. In the activation pathway of p53, heat shock proteins (HSP) function as molecular chaperones [15,16,17] that assist p53 in folding and adopting its active, tetrameric form [18,19,20,21,22], which binds to targeted DNAs for gene transcription (Figure 1).
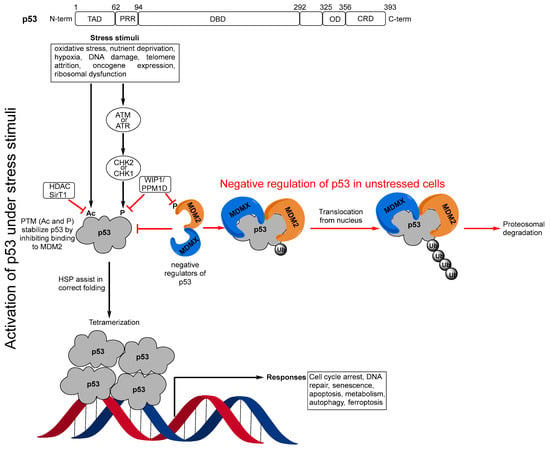
Figure 1.
(p53 domains) Transactivation domain (TAD), proline rich region (PRR), DNA binding domain (DBD), oligomerization domain (OD), C-terminal regulatory domain (CRD); (Negative regulation of p53 in unstressed cells) In unstressed conditions, p53 levels are kept low by binding to its major negative regulators MDM2 and MDMX, which block its transactivation activity and polyubiquitinate it for proteosomal degradation; (Activation of p53 under stress stimuli) Under stress stimuli, the post-translational modifications (PTM), acetylation (Ac), or phosphorylation (P) by checkpoint kinase 1 or 2 (CHK1 or CHK2), which result from activation of the upstream kinases ataxia telangiectasia mutated (ATM), and ataxia telangiectasia and Rad3-related protein (ATR), stabilize and activate p53 by inhibiting its binding to MDM2. Heat shock proteins (HSP) then assist in the correct folding of the p53 monomer, which then forms its active tetrameric form, which binds to DNA for gene transcription. In this activation process, there are also other mechanisms that can result in repression of p53 through removal of the PTMs. The histone deacetylase (HDAC), sirtuin 1 (SirT1), deacetylates p53 and enhances its ubiquitination by MDM2. p53 also induces expression of another of its negative regulators, WIP1/PPM1D, which is a phosphatase 1 that destabilizes p53 by dephosphorylating serine 15 in p53 but also by dephosphorylating MDM2, resulting in stabilization of MDM2.
In essentially almost all human cancers, the powerful tumor suppressor function of p53 is compromised through a variety of mechanisms, such as amplification or upregulation of its negative regulators MDM2 and/or MDMX, or through the mutation or deletion of the TP53 gene. In the last 20 years, significant efforts have been made to reactivate the tumor suppressor function of both wild-type (wt) and mutant p53.
The therapeutic strategies targeting p53 can be broadly divided into two categories: (i) activation of wt-p53 and (ii) restoration of wt-like functions of mutant p53, which have been the subjects of a number of reviews [2,3,23,24,25,26]. In this review, we will focus on recent therapeutic advances in targeting the major negative regulation MDM2–MDMX–p53 axis and activation of one specific p53 Y220C mutant.
2. Targeting the Protein–Protein Interaction between MDM2 and p53
In approximately 50% of human cancers, p53 retains its wild-type (wt) status, but its functions are effectively inhibited by the murine double minute 2 (MDM2) protein through a direct protein–protein interaction [27,28,29,30,31,32,33]. Through direct binding, the MDM2 protein blocks the transactivation domain of p53, mono-ubiquitinates it to transport p53 from the nucleus to the cytoplasm, and then poly-ubiquitinates p53 for proteasomal degradation [27]. The interaction of MDM2 and p53 occurs between the first 120 N-terminal amino acids of MDM2 and the first 30 N-terminal amino acids in the transactivation domain of p53. A high-resolution co-crystal structure of residues 15–29 of p53 bound to MDM2 has been reported [31,34]. In this co-crystal structure, the p53 peptide adopts an α-helical conformation, which brings its three hydrophobic residues Phe19, Trp23, and Leu26 together to form a cluster, which interacts with a well-defined, hydrophobic pocket in MDM2. The discovery of this well-defined pocket in MDM2 suggested that the MDM2–p53 interaction may be effectively targeted by small-molecule inhibitors, which has been the subject of extensive research efforts by the academia and industry in the last 20 years. To date, a number of potent and selective MDM2 inhibitors have been progressed into clinical development (Figure 2). Here, we summarize a number of successful programs that led to the advancement of MDM2 inhibitors into clinical trials.
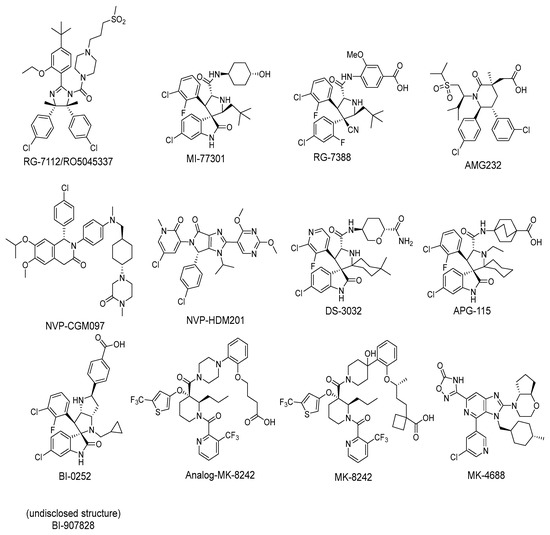
Figure 2.
Chemical structure of MDM2 inhibitors.
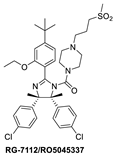
Among the first in class MDM2 inhibitors that were reported are the Nutlins, which were discovered by Hoffmann-La Roche through screening of a library of synthetic chemicals. Further chemical optimization to improve potency and selectivity produced the first lead compound, Nutlin-3a [35]. Further optimization led to the discovery of RG-7112 (Figure 2 and Figure 3A), which has an MDM2 binding affinity with IC50 = 18 nM and an average cellular activity with IC50 = 400 nM in wt-p53 cancer cells (Table 1). RG-7112 was the first MDM2 inhibitor to enter clinical trials [36]. In the reported optimization SAR, substitution of the 4 and 5 positions of the imidazoline core of Nutlin-3a with methyl groups was accomplished to prevent oxidation, which converts this core of the compound into an inactive imidazole. The tert-butyl group in RG7112 was introduced to replace the methoxyl group, which was found to be a major metabolic soft spot, producing the phenol metabolite. The isopropyl group in Nutlin-3a that enters the Phe19 pocket was replaced with an ethyl group to decrease molecular weight, while the piperazine substituent was modified to further improve the binding and pharmacokinetic properties, leading to the optimized RG-7112 that entered clinical trials [36] (Figure 3A). Its clinical trial demonstrated that MDM2 inhibitors can indeed activate p53 signaling in human tumors, validating this approach.
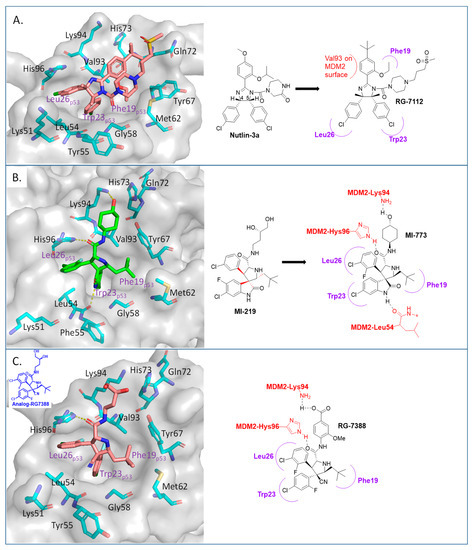
Figure 3.
Co-crystal structures and chemical structures of inhibitors bound to MDM2 protein: (A) RG7112; (B) MI-773; and (C) RG7388. Labels and semicircles in magenta indicate the binding pockets in MDM2 where the p53 residues Phe19, Trp23, and Leu26 bind. Labels in red indicate MDM2 residues.

Table 1.
Available data of MDM2 inhibitors.
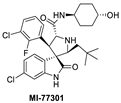
The Wang lab at the University of Michigan, inspired by structures of natural products and rational design, discovered the spiro-oxindoles as a novel class of MDM2 inhibitors, with MI-219 as the initial lead [50,51,52]. Based on the chemistry used to build the spiro-oxindole scaffold, the stereochemistry around the pyrrolidine ring of MI-219 was assigned, but subsequently, it was discovered that a rapid and reversible retro-Mannich and recyclization reaction converted the spiro-oxindoles to their more potent and energetically stable all-trans isomers (Figure 3B) [53,54]. Further optimization of this class of compounds led to the discovery of MI-773, which was entered into clinical trials [37]. The optimization of this compound was mainly achieved by extensive modification of the carboxamide substituent, which improved its potency, pharmacokinetic properties, and in vivo efficacy. Its successful co-crystallization with MDM2 revealed the reason for its superior potency, showing that MI-773 establishes additional interactions with the MDM2 protein, which are not experienced by the Nutlins (Figure 3B). In addition to occupying the Phe19, Trp23, Leu26 pockets in MDM2 through the substituents on the core pyrrolidine of MI-773, the N-H of the oxindole forms a hydrogen bond with the backbone carbonyl of Leu54; the carbonyl of the carboxamide functions as a hydrogen bond acceptor from the NH of Hys96, and the 4-hydroxyl-cyclohexyl-carboxamide substituent has additional hydrophobic interaction with the MDM2 surface, and its hydroxyl group forms a hydrogen bond with Lys94 of the MDM2 protein (Figure 3B). Capturing these additional hydrophobic and polar interactions together with the three key p53 interactions contributes to this class of compounds having a significantly enhanced MDM2 binding affinity (Ki = 0.88 nM) and cellular activity (IC50 < 200 nM) in wt-p53 cancer cells (Table 1). This compound showed good pharmacokinetics and achieved complete tumor regression in mouse xenograft models with no signs of toxicity [37].
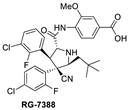
After discovering the Nutlins, the Hoffmann-La Roche team continued their exploration of novel MDM2 inhibitors, including the investigation of spiro-oxindole analogs [55]. Inspired by the highly functionalized pyrrolidine core present in the spiro-oxindole class of MDM2 inhibitors and through de novo design around the pyrrolidine ring, they explored different configurations of the pyrrolidine ring substituents and found that the trans-configuration between the aryl groups gave the best results [38]. It was found that the CN group was critical to achieving this trans-configuration, and this led to the discovery of a new series of MDM2 inhibitors with a novel pyrrolidine core. This exploration yielded RG7388 (Figure 3C), which has an MDM2 binding affinity (IC50 = 6 nM) and an average cellular activity (IC50 = 30 nM) in wt-p53 cancer cells (Table 1). RG7388 is now in clinical trials [38]. The co-crystal structure of RG7388 with MDM2 was not obtained, but that of a close analog shows its likely binding mode (Figure 3C). RG7388 has almost the same substituents as MI-773, which occupy the Phe19, Trp23, and Leu26 pockets, with the exception of the spiro-oxindole portion, which is replaced and mimicked by the quaternary carbon-3, which is substituted with a cyano and a 2-fluoro-4-chlorophenyl group. Furthermore, in RG7388, the carboxamide substituent was switched from an aliphatic group to a meta-methoxylbenzoic acid. Not surprisingly, the binding mode of the RG7388 class to MDM2 was identical to that of MI-773, with the exception that RG7388 does not possess the oxindole, which can form a hydrogen bond with the carbonyl of Leu54 of the MDM2 protein. It was reported, however, that replacement of the aliphatic with an aromatic carboxamide substituent was critical to metabolic stability and, in this class of molecules, significantly improved the cellular potency, PK profile, and in vivo efficacy [38]. The loss of the oxindole hydrogen bonding with Leu54 in RG7388 is probably compensated for by the increased potency from the 3-methoxy substituent on the aromatic carboxamide and the strong electrostatic interaction of the carboxylate with Lys94 of MDM2, which is only a hydrogen bond in the co-crystal structure involving MI-773. RG7388 is the MDM2 inhibitor, which has progressed the furthest and is currently in phase III clinical trials.
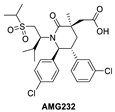
Inspired by available structural information, Amgen scientists used a de novo design approach in their discovery of the trans-piperidone class of MDM2 inhibitors, which produced the clinical compound AMG232 (Figure 4A) [39,40]. Reports from this group revealed the power of rational design using available structural information for the initial design, then further optimization guided by a combination of modeling and co-crystal structures of compounds in the series. As a consequence, each substituent in the structure has a specific interaction with the MDM2 protein and helps maintain adequate physicochemical properties for a drug candidate. AMG232 has an MDM2 binding affinity (IC50 = 0.6 nM) and an EdU cell proliferation (IC50 = 9.1 nM) in SJSA-1 cancer cells (Table 1). The co-crystal structure of AMG232 bound to MDM2 has not been reported, but that of a close analog shows the key interactions (Figure 4A) [39]. This co-crystal structure indicates that the isopropyl, para-chlorophenyl, and meta-chlorophenyl substituents occupy the three key binding pockets on MDM2, mimicking the interactions of the p53 residues Phe19, Trp23, and Leu26, respectively. Its acetic acid functionality captures a strong electrostatic interaction with the imidazole of His96 of MDM2, and this significantly contributes to potency. As explained in their report, a methyl substituent was incorporated at C3 to introduce a 1,3-steric strain and destabilize the undesired anti-like conformation between the two aryl groups, leading to a dominant gauche conformation, which resulted in a further increase in the potency [40]. Unique to this class of compounds is their capture of hydrophobic interactions in the Gly58 shelf with the isopropyl group associated with the sulfone moiety. They explained that the incorporation of this sulfone moiety served two roles. The first role is that this hydrophilic group stabilizes the conformation required to direct the N-alkyl substituent into the Phe19(p53) binding pocket and the second is to capture the interaction in the Gly58 shelf of the MDM2 protein to further improve potency and maintain good metabolic stability [40].
