Drug-Resistant Tuberculosis Treatment Recommendation, and Multi-Class Tuberculosis Detection and Classification Using Ensemble Deep Learning-Based System
Abstract
1. Introduction
- It is the first model that can determine from CXR image whether a patient has tuberculosis and, if so, what kinds of drug resistance they may have.
- The proposed model is the first to employ ensemble deep learning to make decisions in multiclass classifications for tuberculosis and drug-resistance classification.
- For the users, we design computer applications. It is the first application that can determine whether a patient has tuberculosis and, if so, what type of drug resistance they have, using only a CXR image.
- In the application, the recommended regimen that is appropriate for a specific patient will be displayed.
1.1. Related Work
1.1.1. AI for TB Detection
1.1.2. AI in Drug-Resistant Classification
1.1.3. Web Applications in Health Diagnosis
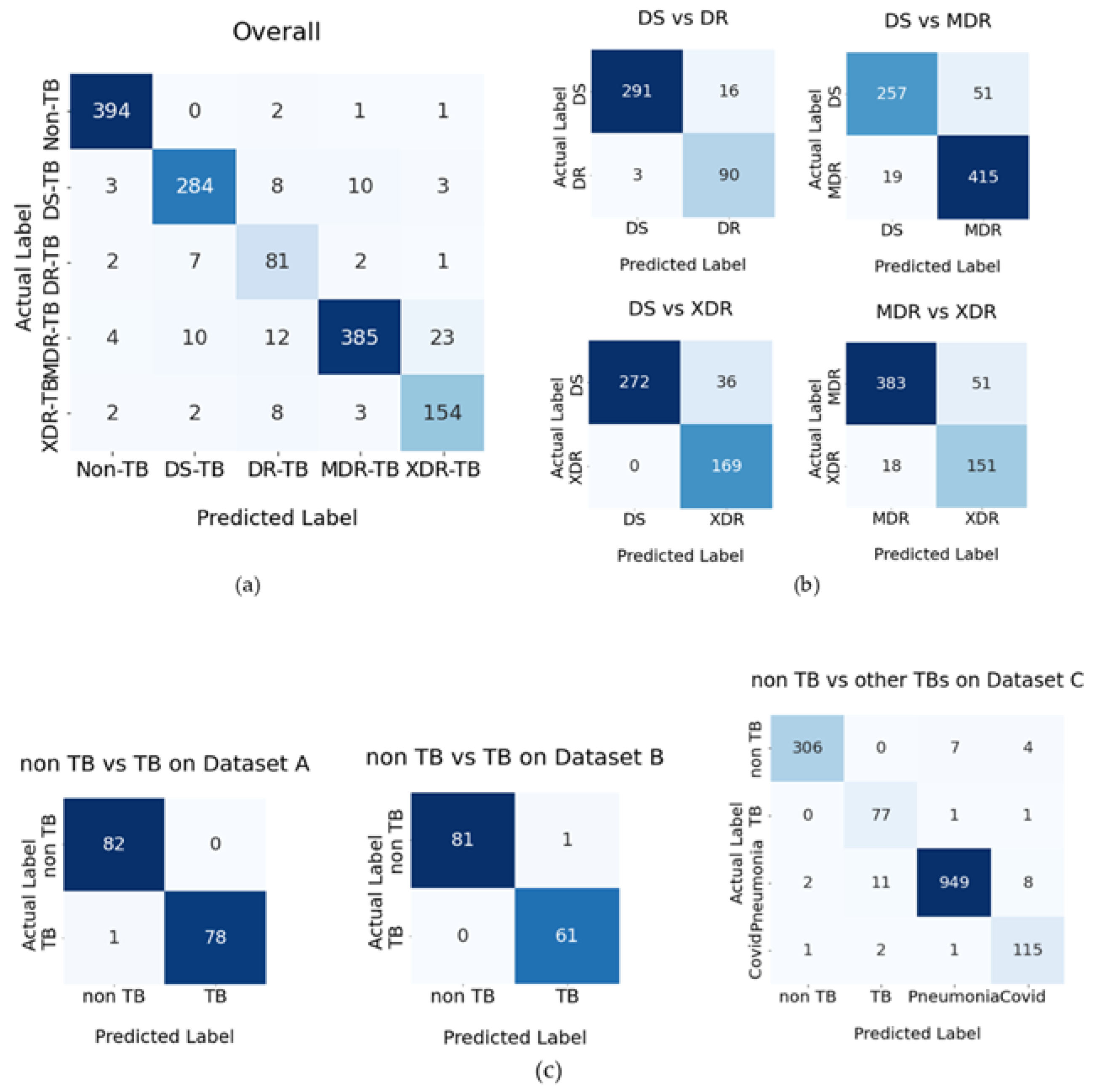
2. Results
2.1. A Test for the Effectiveness of the Proposed Methods
2.1.1. Revealing the Most Effective Proposed Methods