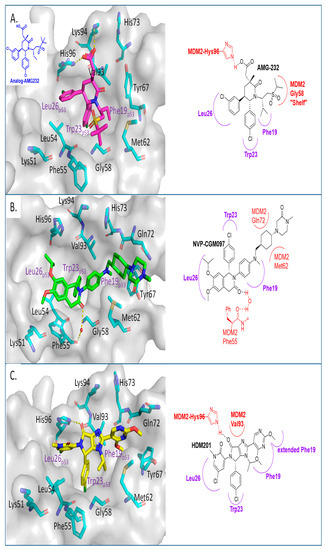
Figure 4.
Co-crystal structures and chemical structures of inhibitors bound to the MDM2 protein: (A) AMG-232; (B) NVP-CGM097; (C) and NVP-HDM201. Labels and semicircles in magenta indicate the binding pockets in MDM2 where the p53 residues Phe19, Trp23, and Leu26 bind. Labels in red indicate MDM2 residues.
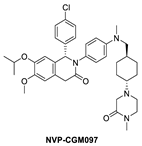
In their pursuit of novel small-molecule MDM2 inhibitors, a Novartis group used a design strategy, which revolves around the “central valine concept”. This concept involves placing a tailored aromatic core close to the Val93 residue in the center of the p53 binding pocket in MDM2 and then substituting positions around the core with appropriate vectors toward the Phe19, Trp23, and Leu26 binding pockets in MDM2 [56]. Virtual screening of their internal collection of compounds resulted in a preliminary selection of around 50,000 compounds, and from these, a dihydroisoquinolinone hit was identified. Through medicinal chemistry exploration, they discovered a new class of dihydroisoquinolinone derivatives as novel MDM2 inhibitors [57]. The major optimization efforts with this class of inhibitors focused on physicochemical properties, which influence in vivo toxicological outcomes, and yielded NVP-CGM097 with an MDM2 binding affinity of IC50 = 1.7 nM and cell activity of IC50 < 450 nM in wt-p53 cancer cells (Table 1). NVP-CGM097 was advanced to clinical trials [41]. Interestingly, the co-crystal structure of NVP-CGM097 revealed an unprecedented binding mode, which is the most distinct binding mode of all the MDM2 inhibitors. Most of its core lies close to and makes hydrophobic contact with residues Ile54, Phe55, and Gly58 of the MDM2 protein. This is the surface wall within the binding cleft and is opposite to that occupied by most of the other MDM2 inhibitors, which interact with the wall containing Val93 (Figure 4B). Even with this unusual binding mode, the substituents of NVP-CGM097 still capture the three key interactions that are necessary for binding to MDM2. The unique features of this scaffold are that it causes the Phe-55 residue of MDM2 to swing up to form a face-to-edge interaction with the dihydroisoquinolinone core and also that the N-methyl piperazinone substituent is nestled between the MDM2 protein walls above the Phe19 binding pocket; these two interactions were reported to contribute significantly to the potency of NVP-CGM097. Interestingly, it was also reported that due to its distinct binding mode to MDM2, a species-dependent binding was observed, which was not seen with Nutlins and presumably not with other MDM2 inhibitors whose binding is similar to that of Nutlins [41]. This difference was attributed to two amino acid residues, Leu54 and Leu57, in the human sequence, which are both switched to isoleucines in rodents, and in the case of dogs, only Leu54 is switched to isoleucine. This additional methyl on isoleucine causes a steric clash with the inhibitor, decreasing its binding in rodents and dogs but retaining its potency in humans and monkeys [41]. NVP-CGM097 was the first MDM2 inhibitor advanced into clinical trials by Novartis.
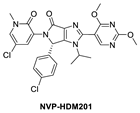
Continuing with their efforts to discover novel inhibitors of MDM2, the Novartis scientists proceeded to re-engineer the dihydroisoquinolinone mainly because its binding mode unexpectedly did not fit the “central valine” binding mode, making it an inefficient binder to MDM2. Deviation from this binding mode was rationalized as the result of a bent core, which results in a conformation energy loss in its binding to MDM2, which is compensated by other van der Waals and hydrogen bonding interactions with the MDM2 binding site [42,58]. This re-engineering produced the imidazolopyrrolidinone analog NVP-HDM201, which is the second MDM2 inhibitor from Novartis that progressed into clinical trials [42,59]. A co-crystal structure of NVP-HDM201 bound to MDM2 was obtained and showed the binding mode of this class of molecules (Figure 4C) [59]. The six-membered lactam fused to an aryl group in NVP-CGM097 was replaced in NVP-HDM201 with a five-membered lactam fused to a five-membered heteroaryl group, essentially removing the methylene group between the carbonyl and aryl, thus flattening the core and forcing the desired conformation of the para-chlorophenyl substituent, and so allowing the molecule to adopt the desired “central valine” binding mode (Figure 4C). In this central valine binding mode, the carbonyl of the lactam hydrogen bonds to His96, and the N-aryl substituent adopts a perpendicular conformation and becomes involved in π-π stacking with His96 of the MDM2 protein, both interactions that are not captured by the NVP-CGM907 inhibitor, which lies on the opposite side of the binding pocket [42]. Although a full report on the discovery of NVP-HDM201 has not been published to date, Novartis summarized their MDM2 program history in a short review article and indicated that NVP-HDM201 possesses advantageous physicochemical and good drug-like properties, and in their in vivo xenograft models, NVP-HDM201 was shown to be 10 times more potent than NVP-CGM097 [42].
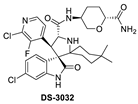
Daiichi-Sankyo also pursued the discovery of novel inhibitors of the p53–MDM2 interaction and advanced DS-3032, a spiro-oxindole MDM2 inhibitor, into clinical trials [43]. No report of their discovery effort that led to this inhibitor has been published, but it is a close analog of other reported spiro-oxindoles. Unique in DS-3032 is the 2-chloro-3-fluoro-pyridine substituent at the C3 of its pyrrolidine core. All the other reported spiro-oxindoles in clinical trials contain a 2-fluoro-3-chlorophenyl substituent at this position (Figure 2). However, since no discovery process was reported, the beneficial properties of this moiety are unknown.
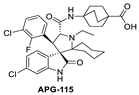
During the exploration by the University of Michigan of the spiro-oxindole small molecules as p53–MDM2 inhibitors, it was discovered that the spiro-oxindoles with mono-substituted C2 of the pyrrolidine ring (now named first-generation spiro-oxindoles) suffered from equilibration through a reversible retro-Mannich recyclization reaction, which allowed the substituents to reorient into an all-trans configuration around the pyrrolidine ring leading to the major isomer [53]. Although this isomer can be isolated in high yield and is stable as a solid, the isomerization in solution caused concern over potential liabilities in development. To overcome this isomerization, the C2 was changed from a chiral center to a symmetrical center, which results in an irreversible conversion to spiro-oxindoles with a trans-configuration between the aryl substituents on the pyrrolidine ring. This yielded the second-generation spiro-oxindole class of MDM2 inhibitors, with MI-1061 as the first lead compound [60]. Further optimization efforts by bio-isosteric replacement of the benzoic acid with a bicyclo [2.2.2]octane-1-carboxylic acid resulted in an initial compound, which showed improved plasma exposure but reduced distribution in a PK study, which was reflected in its poor efficacy in a mouse xenograft model. The exploration of strategies to decrease plasma-protein binding (PPB) and improve distribution resulted in the discovery of an N-ethylated pyrrolidine spiro-oxindole MDM2 inhibitor, APG-115 (Figure 5). This compound has an MDM2 binding affinity of IC50 = 4.8 nM, cell activity of IC50 < 100 nM in wt-p53 cancer cell lines, and it retains good plasma exposure, improved distribution, and in vivo complete tumor regression in a mouse xenograft model (Table 1) [44]. Consequently, APG-115 was selected for progression to clinical trials. Interestingly, DS-3032 and APG-115 are both second-generation spiro-oxindole MDM2 inhibitors, which have a symmetrical C2 on the pyrrolidine core. No co-crystal structure of these second-generation spiro-oxindoles bound to MDM2 has been reported, but, presumably, they have a binding mode similar to that of MI-773.
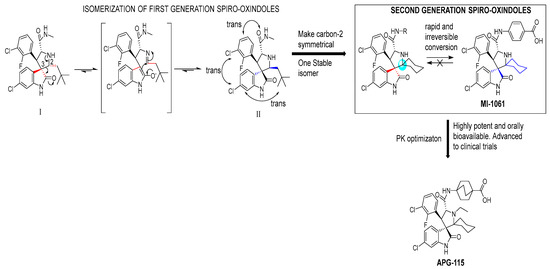
Figure 5.
Discovery of second-generation spiro-oxindole class of MDM2 inhibitors. Red lines indicate the starting stereochemistry and the blue lines indicate the desired ending stereochemistry. The blue circle indicates the symmetrical stereocenter in the second-generation spiro-oxindoles.
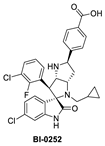
Boehringer Ingelheim (BI) has also advanced an MDM2 inhibitor, BI-907828, with undisclosed structure, into clinical trials [61,62,63]. In the literature, BI has reported a novel class of spiro-oxindoles as p53–MDM2 inhibitors, and their candidate may be an optimized analog belonging to this new class of spiro-oxindoles. In this review, we will discuss insights into the reported SAR of BI-0252, which is the earliest member of this new class of reported spiro-oxindole MDM2 inhibitors [45]. Inspired by the spiro-oxindole inhibitors discovered in the Wang lab at the University of Michigan and natural product architectures, scientists at BI focused on overcoming the chemical instability of the core pyrrolidine ring of the first-generation spiro-oxindoles. Epimerization at C2 and C3 is known to occur through a reversible ring-opening, retro-Mannich reaction, and it was hypothesized that shifting the nitrogen of the pyrrolidine ring one atom closer to the oxindole would prevent this retro-Mannich reaction. Consequently, spiro-oxindoles with this modification, which effectively abolished this phenomenon, were produced [45]. Further rational design and SAR optimization to correctly substitute the pyrrolidine ring and capture the known interactions with the MDM2 pocket resulted in the discovery of BI-0252 as the lead compound. This compound has an MDM2 binding affinity of IC50 = 4 nM and cell activity of IC50 = 471 nM in SJSA-1 cancer cells (Table 1). A co-crystal structure of BI-0252 bound to MDM2 was reported and supports their design rationale (Figure 6A). Interestingly, the interactions with His-96 and Lys94 could only be captured by adding a ring, thus forming a second fused pyrrolidine ring to lock the orientation of the NH and benzoic acid substituents toward His96 and Lys94 residues in MDM2. The NH of the pyrrolidine ring in this case functions as a H-bond donor to His96; in contrast to other inhibitors, this His96 functions as a H-bond donor to a carbonyl or interacts electrostatically with a carboxylic acid. A highlight of this compound is that a high single oral dose achieved tumor regression, and the same was claimed for the clinical candidate BI-907828 [45].
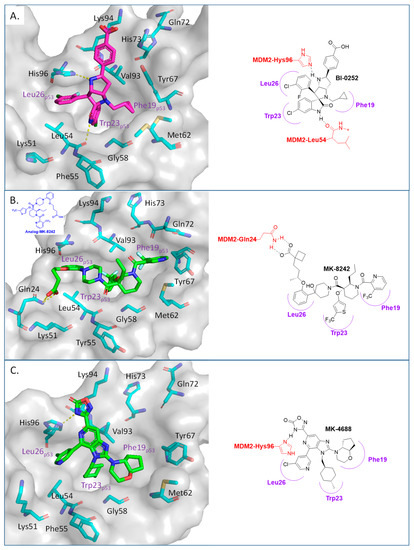
Figure 6.
Co-crystal structures and chemical structures of inhibitors bound to MDM2 protein: (A) BI-0252; (B) MK-8242; (C) and MK-4688. Labels and semicircles in magenta indicate the binding pockets in MDM2 where the p53 residues Phe19, Trp23, and Leu26 bind. Labels in red indicate MDM2 residues.
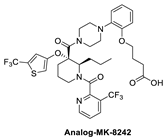
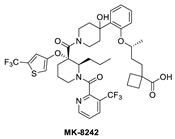
The Merck Company has also worked on the discovery of novel small-molecule inhibitors of the p53–MDM2 interaction and advanced MK-8242 [47,48], a substituted piperidine class of MDM2 inhibitors [46,64,65,66], into clinical trials. Structurally, this molecule appears to be quite different from other MDM2 inhibitors, which have a more condensed core. There is no exact report of the discovery of MK-8242, but a report of a SAR study of a very close analog is available and includes its co-crystal structure bound to MDM2 [46]. This class of compounds was discovered in an in-house high-throughput screen (HTS), which identified a hit that already possessed substituents similar to those in MK-8242 but only had an MDM2 binding IC50 of 2.3 µM [64]. In a subsequent study, it was reported that significant improvement was achieved by adding an n-propyl substituent on position 2 of the piperidine core [66]. Further optimization reports indicate that replacing the trifluoromethylphenyl group, present in the initial hit, with a trifluoromethyl-thiophene moiety improved its potency, and further modification of the alkoxy substituent on the aryl ring with a long-chain carboxylic acid further improved its potency, permeability, and PK properties [46]. The co-crystal structure of this nearly optimized analog of MK-8242 showed that, like NVP-CGM097, this compound lies mainly on the wall opposite the Val93 wall on which most MDM2 inhibitors sit (Figure 6B) [46]. The piperidine core sits close to Gly58. The trifluoromethyl-thiophene effectively occupies the Trp23 pocket; the trifluoromethyl of the pyridyl group is placed deep in the Phe19 pocket; and the alkoxy phenyl group fits in the Leu26 pocket, where it π-π-stacks with His96, and its alkyl acid interacts with Gln24. Presumably, these different interactions, absent in other MDM2 inhibitors, compensate for their different binding modes, which deviate from those of other MDM2 inhibitors. The MK8242 analog has an MDM2 binding of IC50 = 7 nM, cell activity of IC50 ≤ 180 nM in wt-p53 cell lines, and it demonstrates robust in vivo antitumor activity [46], comparable to other MDM2 inhibitors (Table 1).
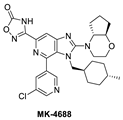
Due to the high molecular weight, high clog P, and high recommended phase II dose of MK-8242, Merck has pursued lower molecular weight, novel MDM2 inhibitors. Using HTS, a purine-derived inhibitor hit was identified and optimized using the available structural information and NMR-based models [49]. Their optimization strategy focused on improving activity without significantly increasing the molecular weight by concentrating the optimization on modifying the volume, shape, and polarization of substituents that occupy each of the three well-defined binding pockets on MDM2. Their efforts yielded MK-4688, which has a MW of 550 and a predicted human dose of 75 mg QD/38 mg BID [49]. A co-crystal structure of MK-4688 bound to MDM2 was obtained, showing it efficiently captures the key Phe19, Trp23, Leu26, p53 interactions with the MDM2 protein (Figure 6C). Interestingly, this compound has a methylcyclohexyl group occupying the Trp23 pocket and is therefore different from other MDM2 inhibitors that generally use an aryl group to mimic the tryptophan residue of p53. They rationalized the use of this methylcyclohexyl group with larger hydrophobic volume to enhance contact in the pocket. Bicyclic morpholine was selected to effectively occupy the Phe19 pocket while maintaining low MW, reduced lipophilicity, and good potency to overcome its metabolic liabilities. Similarly, the 5-chloropyridine group occupying the Leu26 pocket was used to decrease lipophilicity and off-target liabilities. Finally, this molecule has only one polar interaction, an H-bond from the 1,2,4-oxadiazole substituent to the His96 of MDM2. The 1,2,4-oxadiazole was used as bio-isosteric replacement for a carboxylic acid, essentially to maintain the polar interaction with His96, and required tuning of its pKa to obtain a cell-permeable compound. Even though this molecule has only one polar interaction, the increased volume of the substituents that enhance the interactions in the key binding pockets is sufficient to achieve potent binding to MDM2 of IC50 = 0.65 nM, cell activity of IC50 = 122 nM in wt-p53 HCT116 cancer cells, and robust antitumor activity in vivo in an SJSA-1 xenograft model (Table 1). Although this compound is not currently in clinical trials, the authors indicated that it was advanced into preclinical safety studies, where it demonstrated a profile that supported its progression to the clinic.
The plethora of novel small-molecule MDM2 inhibitors discovered and advanced into clinical trials indicates the perceived importance of the p53–MDM2 axis in cancer treatment strategies. However, since p53 also retains its wild-type status in normal cells, some important limitations of MDM2 inhibitors have been observed in the clinic. For example, MDM2 inhibitors have shown hematological toxicities, mainly thrombocytopenia [67,68,69,70,71,72,73], due to activation of p53. Several strategies to circumvent this toxicity have been explored, including use of more potent compounds, which can be administered at lower doses or higher doses with less frequent administration to allow recovery [45,59,62]. Another limitation arising from activation of p53 by an MDM2 inhibitor is the upregulation of MDM2, itself a target gene of p53, as it is part of a natural autoregulation mechanism to prevent aberrant p53 activation in normal cells [27]. Hence, even though MDM2 inhibitors can strongly activate p53, the upregulation of MDM2 itself may limit their activity.
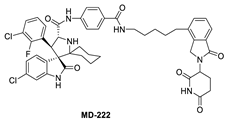
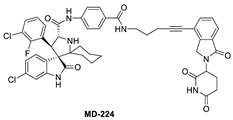
To overcome this feedback limitation, MDM2 degraders have been designed based upon the PROteolysis Targeting Chimera (PROTAC) technology [74,75,76,77,78]. PROTAC degraders are heterobifunctional small molecules, which are composed of three structural components: a ligand for a protein of interest (POI), another ligand for an E3 ubiquitin ligase, and a linker to tether the two ligands together. A PROTAC molecule binds to the POI on one end and to an E3 ubiquitin ligase complex on the other end to bring them to close proximity to facilitate ubiquitination of the POI, leading to proteasomal degradation of the POI [79,80]. A PROTAC degrader functions as a “catalyst” in inducing the degradation of the POI, thus being capable of achieving very high degradation potency [81]. Since a PROTAC MDM2 degrader efficiently reduces the levels of MDM2, they are much more potent and effective in inducing the activation of p53. Considering these potential advantages, the Wang lab at the University of Michigan explored the PROTAC strategy for the design of MDM2 degraders. Using their own MDM2 inhibitors, they reported the first-in-class PROTAC MDM2 degraders exemplified by MD-222 and MD-224, which were designed using a potent MDM2 inhibitor MI-1061 linked to the cereblon ligand lenalidomide (Figure 7 and Figure 8) [82]. As hypothesized, Western blotting revealed complete degradation of the MDM2 protein by its MDM2 degraders at very low concentrations, leading to robust increase in p53 protein and increased expression of p53-targted genes and gene products and apoptosis induction in p53 wild-type tumor cells. PROTAC MDM2 degraders are 100 times more potent than their corresponding MI-1061 inhibitor at inhibition of cell growth. MD-224 achieves much stronger antitumor activity in vivo at much lower doses and lower dosing frequency than its corresponding inhibitor. Further optimization of this class of PROTAC–MDM2 degraders yielded AA-265, which is currently in advanced preclinical evaluation for progression into clinical trials.
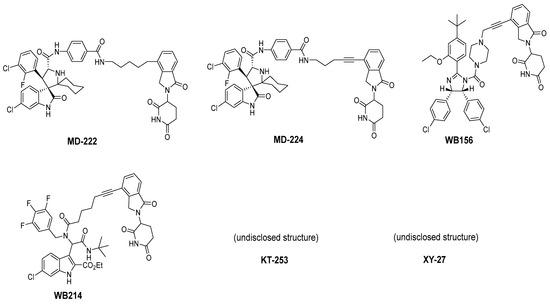
Figure 7.
Chemical structure of PROTAC–MDM2 degraders.
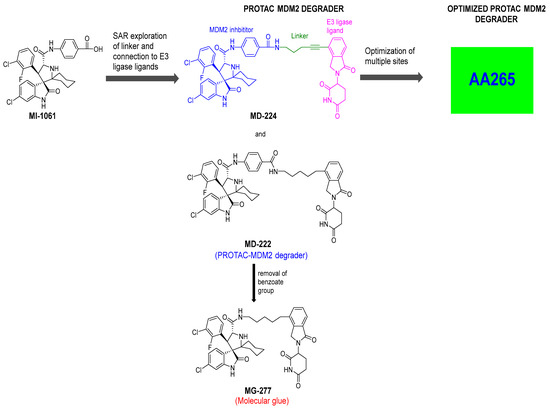
Figure 8.
Discovery of PROTAC–MDM2 degrader and molecular glue that degrades GSPT1.
Notably, during the exploration of PROTAC–MDM2 degraders, it was discovered that removing the benzamide substituent from the MDM2 inhibitor portion resulted in MG277, which showed increased potency even in mutant or deleted p53 cancer cells. Further research revealed that this structural modification converted the chimera into a molecular glue, which degraded the G1 to S phase transition 1 (GSPT-1) protein, a translation termination factor, also known as eRF3a, which mediates stop codon recognition and nascent protein release from the ribosome through the interaction with the release factor, eRF1 (Figure 8) [83]. Thus, the activity in cell lines with compromised p53 was due to GSPT-1 degradation and not to MDM2 degradation. Interestingly, compounds with benzamide or terminal amides, such as MD-222 and MD-224, were all selective MDM2 degraders and did not have off-target GSPT-1 activity [83].
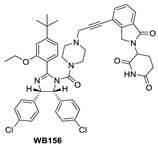
Shortly after the first PROTAC–MDM2 degraders were disclosed by the Wang group at the University of Michigan, the Tang group at the University of Wisconsin-Madison reported the discovery of a PROTAC–MDM2 degrader derived from the Nutlin class of MDM2 inhibitors and the cereblon ligand lenalidomide [84]. The characteristic feature of these PROTAC degraders is that the linker-lenalidomide moiety is tethered from the solvent projecting piperazine substituent of the Nutlin class of MDM2 inhibitors. This class of compounds is exemplified by WB156 (Figure 7), which boasts the shortest linker ever reported and achieves > 1000-fold improvement in cell potency relative to its corresponding inhibitor [84].
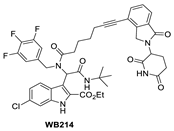
Due to the activity of WB156 on a limited number of leukemia cells, Tang et al. explored the creation of a new class of PROTAC–MDM2 degraders derived from another class of known MDM2 inhibitors, which could be diversified by a four-component Ugi reaction. This produced a new class of heterobifunctional compounds, including WB214 (Figure 7), which displayed the best potency and activity in a broader group of cancer cells [85]. Interestingly, the analysis of its mechanism of action (MOA) revealed that this new class of compounds also functioned as a GSPT-1 degrader, which was independent of MDM2 and p53 degradation. They found that p53 was degraded as a bystander, which is associated with MDM2, and thus suggested that this class of molecules may bind to other parts of MDM2 and not the same pocket occupied by p53 [85].
Two other PROTAC–MDM2 degrader molecules have been reported, but their structures have not been disclosed. Marcellino et al. reported the development of the PROTAC–MDM2 degrader XY-27 using an undisclosed MDM2 inhibitor tethered to a ligand of the VHL E3 ubiquitin ligase [86]. VHL was selected due to its higher expression in acute myelogenous leukemia (AML) compared to other cancers, and its use can potentially improve the potency and specificity in AML. The VHL-based PROTAC–MDM2 degrader demonstrated superior potency over the AMG232 MDM2 inhibitor in MOLM13 and MV4;11 cell lines and showed potent activity when combined with azacytidine or cytarabine. Kymera Therapeutics also reported the development of a PROTAC–MDM2 degrader KT-253 using an undisclosed MDM2 inhibitor tethered to a ligand of an undisclosed E3 ubiquitin ligase [87]. They report picomolar DC50 results (Table 2) and an cell killing activity, which is >200-fold more potent than small-molecule inhibitors of MDM2 in clinical trials and especially compared to DS-3032. Impressively, a single dose of KT-253 at 1, 3, or 10 mg/kg achieved complete tumor regression in an RS4;11 xenograft model in mice and was longer lasting with the 10 mg/kg dose. Kymera Therapeutics indicated that they intend to move this compound into clinical trials, with anticipated IND filing in 2022 [87].

Table 2.
PROTAC–MDM2 degraders.
These MDM2–PROTAC reports indicate that application of the PROTAC strategy for targeted degradation of MDM2 is indeed achievable; however, the molecular glue activity seen in some putative MDM2 degraders indicates that if other MDM2 inhibitors are used to produce PROTAC–MDM2 degraders, a careful profiling of the MOA is necessary to define the mechanism of action. Of significance, bona fide PROTAC–MDM2 degraders have been shown to overcome the MDM2–p53 feedback loop, leading to more robust antitumor activity in vivo than MDM2 inhibitors.
Disrupting MDMX–p53 Protein–Protein Interactions
In some cancers, such as retinoblastomas [88], breast carcinomas [89], and melanomas [90], the inhibition of MDM2 may not be sufficient to stop tumor progression because the cancers highly express MDMX, which is a structural homolog of MDM2. MDMX does not have E3 ligase activity but still participates in the regulation of p53 by binding to the N-terminus of p53 and thus neutralizing its transactivation activity [91]. In addition, MDMX and MDM2 share a comparable sequence in their RING domain, and through their domains, MDMX binds to MDM2 [92,93]. This stabilizes MDM2, resulting in the potentiation of MDM2′s capability for ubiquitination of p53 [94,95]. Furthermore, the same p53 residues that bind to MDM2 also bind to a similar cleft in MDMX, and this indicates that the inhibition of the MDMX–p53 interaction is plausible and may be possible and beneficial for cancer treatment [34,96]. Below, we summarize the progress in the efforts to inhibit the MDMX–p53 interaction, focusing only on the compounds that have been shown to have reasonable binding to MDMX or have co-crystal structures that can provide insights for targeting MDMX (Figure 9).
Figure 9.
Chemical structure of MDMX inhibitors.
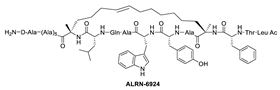
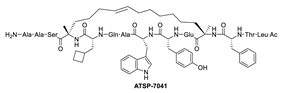
Due to the high similarity of the MDMX and MDM2 binding pockets, the same p53 residues Phe19, Trp23, and Leu26 also bind to MDMX, indicating that selective MDMX inhibition will be challenging [34,96]. Hoffmann-La Roche, after abandoning RO-5963, a small-molecule dual inhibitor of MDMX/MDM2 (discussed below), due to its poor pharmacological characteristics [8], approached dual inhibition by optimization of reported short peptides, which individually potently bind to MDMX/MDM2. It had been shown that appropriate stapling of short peptides resulted in stabilization of the α-helical conformation, improved permeability, and other physicochemical properties, and as a result, it became the optimization strategy employed by Hoffmann-La Roche [97]. Their optimization efforts focused on improving biological and physicochemical properties, and this effort provided ATSP-7041 [97]. Subsequently, Aileron Therapeutics reported ALRN-6924 [98], which is currently in clinical trials (Figure 9). ATSP-7041 is a potent dual inhibitor, which is cell permeable and boasts significantly slow off-rate kinetics compared to small-molecule inhibitors and causes an extended inhibitory effect even in cells. Additionally, its optimized α-helical stability resulted in improvement in cell permeability, in vivo stability, favorable PK properties, and, consequently, potent cellular and in vivo antitumor activity in cancers that overexpress MDM2 or MDMX [97]. The advantages of ATSP-7041 and ALRN-6924 dual inhibition of MDMX and MDM2 were shown by head-to-head comparison with selective MDM2 inhibitors, RG7112 and RG7388, in cancers that overexpress MDMX, where these dual-peptide-based inhibitors showed superior activity [97,98]. These results support the idea that dual inhibition of MDMX and MDM2 may be superior to selective inhibition of only one of these homologs. No co-crystal structure of ALRN-6924 bound to MDMX or MDM2 has been reported, but a similar stapled peptide ATSP-7041 was successfully co-crystallized with MDMX and showed the binding mode of these stapled peptides (Figure 10A) [97]. This structural information indicates that the natural p53 residues, Phe19, Trp23, and Cba26 (a cyclobutyl Leu26 isostere), present in ATSP-7041, occupy the same p53 binding pockets in MDMX. In addition, its Tyr22 residue captures additional hydrophobic interactions and a water-mediated hydrogen bonding interaction with Lys93 and His68. The aliphatic stapled chain also captures additional hydrophobic interactions and potentially a cation–π interaction between its alkene and His51 on the MDMX surface wall. These interactions are the structural basis for the strong binding of these stable stapled peptides to both MDMX and MDM2.