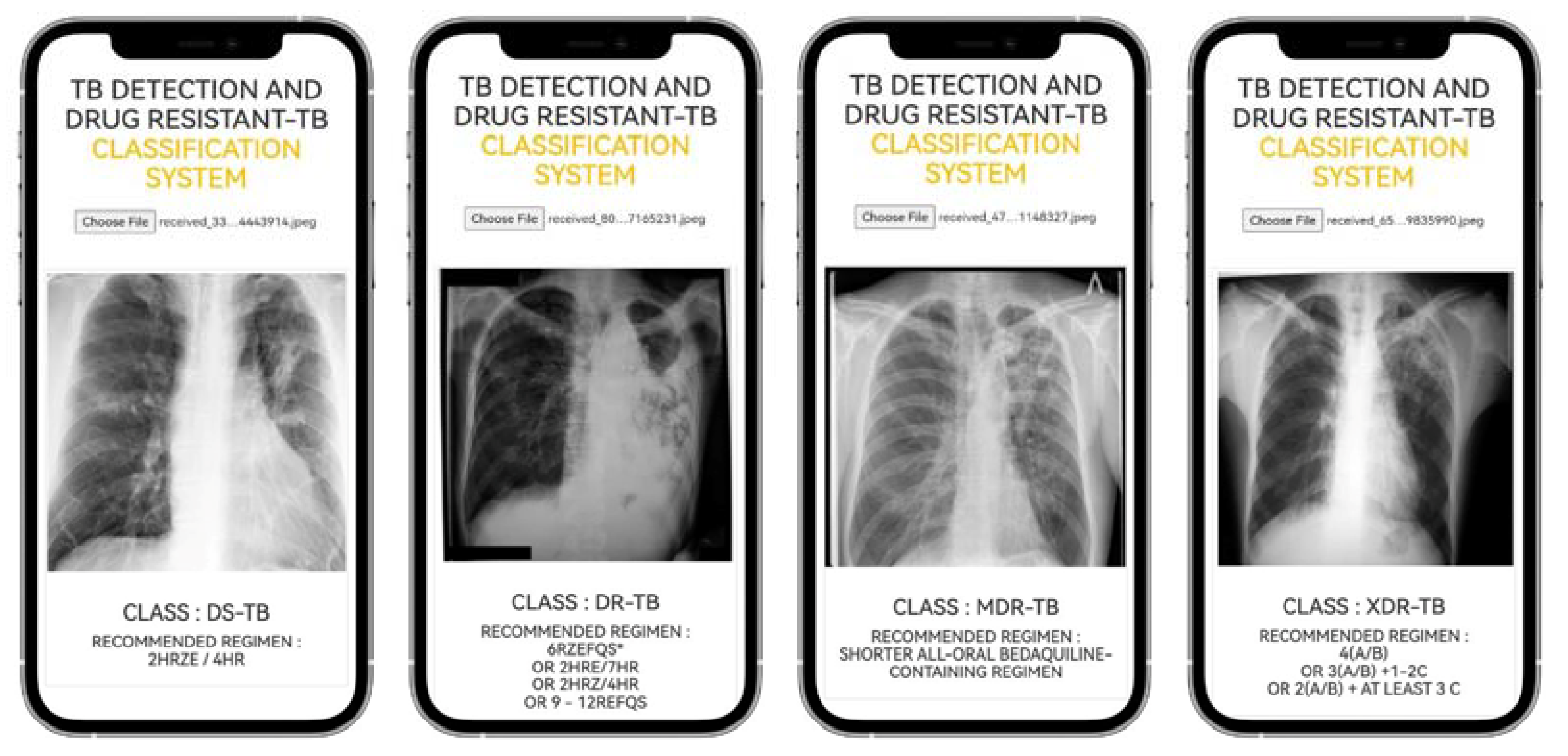
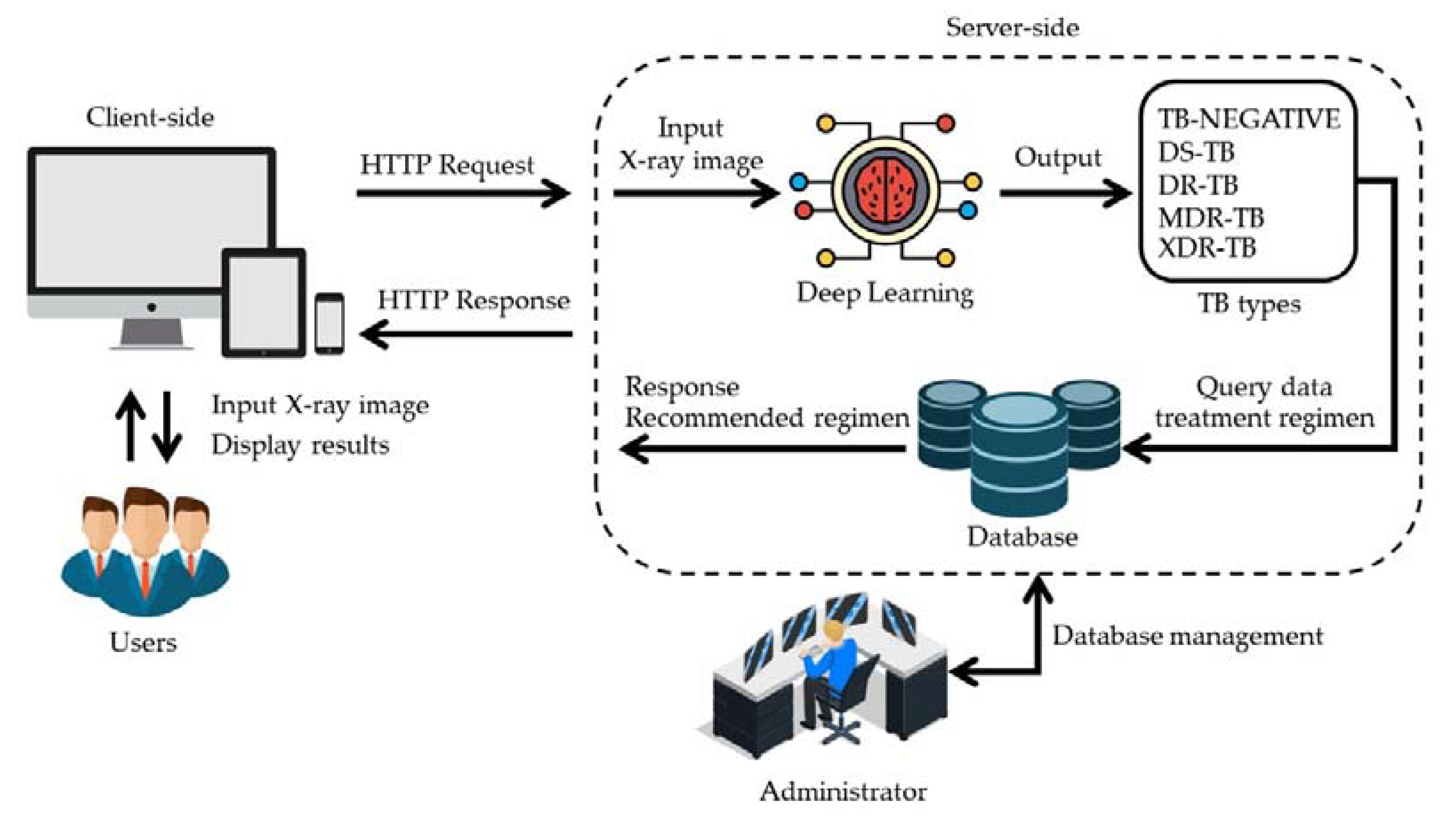
2.1.2. The Web Application Result
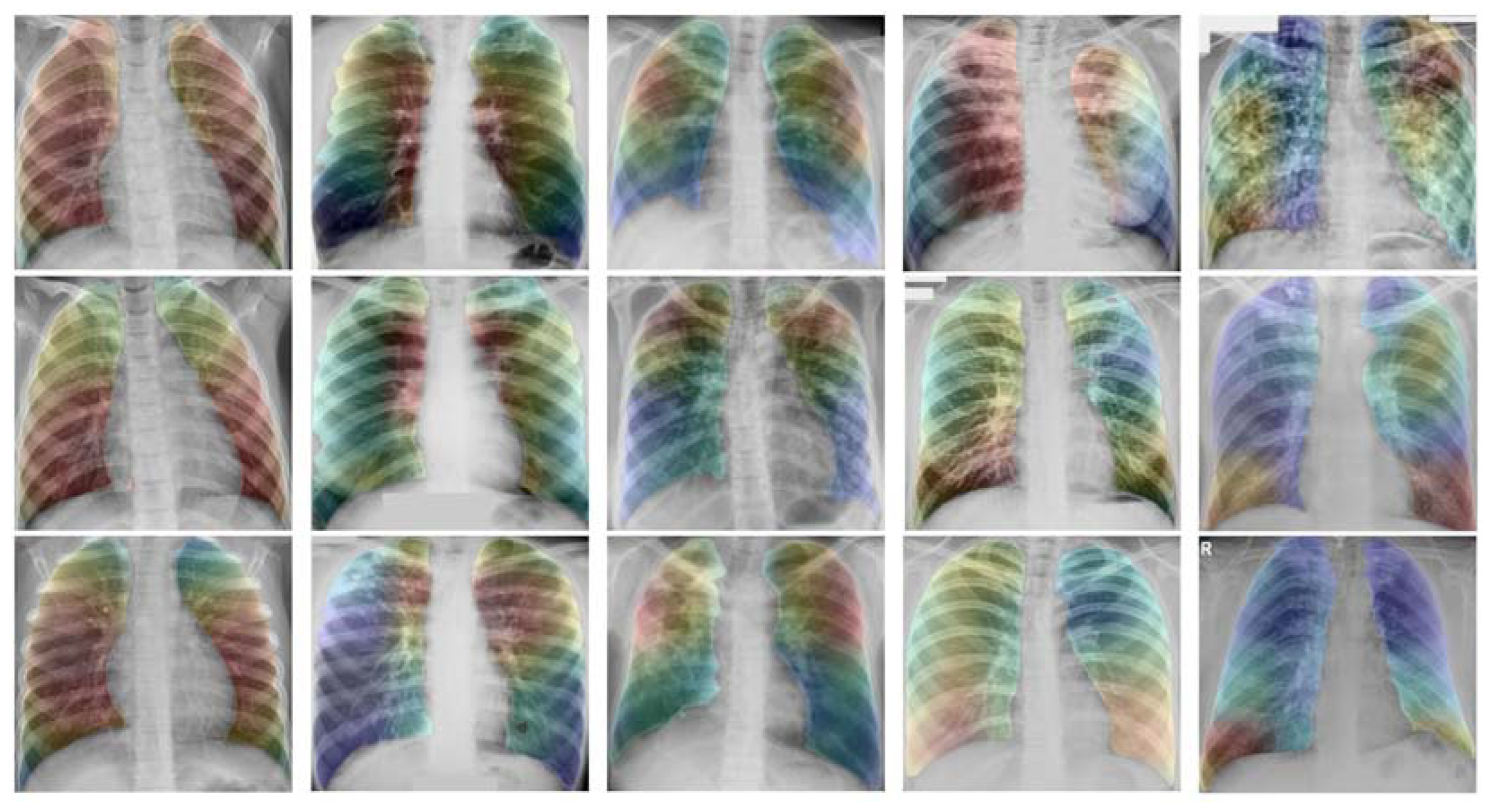
3. Discussion
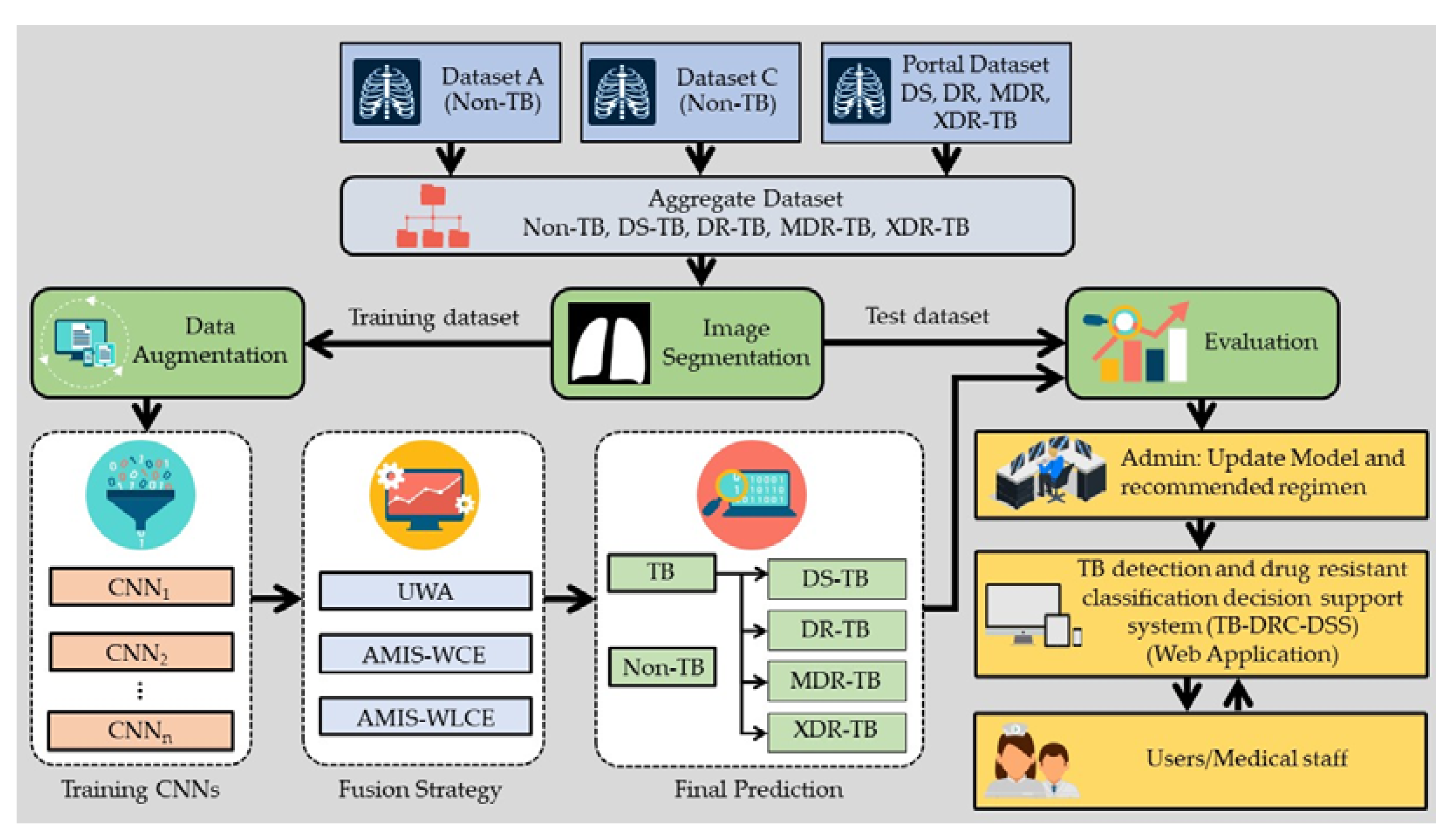
4. Materials and Methods
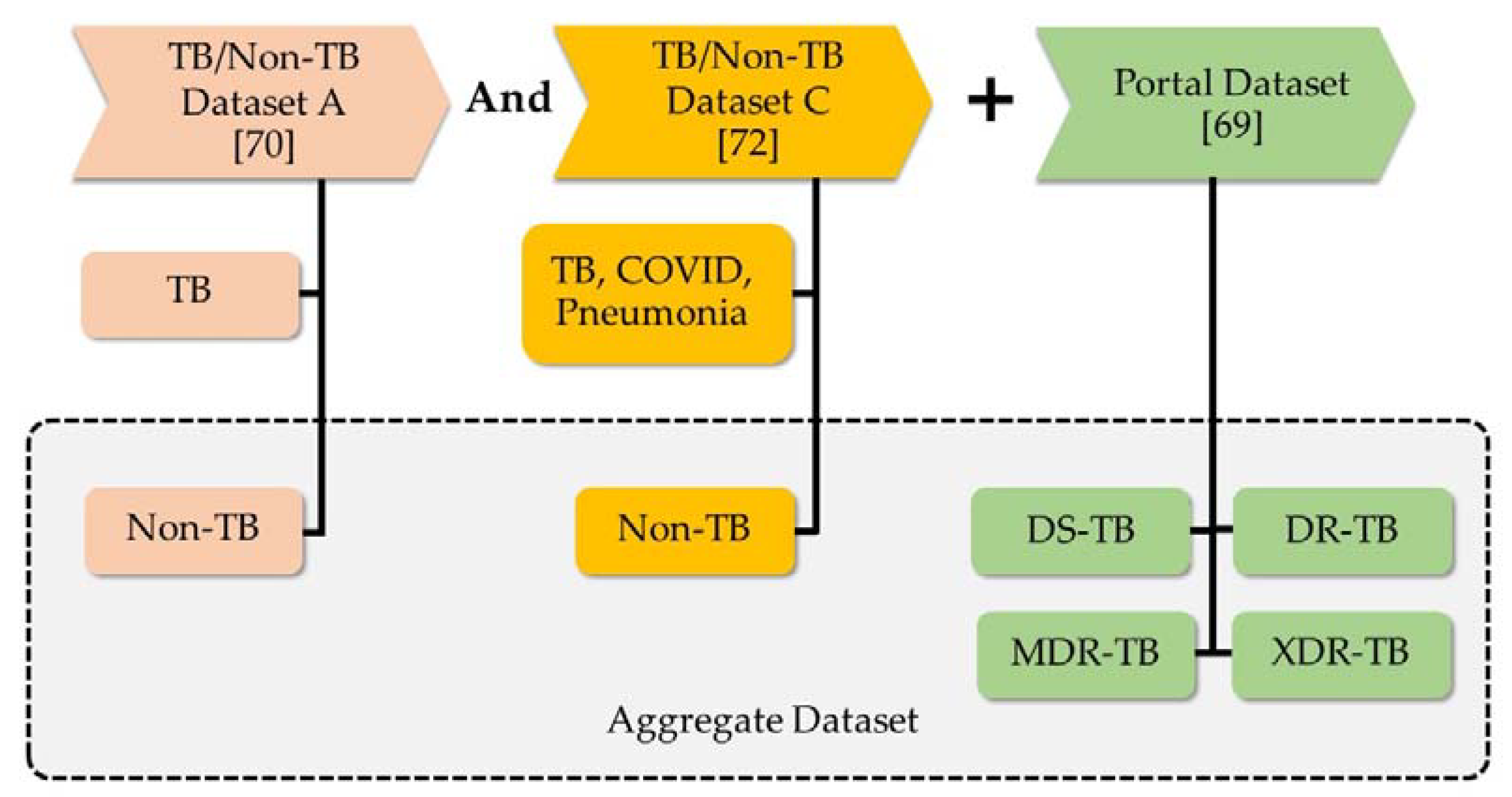
4.1. Revealed Dataset and Compared Methods
4.2. The Development of Effective Methods
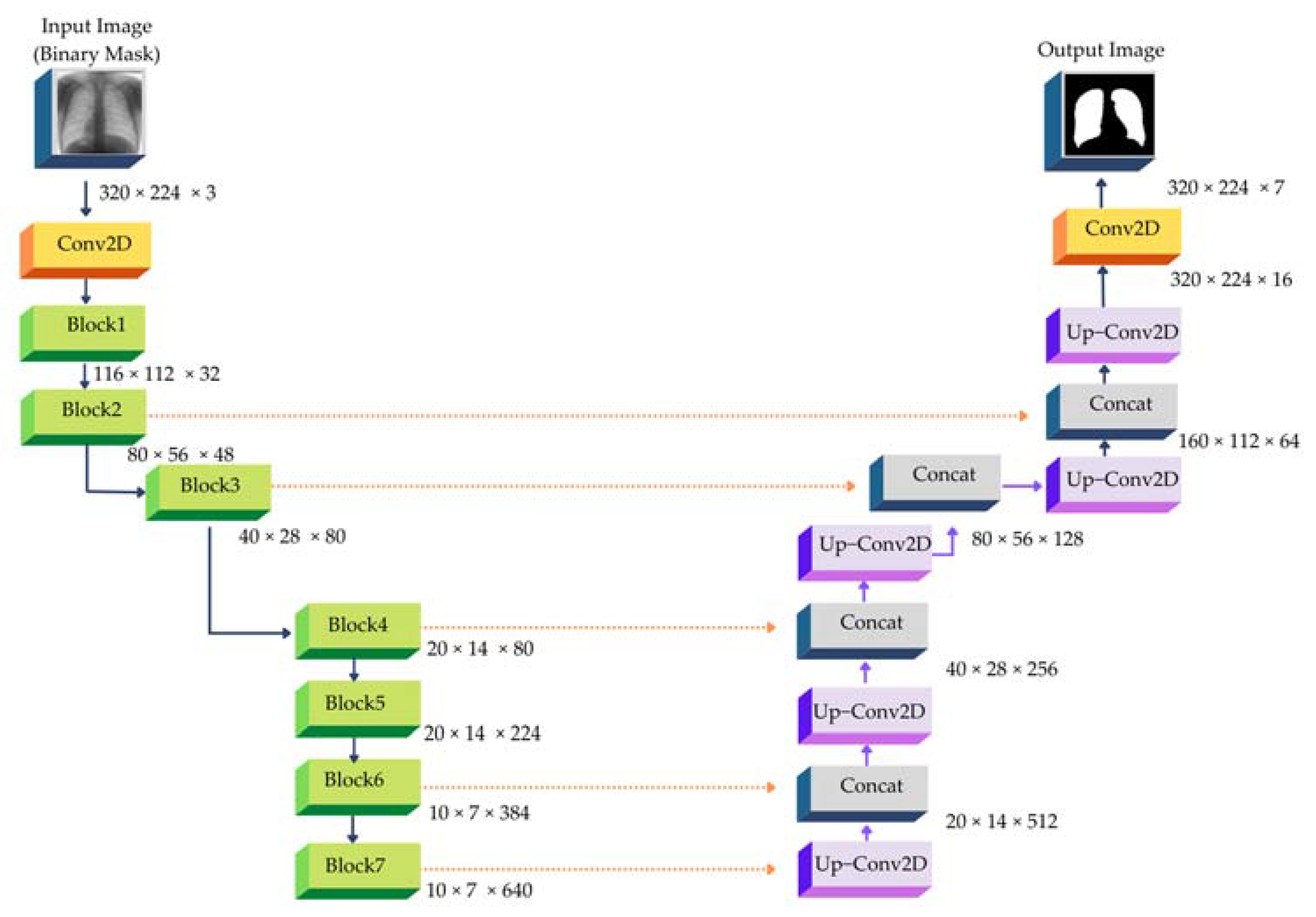