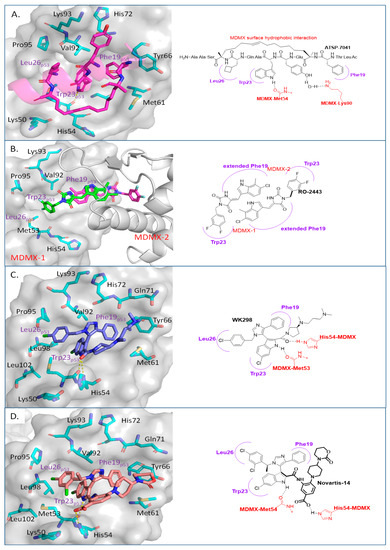
Figure 10.
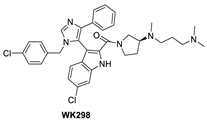
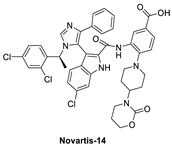
Co-crystal structures and chemical structures of inhibitors bound to MDMX protein: (A) ATSP-7041; (B) RO-2443; (C) WK298; (D) and Novartis-14. Labels and semicircles in magenta indicate the binding pockets in MDMX where the p53 residues Phe19, Trp23, and Leu26 bind. Labels in red indicate MDMX residues.
It is conceivable that if the same residues of p53 are critical for binding to both MDMX and MDM2, then the small molecules that mimic them and have been used as MDM2 inhibitors should also achieve potent MDMX inhibition. Interestingly, this is not the case, and it has been seen that many potent MDM2 inhibitors do not have potent binding to MDMX [99,100]. The enhanced activity observed from the peptide-based dual inhibitors in cancers, which carry high levels of MDMX, supports the development of small-molecule selective or dual MDMX/2 inhibitors, and efforts toward this end will be summarized here—specifically for compounds with confirmed binding to MDMX with co-crystal structures.
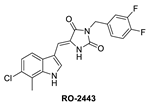
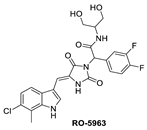
One of the first small molecules that showed nM binding to MDMX and was also equipotent to MDM2 is RO-2443 (Figure 9) [8]. This dual inhibitor was discovered by Hoffmann-La Roche through high-throughput screening for MDMX binding and initially had a predicted binding mode in which its indole moiety fit into and mimicked the Trp23 of p53. However, NMR and size exclusion chromatography studies indicated MDMX dimer formation, and the co-crystallization efforts confirmed an MDMX dimer that sandwiches two molecules of RO-2443 (Figure 10B) [8]. In the dimer crystal structure, the indolyl-hydantoin moieties of each RO-2443 π-π stack on top of each other, and each one occupies an extended Phe19 pocket of one of the MDMX protein monomers. Interestingly, the difluorobenzyl substituent occupies the Trp23 pocket in the opposite MDMX protein monomer, indicating that, essentially, two molecules are required to complement their binding in the MDMX dimers, and this is necessary if potent MDMX and MDM2 binding is to be achieved. Due to the poor solubility of RO-2443, it was not suitable for intracellular evaluation; therefore, its more soluble analog, RO-5963, was synthesized with a diol containing carboxamide substitution at the methylene position of the benzyl group (Figure 9) [8]. This substitution was reported to potentially capture additional interactions in the unoccupied Leu26 pocket and to improve its solubility for evaluation in cell-based assays. In fact, this compound demonstrated superior cell activity over Nutlin-3a in cancer cells that overexpress MDMX. This indicates that dual MDMX/MDM2 inhibitors may be more suitable in cancers that have high levels of MDMX. Its poor pharmacological characteristics prevented the further development of RO-5963 [8].
Although binding to MDMX only weakly, the MDM2 inhibitor WK298 (Figure 9) was the first non-peptide small molecule, which was successfully co-crystalized with MDMX. This showed unambiguously that small molecules that mimic the three key p53 binding residues, Phe19, Trp23, and Leu26, can indeed bind to both MDM2 and MDMX (Figure 10C) [101]. The phenyl, chloroindole, and 4-chlorobenzyl substituents of WK298 effectively occupy the Phe19, Trp23, and Leu26 p53 binding pockets of MDMX, respectively, similar to their binding modes in MDM2. However, in the case of MDMX, these interactions are not sufficient for potent binding and highlight the challenge for the development of potent MDMX inhibitors. This lack of improved binding was attributed to the residues, which are different in MDM2 and MDMX, especially in the Leu26 pocket, which contains most of the differences [101]. In this pocket, the MDM2 residues Leu54, Ile103, Ile99, and His96 are the Met53, Leu102, Leu98, and Pro95 residues in MDMX, respectively. This change makes the MDMX pocket more open than the pocket in MDM2 and presumably decreases the tight binding. Other MDM2 inhibitors have also been co-crystallized with MDMX, but they did not reveal any additional interaction, which is unique to MDMX, and thus failed to achieve improved MDMX inhibition [99].
A more highly substituted analog of WK298, called Novartis-14 (Figure 9), was found to potently bind MDMX with a TR-FRET IC50 of 17 nM (Table 3), and the co-crystal structure of this compound reveals additional interactions that underlie its potent MDMX binding (Figure 10D) [99]. As in WK298, the substituted imidazole core in Novartis-14 effectively occupies the Phe19, Trp23, Leu26 p53 binding pockets in MDMX, but additional interactions add to the tighter binding to MDMX. The 2,4-dichlorobenzyl substituent in Novartis-14 can occupy the more open Leu26 p53 binding pocket in MDMX, and the large amide substituent that hovers above the core projects its hydrophobic oxazinanone into the expanded Phe19 p53 binding pocket in MDMX. These two both contribute to the improved binding to MDMX. More importantly, the carboxylic acid of its benzoic acid substituent is positioned effectively close and captures a strong electrostatic interaction with the His54 residue of MDMX, another of the residues, which distinguishes it from the MDM2 (Phe55) binding pocket. This electrostatic interaction is important for MDMX binding, as its analogs, which lack or have a blocked carboxylic acid, significantly suffer from weaker MDMX binding, as seen in the related patent [102].

Table 3.
MDMX inhibitors.
These reports indicate the potential for obtaining selective MDMX or dual MDMX/MDM2 inhibitors. From the Novartis-14 data, it was revealed that capturing an interaction with the His54 residue of MDMX improved its binding. The crystal structures also revealed that both the Phe19 and especially the Leu26 binding pocket in MDMX are more open, indicating that targeting these pockets may be a potential approach to tuning in MDMX binding and design of selective inhibitors. Additionally, no PROTAC–MDMX degraders have been reported, and the discovery of these MDMX binders may provide an opportunity for exploration of this strategy. To the best of our knowledge, no non-peptide small-molecule dual MDMX/MDM2 inhibitor has advanced to clinical trials, but certainly, these data will give useful insights into improvements of MDMX binding, which can be used in programs with this target.
3. Mutant p53 Binders as Mutant p53 Activators
Reactivating wt-p53 by targeting its negative regulators, MDM2 and MDMX, has seen significant progress. However, approximately 50% of human cancers harbor mutated p53, and both MDM2/MDMX inhibitors and MDM2 degraders are ineffective in inducing activation of mutated p53 [103,104,105,106,107]. Mutations in p53 typically result in loss of the wt-p53 function and, in some cases, even in gain of the oncogenic function [108,109]. The loss of function results from the mutant p53 being unable to bind to DNA either by loss of residue contact to DNA or loss of the ability to fold into its active conformation required for binding to DNA. Interestingly, some cancers with mutant p53, particularly early-stage cancers, are heterozygous, and the mutant p53 dimers form an inactive heterotetramer with wt-p53 dimers, acting as dominant negatives and neutralizing the wt-p53 activity [20,110,111]. On the other hand, mutant p53 can have a gain of function by interacting with other partners, such as the transcription factors p63 and p73, blocking their anticancer activity [112,113]. Furthermore, mutant p53 can also bind to other transcription factors or cofactors to enhance other genes, resulting in tumor cell survival [113] (Figure 11).
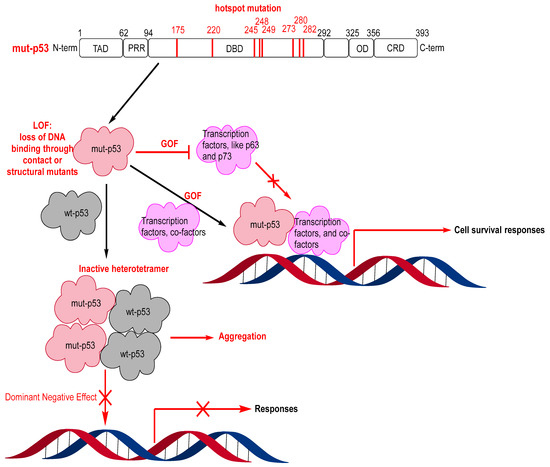
Figure 11.
Mutant p53 hotspot mutations, loss of function (LOF), gain of function (GOF), and dominant negative effect (DNE).
Direct targeting of the p53 protein is therefore a very appealing anticancer therapeutic strategy. Unfortunately, the p53 protein is frequently considered to be “undruggable” because neither wt-p53 nor mutant p53 has binding pockets or allosteric sites, which can be readily targeted by small molecules [23]. This is further complicated by the existence of over 2000 possible mutant p53 variants. However, recent progress has been made in restoration of wt-like activity in mutant p53 [23,107,114,115]. Here, we summarize some of the advances in this research.
Of the >50% of human cancers that have TP53 gene mutations, 75% are missense mutations and mainly occur in the DBD of p53 [116,117,118]. Most of these mutations occur in “hot-spot” residues, and the change in these residues results in either loss of contact with DNA or change in the structural stability of the p53 protein (Figure 11) [23,119,120]. Consequently, the mutants can be divided into contact mutants or structural mutants [2,23,113,121,122,123]. Contact mutants involve residues that interact directly with DNA, and for effective targeting, the introduction of interactions to compensate for missing DNA contacts is necessary [124]. Due to the lack of well-defined pockets in these contact p53 mutants, targeting them with small molecules can be very challenging [121]. On the other hand, structural mutants have residue mutations, which influence the thermal stability and structure of p53, making the mutants unstable at physiological temperatures and preventing them from achieving correct folding and having wt-like functions [119]. Therefore, small molecules, which can bind, stabilize, and restore the level of correctly folded p53, may represent a more achievable strategy in targeting structural p53 mutants. Indeed, several small molecules that stabilize an active p53 conformation and restore wt-like transcriptional function of mutant p53 have been reported [23,107,114,115].
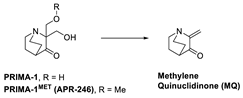
Several reviews on small molecules targeting mutant p53 have been published, but the MOA for some of these small molecules is not fully understood. They can sometimes rescue both contact and structural mutants of p53 but may also have p53-independent activity [23,119]. Here, we focus on those molecules, which have crystal structural information supporting their direct binding to mutant p53. Among these are some small molecules that target the hydrophobic pocket created at the mutation site of the structural mutants of tyrosine 220 (Y220). Small molecules have been designed to target this site and were successful in reactivation of the wt-like p53 function of these p53 mutants. Additionally, the co-crystal structures of these small molecules bound to the Y220C mutant have shown unambiguously that this mutant pocket can be targeted with small molecules. Targeting mutant p53 to rescue its wt-like function has yielded only two molecules, which have entered clinical trials, PRIMA-1MET(APR-246) [125] and COTI-2 [126,127].
APR-246 is a more potent methylated derivative of PRIMA-1 and was discovered by screening for compounds that show mutant-p53-dependent cell growth inhibitory activity and also reactivate mutant p53 (Figure 12) [125,128]. It was found that these compounds restore an active p53 conformation, which can bind to DNA and consequently rescue wt-like transcription activity, which results in induction of apoptosis in human cancer cells. It was found that APR-246 and its parent PRIMA-1 are prodrugs, which are converted to methylene quinuclidinone (MQ)—a very reactive Michael acceptor, which reacts with cysteine residues in “the core domain” of mutant p53 (Figure 13) [129,130]. The exact mechanism of the reactivation of p53 by APR-246/MQ is not fully understood; however, a study of Cys to Ala mutations in the p53 core domain using mass spectrometry identified Cys277 as the most nucleophilic and most solvent-accessible of the 10 cysteines in the core domain. Further studies showed that Cys277 and Cys124 both have an important role in APR-246/MQ-mediated mutant p53 reactivation [129]. Similarly, an independent study using various high-resolution crystal structures of mutant and wt-p53 core domains bound to MQ also identified the same Cys124 and Cys277, and, in addition, Cys229, as contributors to MQ-mediated stabilization of mutant and wt-p53 [131]. These studies concluded that these alkylating agents react with the most exposed cysteine residues in the p53 protein. MQ was shown to be able to rescue wt-p53-like function in both structural and contact mutants. Interestingly, it was also found that APR246/MQ also induces high levels of reactive oxygen species resulting from its depletion of glutathione (GSH), contributing to their anticancer activity [132,133].
Figure 12.
Mutant p53 activators.
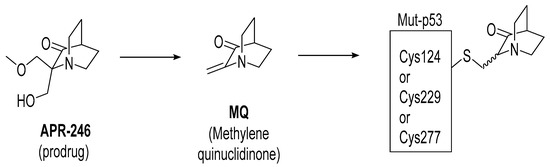
Figure 13.
APR-246 forms MQ, which is a reactive Michael acceptor, which undergoes covalent addition with mutant p53 residues Cys124, Cys229, and Cys277, which are the most exposed and reactive residues.
The clinical compound COTI-2 is a third-generation thiosemicarbazone class molecule, which was identified using CHEMSAS®, a proprietary computational platform that incorporates various drug discovery principles and technologies to optimize novel compounds, which could target various human malignancies [134]. COTI-2 was found to be active against a wide variety of human cancer cell lines and xenografts, which are traditionally difficult to treat; however, its mechanism of action was not known at the time. The initial studies suggested that the MOA of COTI-2 involved negative modulation of the PI3K/AKT/mTOR pathway. COTI-2 was evaluated in a variety of cancer cell lines encompassing various types of human malignancies, including cancers with TP53, KRAS, PIK3CA, APC, and PTEN mutations. While COTI-2 demonstrated potent activity in all of these cancer cell lines, it was especially potent against mutant p53 cancer cell lines [134]. Subsequently, it was reported that COTI-2 restored wt-like p53 function through a mechanism involving zinc chelation, in addition to modulation of the PI3K/AKT/mTOR pathway [135]. While the exact mechanism of COTI-2 is unclear, its potent activity and low toxicity in a variety of difficult-to-treat cancer models has resulted in its progression into human clinical trials [134].
Structural mutants of p53 result in reduced thermal stability of the DBD, causing it to unfold and aggregate at physiological temperatures, thus becoming inactive [120,136,137]. It is estimated that one-third of the p53 mutants have lower melting temperatures, but studies have shown that at temperatures below physiological temperatures, their wt-like conformations are restored. This indicates that if a small molecule can bind to mutant p53 and increase the folded thermal stability of mutant p53, it may also restore wt-like conformation and function of mutant p53 [137]. This approach has been pursued, and one of the most attractive of these structural mutants is Y220C, which is found annually in about 100,000 new cancer cases [138]. This mutation results in a highly destabilized mutant p53 protein and is also known to create a pocket that is distant from the surface involved with DNA binding and is thus an attractive site for targeting by small molecules [138].
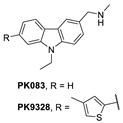
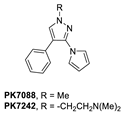
A group at the Medical Research Council, Cambridge, UK, reported a class of carbazoles exemplified by PK083 (Figure 12) as activators of mutant p53 Y220C [139]. This was accomplished by in silico analysis of the T-p53C-Y220C crystal structure by virtual screening, followed by NMR screening of the hits, examining changes in the chemical shift of 15N-labeled-T-p53C-Y220C or T-p53C. This molecule stabilized the wt-like folded conformation of this mutant, and it was shown unambiguously by a high-resolution co-crystal structure that PK-083 bound in the Y220C pocket of p53. Further optimization efforts by the same group led to the carbazole analog PK-9328 (Figure 12 and Figure 14A), whose 4-methylthiazole ring was designed to occupy the pocket formed by Pro152 and Pro153, enabling PK-9328 to improve the Kd by >70-fold from 125 µM in PK-083 to 1.7 µM in PK-9328 [137]. Similarly, the binding of PK-9328 to the Y220C crevice and stabilization of the mutant p53 were demonstrated using biophysical and cell-based techniques and by its co-crystal structure bound to the Y220C-p53 mutant protein, confirming the additional interactions in the Pro152, Pro153 pocket (Figure 14A) [137].
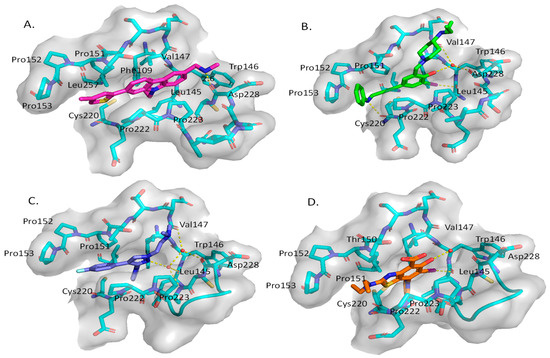
Figure 14.
Co-crystal structure of binders of Y220C: (A) PK9328; (B) PK5196; (C) PK7242; (D) and MB710.
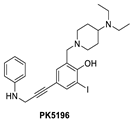
Using the same strategy, the Cambridge (UK) group reported the discovery of three other classes of Y220C-p53 mutant activators, which function by binding to the Y220C pocket. The 2-(aminomethyl)-4-ethynyl-6-iodophenols exemplified by PK-5196 [140], the substituted 4-phenyl-3-(1H-pyrrol-1-yl)-1H-pyrazole class exemplified by the soluble analog PK-7242 [141], and the aminobenzothiazole derivatives exemplified by MB-710 [142] were all shown by biophysical and cell-based methods to stabilize the Y220C-p53 mutant, and the co-crystal structures showed that they all bind in the Y220C pocket (Figure 14B–D). In comparison, their crystal structures showed that the cores of these three molecules are mainly sandwiched between Val147 and Pro151 on one side, and Pro222 and Pro223 on the other side of the pocket, and similarly to PK-9328, they all have substituents that extend into the pocket surrounded by Pro152 and Pro153 (Figure 14A,B). However, these three molecules do not extend so far toward the Trp146 side of the pocket. In fact, their substituents, which face this side of the pocket, have structural water-mediated polar interactions with backbone carbonyls of Val147, Leu145, and Asp228 (Figure 14B–D) [140,141,142]. In PK-9328, one of the aryl rings displaces these structural water molecules, and the methylenemethylamine chain, which is extended from this aryl ring, forms a hydrogen bond with the backbone carbonyl of Asp228 (Figure 14A) [137]. This additional occupancy of the pocket may be the reason why PK9328 has a better binding Kd than the other three classes of compounds. Even considering the better binding Kd of PK-9328, all these small molecules still only have µM potency, which may be inadequate for drug candidates. However, the significant improvement in potency compared to the initial lead compound, PK-083, indicates that there may still be opportunity for improvement. Potentially, this can be achieved by capturing additional hydrophobic and polar interactions with the surface residues, such as those seen with the structural waters, or by the selective covalent binding to the Cys220 residue, which is unique to this mutant and forms part of the pocket.
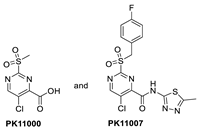
In their continued pursuit of mutant p53 Y220C binders and activators, the same group discovered the 2-sulfonylpyrimidine PK-11000 series (Figure 12) [143]. Through their NMR, mass spectrometry, and crystallographic studies, they discovered that this compound did not bind in the Y220C crevice but instead added covalently to the two most reactive and exposed cysteine residues, Cys182 and Cys277, and this addition was selective even at high concentrations. Although PK-11007, a more reactive analog, was found to increase the thermal stability of mutant p53 and restore the wt-like activity by upregulation of p53 target genes, such as p21 and PUMA, in some cells, the major cell death activity was found to implicate two other mechanisms, namely, induction of high levels of reactive oxygen species, which resulted from glutathione depletion, and also an unfolded protein response (UPR) triggered by endoplasmic reticulum (ER) stress, both of which are similar to that observed in APR-246 [143]. In their crystallization efforts, high-resolution data are available only for the Cys182-PK-11000 adduct and confirm the structure of the SNAr adduct (Figure 15). In the crystal structure of this adduct, the alkylating agent is not located in any crevice, signifying that the covalent bonding is not facilitated by other interactions with the protein. Instead, the conclusion supports the finding that alkylating agents probe and react with the most exposed and reactive cysteines, among which Cys182 and Cys277 are the most reactive in the DBD. This study indicates that alkylating agents may not have an exclusive MOA of p53 alkylation but may also be subject to consequences of alkylation of other reactive thiol-containing protein targets.
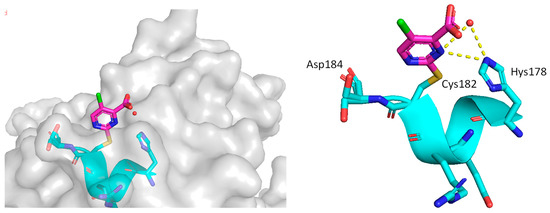
Figure 15.
Co-crystal structure of PK11000 covalently bound to Cys182 of a p53 mutant.
These studies have shown that the structural mutant Y220C, which creates a pocket in mutant p53, can be successfully targeted by various classes of small molecules. Importantly, the binding of small molecules in this pocket results in stabilization of the wt-like conformation and rescues the wt-like activity of this mutant. Recently, PMV Pharma reported PC14586, a small molecule whose structure is undisclosed but which binds specifically to the Y220C pocket and is the first-in-class Y220C reactivator to enter clinical trials [144]. They report tumor regression in xenograft models of Y220C p53 mutant tumors and indicate an IC50 of ~192–722 nM in cell lines generated from Y220C mutant lymphoma and sarcoma tumors (Table 4). In addition, they showed a synergistic antitumor effect when the Y220C activator was combined with anti-PD-1 checkpoint therapy and are currently investigating this in clinical trials [144].

Table 4.
Mutant p53 activators.
4. Conclusions
The tremendous efforts at reactivation of wild-type and mutant p53 clearly demonstrate their unmistakable importance as novel cancer therapeutic strategies. Successful activation of wt-p53 through disruption of its principal negative regulators, MDM2 and MDMX, has yielded several molecules, which are being clinically evaluated and have shown some success. However, this accounts for only half of all cancers, and in the other half, p53 is compromised by mutations that can also occur from treatment with MDM2 inhibitors. Ideally, it would be beneficial to achieve similar success in the reactivation of mutant p53, but this has been more challenging than in the MDM2/MDMX field. The progress seen in targeting the p53 mutant Y220C with small molecules, along with their structural information and successful rescuing of wt-like functionality, provides the proof of concept and a tremendous opportunity to develop a new class of anticancer drugs for this mutant, which has a high rate of incidence in human cancers. It is exciting that the Y220C binder PC14586 has recently progressed into clinical trials.
Activation of wt-p53, through targeting the MDM2/MDMX–p53 interactions, and rescue of wt-like activity of Y220C mutant p53, through selective mutant Y220C binders, have successfully yielded molecules, which have advanced to clinical trials. Both have shown that, in fact, wild-type and mutant p53 activation leads to powerful anticancer activity, albeit with some limitations. Of the clinical trials in progress, with the exception of those of single agents, the majority are studies in combination with other therapeutic agents, which, preclinically, have shown synergy and the ability to potentially circumvent the limitations by attacking related and unrelated mechanisms, giving cancer cells less chance to develop resistance. These clinical trials are summarized in Table 5. Although none of these programs has an approved compound, the activation of p53 is still an attractive cancer therapeutic strategy.

Table 5.
Clinical trials of wild-type and mutant p53 activators.
Author Contributions
A.A. and S.W. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the National Cancer Institute, National Institutes of Health (R01CA219345), and from Ascentage Pharma Group.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank the past and present members of the Wang Laboratory at the University of Michigan for their contributions to our research on MDM2 inhibitors and degraders. We are grateful to our development partner Ascentage Pharma Group for their tremendous efforts in the development of APG-115 (Alrizomadlin) as a new anticancer agent.
Conflicts of Interest
A.A. and S.W. are co-inventors of APG-115 (Alrizomadlin), which has been licensed by Ascentage Pharma Group from the University of Michigan. A.A. and S.W. receive royalties from APG-115. S.W. is a co-founder and a paid consultant of Ascentage and owns equity in Ascentage and serves on its board of directors. The University of Michigan has received a research contract from Ascentage Pharma Group.