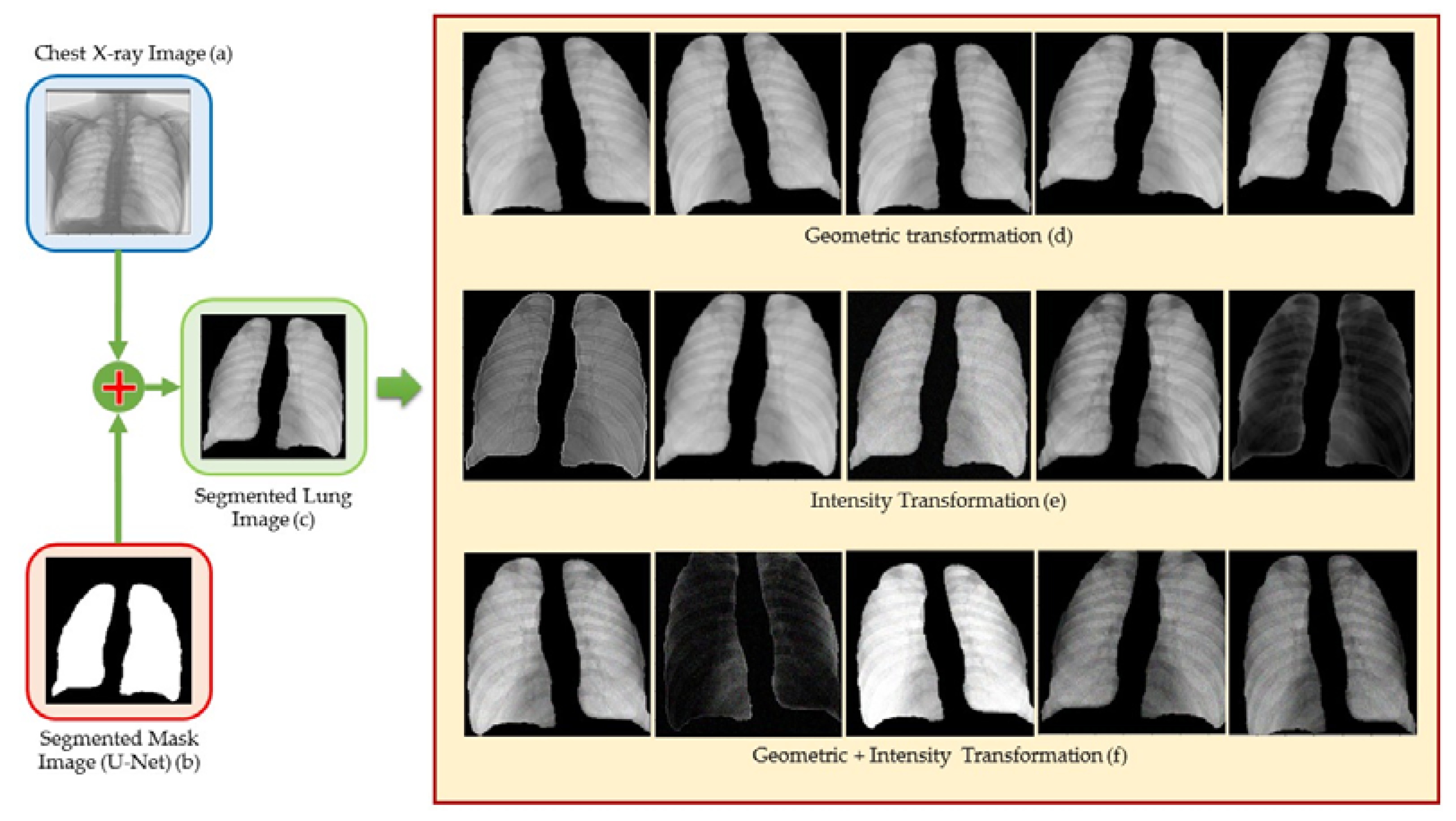
4.3. Image Segmentation
4.4. Data Augmentation
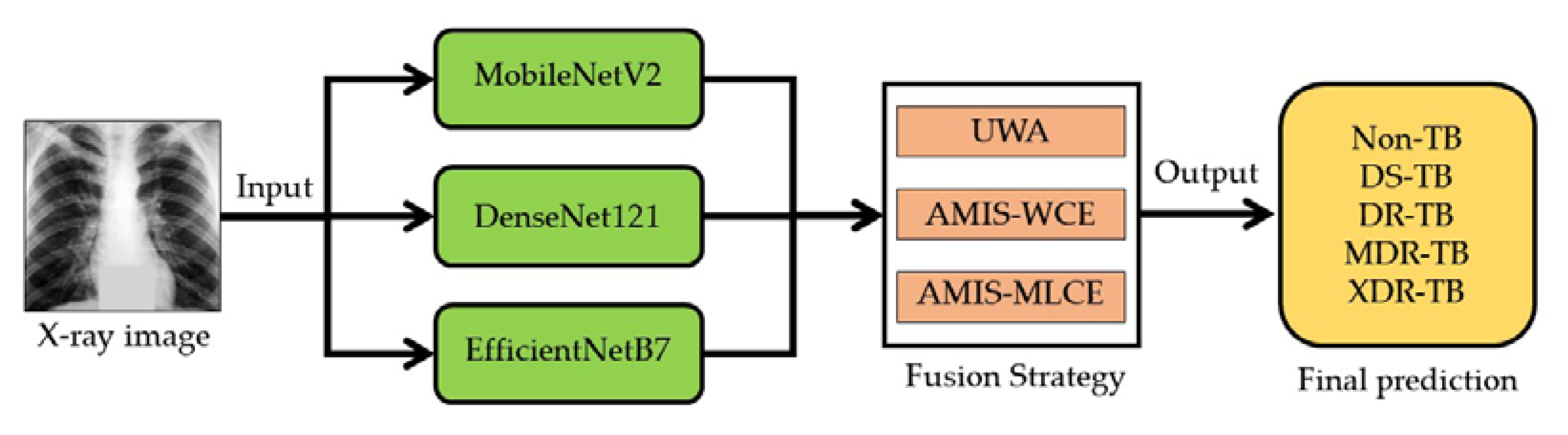
4.5. CNN Architectures
4.6. Decision Fusion Strategy
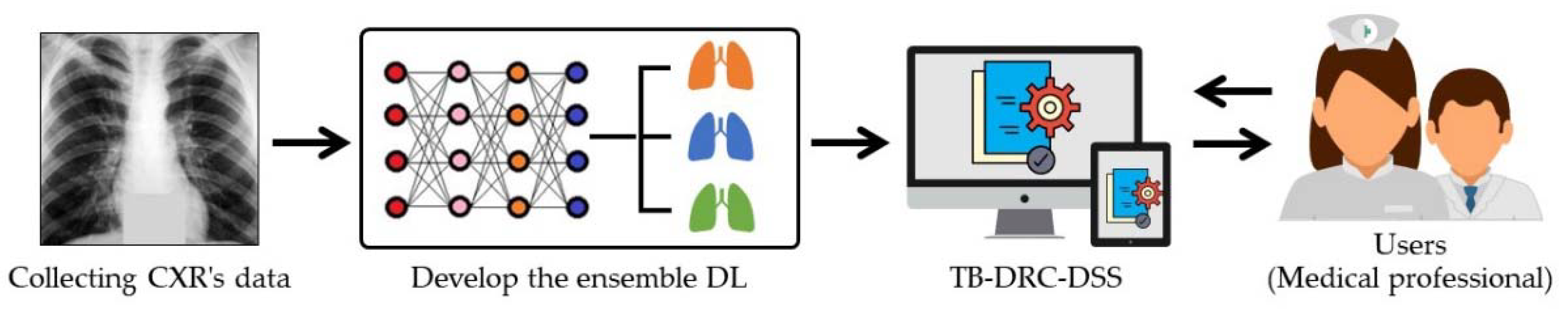
4.7. TB-DRC-DSS Design
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Fekadu, G.; Tolossa, T.; Turi, E.; Bekele, F.; Fetensa, G. Pretomanid