References
- Lane, D.P. Cancer. P53, Guardian of the Genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Nguyen, D.; Liao, W.; Zeng, S.X.; Lu, H. Reviving the Guardian of the Genome: Small Molecule Activators of P53. Pharmacol. Ther. 2017, 178, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Ladds, M.; Lain, S. Small Molecule Activators of the P53 Response. J. Mol. Cell Biol. 2019, 11, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Reed, M.; Wang, P.; Stenger, J.E.; Mayr, G.; Anderson, M.E.; Schwedes, J.F.; Tegtmeyer, P. P53 Domains: Identification and Characterization of Two Autonomous DNA-Binding Regions. Genes Dev. 1993, 7, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.; Klein, C.; Muller, L.; Hansen, S.; Buchner, J. P53 Contains Large Unstructured Regions in Its Native State. J. Mol. Biol. 2002, 322, 917–927. [Google Scholar] [CrossRef]
- Hock, A.; Vousden, K.H. Regulation of the P53 Pathway by Ubiquitin and Related Proteins. Int. J. Biochem. Cell Biol. 2010, 42, 1618–1621. [Google Scholar] [CrossRef]
- Huang, L.; Yan, Z.; Liao, X.; Li, Y.; Yang, J.; Wang, Z.G.; Zuo, Y.; Kawai, H.; Shadfan, M.; Ganapathy, S.; et al. The P53 Inhibitors Mdm2/Mdmx Complex Is Required for Control of P53 Activity in Vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 12001–12006. [Google Scholar] [CrossRef]
- Graves, B.; Thompson, T.; Xia, M.; Janson, C.; Lukacs, C.; Deo, D.; Di Lello, P.; Fry, D.; Garvie, C.; Huang, K.S.; et al. Activation of the P53 Pathway by Small-Molecule-Induced Mdm2 and Mdmx Dimerization. Proc. Natl. Acad. Sci. USA 2012, 109, 11788–11793. [Google Scholar] [CrossRef]
- Shieh, S.Y.; Ahn, J.; Tamai, K.; Taya, Y.; Prives, C. The Human Homologs of Checkpoint Kinases Chk1 and Cds1 (Chk2) Phosphorylate P53 at Multiple DNA Damage-Inducible Sites. Genes Dev. 2000, 14, 289–300. [Google Scholar] [CrossRef]
- Shiloh, Y. Atm and Related Protein Kinases: Safeguarding Genome Integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Li, M.; Luo, J.; Brooks, C.L.; Gu, W. Acetylation of P53 Inhibits Its Ubiquitination by Mdm2. J. Biol. Chem. 2002, 277, 50607–50611. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Li, M.; Tang, Y.; Laszkowska, M.; Roeder, R.G.; Gu, W. Acetylation of P53 Augments Its Site-Specific DNA Binding Both in Vitro and in Vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 2259–2264. [Google Scholar] [CrossRef] [PubMed]
- Bullock, A.N.; Henckel, J.; DeDecker, B.S.; Johnson, C.M.; Nikolova, P.V.; Proctor, M.R.; Lane, D.P.; Fersht, A.R. Thermodynamic Stability of Wild-Type and Mutant P53 Core Domain. Proc. Natl. Acad. Sci. USA 1997, 94, 14338–14342. [Google Scholar] [CrossRef] [PubMed]
- Canadillas, J.M.; Tidow, H.; Freund, S.M.; Rutherford, T.J.; Ang, H.C.; Fersht, A.R. Solution Structure of P53 Core Domain: Structural Basis for Its Instability. Proc. Natl. Acad. Sci. USA 2006, 103, 2109–2114. [Google Scholar] [CrossRef] [PubMed]
- Walerych, D.; Olszewski, M.B.; Gutkowska, M.; Helwak, A.; Zylicz, M.; Zylicz, A. Hsp70 Molecular Chaperones Are Required to Support P53 Tumor Suppressor Activity under Stress Conditions. Oncogene 2009, 28, 4284–4294. [Google Scholar] [CrossRef] [PubMed]
- Walerych, D.; Gutkowska, M.; Klejman, M.P.; Wawrzynow, B.; Tracz, Z.; Wiech, M.; Zylicz, M.; Zylicz, A. Atp Binding to Hsp90 Is Sufficient for Effective Chaperoning of P53 Protein. J. Biol. Chem. 2010, 285, 32020–32028. [Google Scholar] [CrossRef]
- Wawrzynow, B.; Zylicz, A.; Zylicz, M. Chaperoning the Guardian of the Genome. The Two-Faced Role of Molecular Chaperones in P53 Tumor Suppressor Action. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 161–174. [Google Scholar] [CrossRef]
- Tidow, H.; Melero, R.; Mylonas, E.; Freund, S.M.; Grossmann, J.G.; Carazo, J.M.; Svergun, D.I.; Valle, M.; Fersht, A.R. Quaternary Structures of Tumor Suppressor P53 and a Specific P53 DNA Complex. Proc. Natl. Acad. Sci. USA 2007, 104, 12324–12329. [Google Scholar] [CrossRef]
- Melero, R.; Rajagopalan, S.; Lazaro, M.; Joerger, A.C.; Brandt, T.; Veprintsev, D.B.; Lasso, G.; Gil, D.; Scheres, S.H.; Carazo, J.M.; et al. Electron Microscopy Studies on the Quaternary Structure of P53 Reveal Different Binding Modes for P53 Tetramers in Complex with DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 557–562. [Google Scholar] [CrossRef]
- Nicholls, C.D.; McLure, K.G.; Shields, M.A.; Lee, P.W. Biogenesis of P53 Involves Cotranslational Dimerization of Monomers and Posttranslational Dimerization of Dimers. Implications on the Dominant Negative Effect. J. Biol. Chem. 2002, 277, 12937–12945. [Google Scholar] [CrossRef]
- Weinberg, R.L.; Veprintsev, D.B.; Fersht, A.R. Cooperative Binding of Tetrameric P53 to DNA. J. Mol. Biol. 2004, 341, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Kitayner, M.; Rozenberg, H.; Kessler, N.; Rabinovich, D.; Shaulov, L.; Haran, T.E.; Shakked, Z. Structural Basis of DNA Recognition by P53 Tetramers. Mol. Cell 2006, 22, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.S.; Ramos, H.; Inga, A.; Sousa, E.; Saraiva, L. Structural and Drug Targeting Insights on Mutant P53. Cancers 2021, 13, 3344. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Singh, M.; Peuget, S.; Selivanova, G. Inhibition of P53 Inhibitors: Progress, Challenges and Perspectives. J. Mol. Cell Biol. 2019, 11, 586–599. [Google Scholar] [CrossRef]
- Yu, X.; Narayanan, S.; Vazquez, A.; Carpizo, D.R. Small Molecule Compounds Targeting the P53 Pathway: Are We Finally Making Progress? Apoptosis 2014, 19, 1055–1068. [Google Scholar] [CrossRef]
- Silva, J.L.; Lima, C.G.S.; Rangel, L.P.; Ferretti, G.D.S.; Pauli, F.P.; Ribeiro, R.C.B.; da Silva, T.B.; da Silva, F.C.; Ferreira, V.F. Recent Synthetic Approaches Towards Small Molecule Reactivators of P53. Biomolecules 2020, 10, 635. [Google Scholar] [CrossRef]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The P53-Mdm-2 Autoregulatory Feedback Loop. Genes Dev. 1993, 7, 1126–1132. [Google Scholar] [CrossRef]
- Momand, J.; Zambetti, G.P.; Olson, D.C.; George, D.; Levine, A.J. The Mdm-2 Oncogene Product Forms a Complex with the P53 Protein and Inhibits P53-Mediated Transactivation. Cell 1992, 69, 1237–1245. [Google Scholar] [CrossRef]
- Freedman, D.A.; Wu, L.; Levine, A.J. Functions of the Mdm2 Oncoprotein. Cell Mol. Life Sci. 1999, 55, 96–107. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Oren, M. Mdm2: The Ups and Downs. Mol. Med. 1999, 5, 71–83. [Google Scholar] [CrossRef]
- Momand, J.; Wu, H.H.; Dasgupta, G. Mdm2—Master Regulator of the P53 Tumor Suppressor Protein. Gene 2000, 242, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Hu, W.W.; Levine, A.J. Mdm2 Is a Central Node in the P53 Pathway: 12 Years and Counting. Curr. Cancer Drug Targets 2005, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. P53 in Health and Disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Elenbaas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the Mdm2 Oncoprotein Bound to the P53 Tumor Suppressor Transactivation Domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In Vivo Activation of the P53 Pathway by Small-Molecule Antagonists of Mdm2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.; Wovkulich, P.; Pizzolato, G.; Lovey, A.; Ding, Q.; Jiang, N.; Liu, J.J.; Zhao, C.; Glenn, K.; Wen, Y.; et al. Discovery of Rg7112: A Small-Molecule Mdm2 Inhibitor in Clinical Development. ACS Med. Chem. Lett. 2013, 4, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Qiu, S.; Ding, Y.; Gao, W.; Stuckey, J.; Krajewski, K.; Roller, P.P.; Tomita, Y.; et al. Structure-Based Design of Potent Non-Peptide Mdm2 Inhibitors. J. Am. Chem. Soc. 2005, 127, 10130–10131. [Google Scholar] [CrossRef]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Wang, G.; Qiu, S.; Shangary, S.; Gao, W.; Qin, D.; Stuckey, J.; Krajewski, K.; et al. Structure-Based Design of Spiro-Oxindoles as Potent, Specific Small-Molecule Inhibitors of the Mdm2-P53 Interaction. J. Med. Chem. 2006, 49, 3432–3435. [Google Scholar] [CrossRef]
- Shangary, S.; Qin, D.; McEachern, D.; Liu, M.; Miller, R.S.; Qiu, S.; Nikolovska-Coleska, Z.; Ding, K.; Wang, G.; Chen, J.; et al. Temporal Activation of P53 by a Specific Mdm2 Inhibitor Is Selectively Toxic to Tumors and Leads to Complete Tumor Growth Inhibition. Proc. Natl. Acad. Sci. USA 2008, 105, 3933–3938. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, L.; Sun, W.; Lu, J.; McEachern, D.; Li, X.; Yu, S.; Bernard, D.; Ochsenbein, P.; Ferey, V.; et al. Diastereomeric Spirooxindoles as Highly Potent and Efficacious Mdm2 Inhibitors. J. Am. Chem. Soc. 2013, 135, 7223–7234. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, S.; Sun, W.; Liu, L.; Lu, J.; McEachern, D.; Shargary, S.; Bernard, D.; Li, X.; Zhao, T.; et al. A Potent Small-Molecule Inhibitor of the Mdm2-P53 Interaction (Mi-888) Achieved Complete and Durable Tumor Regression in Mice. J. Med. Chem. 2013, 56, 5553–5561. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Sun, W.; Zhao, Y.; McEachern, D.; Meaux, I.; Barriere, C.; Stuckey, J.A.; Meagher, J.L.; Bai, L.; Liu, L.; et al. Sar405838: An Optimized Inhibitor of Mdm2-P53 Interaction That Induces Complete and Durable Tumor Regression. Cancer Res. 2014, 74, 5855–5865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chu, X.J.; Liu, J.J.; Ding, Q.; Zhang, J.; Bartkovitz, D.; Jiang, N.; Karnachi, P.; So, S.S.; Tovar, C.; et al. Discovery of Potent and Orally Active P53-Mdm2 Inhibitors Ro5353 and Ro2468 for Potential Clinical Development. ACS Med. Chem. Lett. 2014, 5, 124–127. [Google Scholar] [CrossRef]
- Ding, Q.; Zhang, Z.; Liu, J.J.; Jiang, N.; Zhang, J.; Ross, T.M.; Chu, X.J.; Bartkovitz, D.; Podlaski, F.; Janson, C.; et al. Discovery of Rg7388, a Potent and Selective P53-Mdm2 Inhibitor in Clinical Development. J. Med. Chem. 2013, 56, 5979–5983. [Google Scholar] [CrossRef]
- Sun, D.; Li, Z.; Rew, Y.; Gribble, M.; Bartberger, M.D.; Beck, H.P.; Canon, J.; Chen, A.; Chen, X.; Chow, D.; et al. Discovery of Amg 232, a Potent, Selective, and Orally Bioavailable Mdm2-P53 Inhibitor in Clinical Development. J. Med. Chem. 2014, 57, 1454–1472. [Google Scholar] [CrossRef]
- Rew, Y.; Sun, D. Discovery of a Small Molecule Mdm2 Inhibitor (Amg 232) for Treating Cancer. J. Med. Chem. 2014, 57, 6332–6341. [Google Scholar] [CrossRef] [PubMed]
- Furet, P.; Chene, P.; De Pover, A.; Valat, T.S.; Lisztwan, J.H.; Kallen, J.; Masuya, K. The Central Valine Concept Provides an Entry in a New Class of Non Peptide Inhibitors of the P53-Mdm2 Interaction. Bioorg. Med. Chem. Lett. 2012, 22, 3498–3502. [Google Scholar] [CrossRef] [PubMed]
- Gessier, F.; Kallen, J.; Jacoby, E.; Chene, P.; Stachyra-Valat, T.; Ruetz, S.; Jeay, S.; Holzer, P.; Masuya, K.; Furet, P. Discovery of Dihydroisoquinolinone Derivatives as Novel Inhibitors of the P53-Mdm2 Interaction with a Distinct Binding Mode. Bioorg. Med. Chem. Lett. 2015, 25, 3621–3625. [Google Scholar] [CrossRef]
- Holzer, P.; Masuya, K.; Furet, P.; Kallen, J.; Valat-Stachyra, T.; Ferretti, S.; Berghausen, J.; Bouisset-Leonard, M.; Buschmann, N.; Pissot-Soldermann, C.; et al. Discovery of a Dihydroisoquinolinone Derivative (Nvp-Cgm097): A Highly Potent and Selective Mdm2 Inhibitor Undergoing Phase 1 Clinical Trials in P53wt Tumors. J. Med. Chem. 2015, 58, 6348–6358. [Google Scholar] [CrossRef]
- Furet, P.; Masuya, K.; Kallen, J.; Stachyra-Valat, T.; Ruetz, S.; Guagnano, V.; Holzer, P.; Mah, R.; Stutz, S.; Vaupel, A.; et al. Discovery of a Novel Class of Highly Potent Inhibitors of the P53-Mdm2 Interaction by Structure-Based Design Starting from a Conformational Argument. Bioorg. Med. Chem. Lett. 2016, 26, 4837–4841. [Google Scholar] [CrossRef]
- Holzer, P. Discovery of Potent and Selective P53-Mdm2 Protein-Protein Interaction Inhibitors as Anticancer Drugs. Chimia 2017, 71, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Jeay, S.; Ferretti, S.; Holzer, P.; Fuchs, J.; Chapeau, E.A.; Wartmann, M.; Sterker, D.; Romanet, V.; Murakami, M.; Kerr, G.; et al. Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the P53-Mdm2 Inhibitor Hdm201. Cancer Res. 2018, 78, 6257–6267. [Google Scholar] [CrossRef] [PubMed]
- Arnhold, V.; Schmelz, K.; Proba, J.; Winkler, A.; Wunschel, J.; Toedling, J.; Deubzer, H.E.; Kunkele, A.; Eggert, A.; Schulte, J.H.; et al. Reactivating Tp53 Signaling by the Novel Mdm2 Inhibitor Ds-3032b as a Therapeutic Option for High-Risk Neuroblastoma. Oncotarget 2018, 9, 2304–2319. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Sun, W.; Liu, L.; Lu, J.; McEachern, D.; Bernard, D.; Deschamps, J.R.; Wang, S. Design of Chemically Stable, Potent, and Efficacious Mdm2 Inhibitors That Exploit the Retro-Mannich Ring-Opening-Cyclization Reaction Mechanism in Spiro-Oxindoles. J. Med. Chem. 2014, 57, 10486–10498. [Google Scholar] [CrossRef]
- Aguilar, A.; Lu, J.; Liu, L.; Du, D.; Bernard, D.; McEachern, D.; Przybranowski, S.; Li, X.; Luo, R.; Wen, B.; et al. Discovery of 4-((3′r,4′s,5′r)-6″-Chloro-4′-(3-Chloro-2-Fluorophenyl)-1′-Ethyl-2″-Oxodispiro[ Cyclohexane-1,2′-Pyrrolidine-3′,3″-Indoline]-5′-Carboxamido)Bicyclo[2.2.2]Octane -1-Carboxylic Acid (Aa-115/Apg-115): A Potent and Orally Active Murine Double Minute 2 (Mdm2) Inhibitor in Clinical Development. J. Med. Chem. 2017, 60, 2819–2839. [Google Scholar]
- Rudolph, D.; Reschke, M.; Blake, S.; Rinnenthal, J.; Wernitznig, A.; Weyer-Czernilofsky, U.; Gollner, A.; Haslinger, C.; Garin-Chesa, P.; Quant, J.; et al. Bi 907828: A Novel, Potent Mdm2 Inhibitor That Induces Antitumor Immunologic Memory and Acts Synergistically with an Anti-Pd-1 Antibody in Syngeneic Mouse Models of Cancer. Cancer Res. 2018, 78, 4866. [Google Scholar] [CrossRef]
- Cornillie, J.; Wozniak, A.; Li, H.; Gebreyohannes, Y.K.; Wellens, J.; Hompes, D.; Debiec-Rychter, M.; Sciot, R.; Schoffski, P. Anti-Tumor Activity of the Mdm2-Tp53 Inhibitor Bi-907828 in Dedifferentiated Liposarcoma Patient-Derived Xenograft Models Harboring Mdm2 Amplification. Clin. Transl. Oncol. 2020, 22, 546–554. [Google Scholar] [CrossRef]
- Rinnenthal, J.; Rudolph, D.; Blake, S.; Gollner, A.; Wernitznig, A.; Weyer-Czernilofsky, U.; Haslinger, C.; Garin-Chesa, P.; Moll, J.; Kraut, N.; et al. Bi 907828: A Highly Potent Mdm2 Inhibitor with Low Human Dose Estimation, Designed for High-Dose Intermittent Schedules in the Clinic. Cancer Res. 2018, 78, 4865. [Google Scholar] [CrossRef]
- Gollner, A.; Rudolph, D.; Arnhof, H.; Bauer, M.; Blake, S.M.; Boehmelt, G.; Cockroft, X.L.; Dahmann, G.; Ettmayer, P.; Gerstberger, T.; et al. Discovery of Novel Spiro[3h-Indole-3,2′-Pyrrolidin]-2(1h)-One Compounds as Chemically Stable and Orally Active Inhibitors of the Mdm2-P53 Interaction. J. Med. Chem. 2016, 59, 10147–10162. [Google Scholar] [CrossRef]
- Kang, M.H.; Reynolds, C.P.; Kolb, E.A.; Gorlick, R.; Carol, H.; Lock, R.; Keir, S.T.; Maris, J.M.; Wu, J.; Lyalin, D.; et al. Initial Testing (Stage 1) of Mk-8242-a Novel Mdm2 Inhibitor-by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer 2016, 63, 1744–1752. [Google Scholar] [CrossRef]
- Ravandi, F.; Gojo, I.; Patnaik, M.M.; Minden, M.D.; Kantarjian, H.; Johnson-Levonas, A.O.; Fancourt, C.; Lam, R.; Jones, M.B.; Knox, C.D.; et al. A Phase I Trial of the Human Double Minute 2 Inhibitor (Mk-8242) in Patients with Refractory/Recurrent Acute Myelogenous Leukemia (Aml). Leuk. Res. 2016, 48, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lahue, B.R.; Shipps, G.W., Jr.; Brookes, J.; Wang, Y. Substituted Piperidines as Hdm2 Inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Lahue, B.R.; Ma, Y.; Nair, L.G.; Shipps, G.W., Jr.; Wang, Y.; Doll, R.; Bogen, S.L. Core Modification of Substituted Piperidines as Novel Inhibitors of Hdm2-P53 Protein-Protein Interaction. Bioorg. Med. Chem. Lett. 2014, 24, 1983–1986. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lahue, B.R.; Gibeau, C.R.; Shipps, G.W., Jr.; Bogen, S.L.; Wang, Y.; Guo, Z.; Guzi, T.J. Pivotal Role of an Aliphatic Side Chain in the Development of an Hdm2 Inhibitor. ACS Med. Chem. Lett. 2014, 5, 572–575. [Google Scholar] [CrossRef] [PubMed]
- Bogen, S.L.; Pan, W.; Gibeau, C.R.; Lahue, B.R.; Ma, Y.; Nair, L.G.; Seigel, E.; Shipps, G.W., Jr.; Tian, Y.; Wang, Y.; et al. Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious Hdm2-P53 Inhibitors. ACS Med. Chem. Lett. 2016, 7, 324–329. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reutershan, M.H.; Machacek, M.R.; Altman, M.D.; Bogen, S.; Cai, M.; Cammarano, C.; Chen, D.; Christopher, M.; Cryan, J.; Daublain, P.; et al. Discovery of Mk-4688: An Efficient Inhibitor of the Hdm2-P53 Protein-Protein Interaction. J. Med. Chem. 2021, 64, 16213–16241. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Blay, J.Y.; Italiano, A.; Le Cesne, A.; Penel, N.; Zhi, J.; Heil, F.; Rueger, R.; Graves, B.; Ding, M.; et al. Effect of the Mdm2 Antagonist Rg7112 on the P53 Pathway in Patients with Mdm2-Amplified, Well-Differentiated or Dedifferentiated Liposarcoma: An Exploratory Proof-of-Mechanism Study. Lancet Oncol. 2012, 13, 1133–1140. [Google Scholar] [CrossRef]
- Iancu-Rubin, C.; Mosoyan, G.; Glenn, K.; Gordon, R.E.; Nichols, G.L.; Hoffman, R. Activation of P53 by the Mdm2 Inhibitor Rg7112 Impairs Thrombopoiesis. Exp. Hematol. 2014, 42, 137–145.e5. [Google Scholar] [CrossRef]
- Siu, L.L.; Italiano, A.; Miller, W.H.; Blay, J.Y.; Gietema, J.A.; Bang, Y.J.; Mileshkin, L.R.; Hirte, H.W.; Reckner, M.; Higgins, B.; et al. Phase 1 Dose Escalation, Food Effect, and Biomarker Study of Rg7388, a More Potent Second-Generation Mdm2 Antagonist, in Patients (Pts) with Solid Tumors. J. Clin. Oncol. 2014, 32, 2535. [Google Scholar] [CrossRef]
- De Weger, V.; Lolkema, M.P.; Dickson, M.; Le Cesne, A.; Wagner, A.; Merqui-Roelvink, M.; Varga, A.; Tap, W.; Schwartz, G.; Demetri, G.; et al. A First-in-Human (Fih) Safety and Pharmacological Study of Sar405838, a Novel Hdm2 Antagonist, in Patients with Solid Malignancies. Eur. J. Cancer 2014, 50, 121–122. [Google Scholar] [CrossRef]
- de Jonge, M.; de Weger, V.A.; Dickson, M.A.; Langenberg, M.; Le Cesne, A.; Wagner, A.J.; Hsu, K.; Zheng, W.; Mace, S.; Tuffal, G.; et al. A Phase I Study of Sar405838, a Novel Human Double Minute 2 (Hdm2) Antagonist, in Patients with Solid Tumours. Eur. J. Cancer 2017, 76, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.J.; Banerji, U.; Mahipal, A.; Somaiah, N.; Hirsch, H.A.; Fancourt, C.; Levonas, A.; Lam, R.; Meister, A.; Kemp, R.K.; et al. A Phase I Trial of the Human Double Minute 2 (Hdm2) Inhibitor Mk-8242 in Patients (Pts) with Advanced Solid Tumors. J. Clin. Oncol. 2015, 33, 10564. [Google Scholar] [CrossRef]
- Wagner, A.J.; Banerji, U.; Mahipal, A.; Somaiah, N.; Hirsch, H.; Fancourt, C.; Johnson-Levonas, A.O.; Lam, R.; Meister, A.K.; Russo, G.; et al. Phase I Trial of the Human Double Minute 2 Inhibitor Mk-8242 in Patients with Advanced Solid Tumors. J. Clin. Oncol. 2017, 35, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Schneekloth, J.S., Jr.; Fonseca, F.N.; Koldobskiy, M.; Mandal, A.; Deshaies, R.; Sakamoto, K.; Crews, C.M. Chemical Genetic Control of Protein Levels: Selective in Vivo Targeted Degradation. J. Am. Chem. Soc. 2004, 126, 3748–3754. [Google Scholar] [CrossRef]
- Lai, A.C.; Crews, C.M. Induced Protein Degradation: An Emerging Drug Discovery Paradigm. Nat. Rev. Drug Discov. 2017, 16, 101–114. [Google Scholar] [CrossRef]
- Crews, C.M.; Georg, G.; Wang, S.M. Inducing Protein Degradation as a Therapeutic Strategy. J. Med. Chem. 2016, 59, 5129–5130. [Google Scholar] [CrossRef][Green Version]
- Bekes, M.; Langley, D.R.; Crews, C.M. Protac Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Chan, K.H.; Zengerle, M.; Testa, A.; Ciulli, A. Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (Bet) Degraders Derived from Triazolodiazepine (Jq1) and Tetrahydroquinoline (I-Bet726) Bet Inhibitor Scaffolds. J. Med. Chem. 2018, 61, 504–513. [Google Scholar] [CrossRef]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural Basis of Protac Cooperative Recognition for Selective Protein Degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Mares, A.; Smith, I.E.; Ko, E.; Campos, S.; Miah, A.H.; Mulholland, K.E.; Routly, N.; Buckley, D.L.; Gustafson, J.L.; et al. Catalytic in Vivo Protein Knockdown by Small-Molecule Protacs. Nat. Chem. Biol. 2015, 11, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Aguilar, A.; McEachern, D.; Przybranowski, S.; Liu, L.; Yang, C.Y.; Wang, M.; Han, X.; Wang, S. Discovery of Md-224 as a First-in-Class, Highly Potent, and Efficacious Proteolysis Targeting Chimera Murine Double Minute 2 Degrader Capable of Achieving Complete and Durable Tumor Regression. J. Med. Chem. 2019, 62, 448–466. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Aguilar, A.; Liu, Z.; Yang, C.Y.; Wang, S. Simple Structural Modifications Converting a Bona Fide Mdm2 Protac Degrader into a Molecular Glue Molecule: A Cautionary Tale in the Design of Protac Degraders. J. Med. Chem. 2019, 62, 9471–9487. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, S.; Liu, J.; Yang, K.; Xie, H.; Tang, W. Development of Selective Small Molecule Mdm2 Degraders Based on Nutlin. Eur. J. Med. Chem. 2019, 176, 476–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, J.; Tandon, I.; Wu, S.; Teng, P.; Liao, J.; Tang, W. Development of Mdm2 Degraders Based on Ligands Derived from Ugi Reactions: Lessons and Discoveries. Eur. J. Med. Chem. 2021, 219, 113425. [Google Scholar] [CrossRef]
- Marcellino, B.; Yang, X.B.; Chen, H.; Chen, K.; Brady, C.; Clementelli, C.; Li, Z.J.; Hoffman, R.; Liu, J.; Kaniskan, H.U.; et al. Development of an Mdm2 Degrader for Treatment of Acute Leukemias. Blood 2021, 138, 1866. [Google Scholar] [CrossRef]
- Chutake, Y.; Mayo, M.; Chen, D.; Enerson, B.; Cho, P.; Filiatrault, J.; Brown, C.; Placke, M.; Adams, M.; Karnik, R.; et al. Abstract 3934: Kt-253, a Highly Potent and Selective Heterobifunctional Mdm2 Degrader for the Treatment of Wildtype P53 Tumors with Superior Potency and Differentiated Biological Activity Compared to Small Molecule Inhibitors (Smi). Cancer Res. 2022, 82, 3934. [Google Scholar] [CrossRef]
- Laurie, N.A.; Donovan, S.L.; Shih, C.S.; Zhang, J.; Mills, N.; Fuller, C.; Teunisse, A.; Lam, S.; Ramos, Y.; Mohan, A.; et al. Inactivation of the P53 Pathway in Retinoblastoma. Nature 2006, 444, 61–66. [Google Scholar] [CrossRef]
- Danovi, D.; Meulmeester, E.; Pasini, D.; Migliorini, D.; Capra, M.; Frenk, R.; de Graaf, P.; Francoz, S.; Gasparini, P.; Gobbi, A.; et al. Amplification of Mdmx (or Mdm4) Directly Contributes to Tumor Formation by Inhibiting P53 Tumor Suppressor Activity. Mol. Cell Biol. 2004, 24, 5835–5843. [Google Scholar] [CrossRef]
- Gembarska, A.; Luciani, F.; Fedele, C.; Russell, E.A.; Dewaele, M.; Villar, S.; Zwolinska, A.; Haupt, S.; de Lange, J.; Yip, D.; et al. Mdm4 Is a Key Therapeutic Target in Cutaneous Melanoma. Nat. Med. 2012, 18, 1239–1247. [Google Scholar] [CrossRef]
- Shvarts, A.; Steegenga, W.T.; Riteco, N.; van Laar, T.; Dekker, P.; Bazuine, M.; van Ham, R.C.; van der Houven van Oordt, W.; Hateboer, G.; van der Eb, A.J.; et al. Mdmx: A Novel P53-Binding Protein with Some Functional Properties of Mdm2. EMBO J. 1996, 15, 5349–5357. [Google Scholar] [CrossRef] [PubMed]
- Tanimura, S.; Ohtsuka, S.; Mitsui, K.; Shirouzu, K.; Yoshimura, A.; Ohtsubo, M. Mdm2 Interacts with Mdmx through Their Ring Finger Domains. FEBS Lett. 1999, 447, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Leslie, P.L.; Ke, H.; Zhang, Y. The Mdm2 Ring Domain and Central Acidic Domain Play Distinct Roles in Mdm2 Protein Homodimerization and Mdm2-Mdmx Protein Heterodimerization. J. Biol. Chem. 2015, 290, 12941–12950. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Kawai, H.; Nie, L.; Kitao, H.; Wiederschain, D.; Jochemsen, A.G.; Parant, J.; Lozano, G.; Yuan, Z.M. Mutual Dependence of Mdm2 and Mdmx in Their Functional Inactivation of P53. J. Biol. Chem. 2002, 277, 19251–19254. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.C.; Francoz, S.; Maetens, M.; Wahl, G.; Toledo, F.; Lozano, G. Keeping P53 in Check: Essential and Synergistic Functions of Mdm2 and Mdm4. Cell Death Differ. 2006, 13, 927–934. [Google Scholar] [CrossRef]
- Popowicz, G.M.; Czarna, A.; Holak, T.A. Structure of the Human Mdmx Protein Bound to the P53 Tumor Suppressor Transactivation Domain. Cell Cycle 2008, 7, 2441–2443. [Google Scholar] [CrossRef]
- Chang, Y.S.; Graves, B.; Guerlavais, V.; Tovar, C.; Packman, K.; To, K.H.; Olson, K.A.; Kesavan, K.; Gangurde, P.; Mukherjee, A.; et al. Stapled Alpha-Helical Peptide Drug Development: A Potent Dual Inhibitor of Mdm2 and Mdmx for P53-Dependent Cancer Therapy. Proc. Natl. Acad. Sci. USA 2013, 110, E3445–E3454. [Google Scholar] [CrossRef]
- Carvajal, L.A.; Neriah, D.B.; Senecal, A.; Benard, L.; Thiruthuvanathan, V.; Yatsenko, T.; Narayanagari, S.R.; Wheat, J.C.; Todorova, T.I.; Mitchell, K.; et al. Dual Inhibition of Mdmx and Mdm2 as a Therapeutic Strategy in Leukemia. Sci. Transl. Med. 2018, 10, eaao3003. [Google Scholar] [CrossRef]
- Kallen, J.; Izaac, A.; Chau, S.; Wirth, E.; Schoepfer, J.; Mah, R.; Schlapbach, A.; Stutz, S.; Vaupel, A.; Guagnano, V.; et al. Structural States of Hdm2 and Hdmx: X-Ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes. ChemMedChem 2019, 14, 1305–1314. [Google Scholar] [CrossRef]
- Zhang, S.; Lou, J.; Li, Y.; Zhou, F.; Yan, Z.; Lyu, X.; Zhao, Y. Recent Progress and Clinical Development of Inhibitors That Block Mdm4/P53 Protein-Protein Interactions. J. Med. Chem. 2021, 64, 10621–10640. [Google Scholar] [CrossRef]
- Popowicz, G.M.; Czarna, A.; Wolf, S.; Wang, K.; Wang, W.; Domling, A.; Holak, T.A. Structures of Low Molecular Weight Inhibitors Bound to Mdmx and Mdm2 Reveal New Approaches for P53-Mdmx/Mdm2 Antagonist Drug Discovery. Cell Cycle 2010, 9, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Furet, P.; Joerg, K.; Julien, L.; Keiichi, M. 3-Imidazolylindoles as Mdm2 and Mdm4 Inhibitors and Their Preparation; Novartis: Basel, Switzerland, 2012; 333p. [Google Scholar]
- Jung, J.; Lee, J.S.; Dickson, M.A.; Schwartz, G.K.; Le Cesne, A.; Varga, A.; Bahleda, R.; Wagner, A.J.; Choy, E.; de Jonge, M.J.; et al. Tp53 Mutations Emerge with Hdm2 Inhibitor Sar405838 Treatment in De-Differentiated Liposarcoma. Nat. Commun. 2016, 7, 12609. [Google Scholar] [CrossRef] [PubMed]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. P53 Mutations in Human Cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Momand, J.; Finlay, C.A. The P53 Tumour Suppressor Gene. Nature 1991, 351, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, M.S.; Bennett, W.P.; Hollstein, M.; Harris, C.C. Mutations in the P53 Tumor Suppressor Gene: Clues to Cancer Etiology and Molecular Pathogenesis. Cancer Res. 1994, 54, 4855–4878. [Google Scholar]
- Leroy, B.; Fournier, J.L.; Ishioka, C.; Monti, P.; Inga, A.; Fronza, G.; Soussi, T. The Tp53 Website: An Integrative Resource Centre for the Tp53 Mutation Database and Tp53 Mutant Analysis. Nucleic Acids Res. 2013, 41, D962–D969. [Google Scholar] [CrossRef]
- Sabapathy, K.; Lane, D.P. Therapeutic Targeting of P53: All Mutants Are Equal, but Some Mutants Are More Equal Than Others. Nat. Rev. Clin. Oncol. 2018, 15, 13–30. [Google Scholar] [CrossRef]
- Schulz-Heddergott, R.; Moll, U.M. Gain-of-Function (Gof) Mutant P53 as Actionable Therapeutic Target. Cancers 2018, 10, 188. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. P53 from Complexity to Simplicity: Mutant P53 Stabilization, Gain-of-Function, and Dominant-Negative Effect. FASEB J. 2000, 14, 1901–1907. [Google Scholar] [CrossRef]
- Dearth, L.R.; Qian, H.; Wang, T.; Baroni, T.E.; Zeng, J.; Chen, S.W.; Yi, S.Y.; Brachmann, R.K. Inactive Full-Length P53 Mutants Lacking Dominant Wild-Type P53 Inhibition Highlight Loss of Heterozygosity as an Important Aspect of P53 Status in Human Cancers. Carcinogenesis 2007, 28, 289–298. [Google Scholar] [CrossRef]
- Xu, J.; Reumers, J.; Couceiro, J.R.; De Smet, F.; Gallardo, R.; Rudyak, S.; Cornelis, A.; Rozenski, J.; Zwolinska, A.; Marine, J.C.; et al. Gain of Function of Mutant P53 by Coaggregation with Multiple Tumor Suppressors. Nat. Chem. Biol. 2011, 7, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Strano, S.; Dell’Orso, S.; Di Agostino, S.; Fontemaggi, G.; Sacchi, A.; Blandino, G. Mutant P53: An Oncogenic Transcription Factor. Oncogene 2007, 26, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of P53 Function Leads to Tumour Regression in Vivo. Nature 2007, 445, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cao, J.; Topatana, W.; Juengpanich, S.; Li, S.; Zhang, B.; Shen, J.; Cai, L.; Cai, X.; Chen, M. Targeting Mutant P53 for Cancer Therapy: Direct and Indirect Strategies. J. Hematol. Oncol. 2021, 14, 157. [Google Scholar] [CrossRef]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of Mutant P53 Functional Properties on Tp53 Mutation Patterns and Tumor Phenotype: Lessons from Recent Developments in the Iarc Tp53 Database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. Tp53 Variations in Human Cancers: New Lessons from the Iarc Tp53 Database and Genomics Data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef]
- Petitjean, A.; Achatz, M.I.; Borresen-Dale, A.L.; Hainaut, P.; Olivier, M. Tp53 Mutations in Human Cancers: Functional Selection and Impact on Cancer Prognosis and Outcomes. Oncogene 2007, 26, 2157–2165. [Google Scholar] [CrossRef]
- Joerger, A.C.; Fersht, A.R. Structure-Function-Rescue: The Diverse Nature of Common P53 Cancer Mutants. Oncogene 2007, 26, 2226–2242. [Google Scholar] [CrossRef]
- Bullock, A.N.; Henckel, J.; Fersht, A.R. Quantitative Analysis of Residual Folding and DNA Binding in Mutant P53 Core Domain: Definition of Mutant States for Rescue in Cancer Therapy. Oncogene 2000, 19, 1245–1256. [Google Scholar] [CrossRef]
- Cho, Y.; Gorina, S.; Jeffrey, P.D.; Pavletich, N.P. Crystal Structure of a P53 Tumor Suppressor-DNA Complex: Understanding Tumorigenic Mutations. Science 1994, 265, 346–355. [Google Scholar] [CrossRef]
- Bullock, A.N.; Fersht, A.R. Rescuing the Function of Mutant P53. Nat. Rev. Cancer 2001, 1, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor P53. Annu. Rev. Biochem. 2008, 77, 557–582. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. The P53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu. Rev. Biochem. 2016, 85, 375–404. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.; Zache, N.; Stridh, H.; Westman, J.; Bergman, J.; Selivanova, G.; Wiman, K.G. Prima-1(Met) Synergizes with Cisplatin to Induce Tumor Cell Apoptosis. Oncogene 2005, 24, 3484–3491. [Google Scholar] [CrossRef] [PubMed]
- Salim, K.Y.; Vareki, S.M.; Danter, W.R.; Koropatnick, J. Coti-2, a New Anticancer Drug Currently under Clinical Investigation, Targets Mutant P53 and Negatively Modulates the Pi3k/Akt/Mtor Pathway. Eur. J. Cancer 2016, 69, S19. [Google Scholar] [CrossRef]
- Westin, S.N.; Nieves-Neira, W.; Lynam, C.; Salim, K.Y.; Silva, A.D.; Ho, R.T.; Mills, G.B.; Coleman, R.L.; Janku, F.; Matei, D. Safety and Early Efficacy Signals for Coti-2, an Orally Available Small Molecule Targeting P53, in a Phase I Trial of Recurrent Gynecologic Cancer. Cancer Res. 2018, 78, 615. [Google Scholar] [CrossRef]
- Bykov, V.J.N.; Issaeva, N.; Shilov, A.; Hultcrantz, M.; Pugacheva, E.; Chumakov, P.; Bergman, J.; Wiman, K.G.; Selivanova, G. Restoration of the Tumor Suppressor Function to Mutant P53 by a Low-Molecular-Weight Compound. Nat. Med. 2002, 8, 282–288. [Google Scholar] [CrossRef]
- Zhang, Q.; Bykov, V.J.N.; Wiman, K.G.; Zawacka-Pankau, J. Apr-246 Reactivates Mutant P53 by Targeting Cysteines 124 and 277. Cell Death Dis. 2018, 9, 439. [Google Scholar] [CrossRef]
- Lambert, J.M.; Gorzov, P.; Veprintsev, D.B.; Soderqvist, M.; Segerback, D.; Bergman, J.; Fersht, A.R.; Hainaut, P.; Wiman, K.G.; Bykov, V.J. Prima-1 Reactivates Mutant P53 by Covalent Binding to the Core Domain. Cancer Cell 2009, 15, 376–388. [Google Scholar] [CrossRef]
- Degtjarik, O.; Golovenko, D.; Diskin-Posner, Y.; Abrahmsen, L.; Rozenberg, H.; Shakked, Z. Structural Basis of Reactivation of Oncogenic P53 Mutants by a Small Molecule: Methylene Quinuclidinone (Mq). Nat. Commun. 2021, 12, 7057. [Google Scholar] [CrossRef]
- Tessoulin, B.; Descamps, G.; Moreau, P.; Maiga, S.; Lode, L.; Godon, C.; Marionneau-Lambot, S.; Oullier, T.; Le Gouill, S.; Amiot, M.; et al. Prima-1met Induces Myeloma Cell Death Independent of P53 by Impairing the Gsh/Ros Balance. Blood 2014, 124, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.S.; Duong, C.P.; Haupt, S.; Montgomery, K.G.; House, C.M.; Azar, W.J.; Pearson, H.B.; Fisher, O.M.; Read, M.; Guerra, G.R.; et al. Inhibiting the System Xc(-)/Glutathione Axis Selectively Targets Cancers with Mutant-P53 Accumulation. Nat. Commun. 2017, 8, 14844. [Google Scholar] [CrossRef] [PubMed]
- Salim, K.Y.; Vareki, S.M.; Danter, W.R.; Koropatnick, J. Coti-2, a Novel Small Molecule That Is Active against Multiple Human Cancer Cell Lines in Vitro and in Vivo. Oncotarget 2016, 7, 41363–41379. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.L.; Osman, A.A.; Patel, A.A.; Tanaka, N.; Tang, L.; Zhou, G.; Myers, J.N. A Novel Third Generation Thiosemicarbazone, Coti-2, Is Highly Effective in Killing Head and Neck Squamous Cell Carcinomas (Hnscc) Bearing a Variety of Tp53 Mutations. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 942. [Google Scholar] [CrossRef]
- Wang, G.; Fersht, A.R. Multisite Aggregation of P53 and Implications for Drug Rescue. Proc. Natl. Acad. Sci. USA 2017, 114, E2634–E2643. [Google Scholar] [CrossRef]
- Bauer, M.R.; Jones, R.N.; Tareque, R.K.; Springett, B.; Dingler, F.A.; Verduci, L.; Patel, K.J.; Fersht, A.R.; Joerger, A.C.; Spencer, J. A Structure-Guided Molecular Chaperone Approach for Restoring the Transcriptional Activity of the P53 Cancer Mutant Y220c. Future Med. Chem. 2019, 11, 2491–2504. [Google Scholar] [CrossRef]
- Joerger, A.C.; Ang, H.C.; Fersht, A.R. Structural Basis for Understanding Oncogenic P53 Mutations and Designing Rescue Drugs. Proc. Natl. Acad. Sci. USA 2006, 103, 15056–15061. [Google Scholar] [CrossRef]
- Boeckler, F.M.; Joerger, A.C.; Jaggi, G.; Rutherford, T.J.; Veprintsev, D.B.; Fersht, A.R. Targeted Rescue of a Destabilized Mutant of P53 by an in Silico Screened Drug. Proc. Natl. Acad. Sci. USA 2008, 105, 10360–10365. [Google Scholar] [CrossRef]
- Wilcken, R.; Liu, X.R.; Zimmermann, M.O.; Rutherford, T.J.; Fersht, A.R.; Joerger, A.C.; Boeckler, F.M. Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53. J. Am. Chem. Soc. 2012, 134, 6810–6818. [Google Scholar] [CrossRef]
- Liu, X.; Wilcken, R.; Joerger, A.C.; Chuckowree, I.S.; Amin, J.; Spencer, J.; Fersht, A.R. Small Molecule Induced Reactivation of Mutant P53 in Cancer Cells. Nucleic Acids Res. 2013, 41, 6034–6044. [Google Scholar] [CrossRef]
- Baud, M.G.J.; Bauer, M.R.; Verduci, L.; Dingler, F.A.; Patel, K.J.; Roy, D.H.; Joerger, A.C.; Fersht, A.R. Aminobenzothiazole Derivatives Stabilize the Thermolabile P53 Cancer Mutant Y220c and Show Anticancer Activity in P53-Y220c Cell Lines. Eur. J. Med. Chem. 2018, 152, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.R.; Joerger, A.C.; Fersht, A.R. 2-Sulfonylpyrimidines: Mild Alkylating Agents with Anticancer Activity toward P53-Compromised Cells. Proc. Natl. Acad. Sci. USA 2016, 113, E5271–E5280. [Google Scholar] [CrossRef] [PubMed]
- Puzio-Kuter, A.M.; Mulligan, C.; Russo, B.; Wiebesiek, A.; Xu, L.; Yang, H.; Vu, B.; Dumble, M. Abstract 1295: Small Molecule Reactivators of Y220c Mutant P53 Modulate Tumor Infiltrating Leukocytes and Synergize with Immune Checkpoint Inhibitors. Cancer Res. 2022, 82, 1295. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).