Development and Its Clinical Roles in Treating Tuberculosis. J. Glob. Antimicrob. Resist. 2022, 31, 175–184. [Google Scholar] [CrossRef]
- Faddoul, D. Childhood Tuberculosis: An Overview. Adv. Pediatr. 2015, 62, 59–90. [Google Scholar] [CrossRef]
- Farrar, J.; Hotez, P.J.; Junghanss, T.; Kang, G.; Lalloo, D.; White, N.J. Manson’s Tropical Diseases, 23rd ed.; Saunders Ltd.: London, UK, 2013. [Google Scholar]
- Sellami, M.; Fazaa, A.; Cheikh, M.; Miladi, S.; Ouenniche, K.; Ennaifer, R.; ben Abdelghani, K.; Laatar, A. Screening for Latent Tuberculosis Infection Prior to Biologic Therapy in Patients with Chronic Immune-Mediated Inflammatory Diseases (IMID): Interferon-Gamma Release Assay (IGRA) versus Tuberculin Skin Test (TST). Egypt. Rheumatol. 2019, 41, 225–230. [Google Scholar] [CrossRef]
- Soares, T.R.; de Oliveira, R.D.; Liu, Y.E.; da Silva Santos, A.; dos Santos, P.C.P.; Monte, L.R.S.; de Oliveira, L.M.; Park, C.M.; Hwang, E.J.; Andrews, J.R.; et al. Evaluation of Chest X-Ray with Automated Interpretation Algorithms for Mass Tuberculosis Screening in Prisons: A Cross-Sectional Study. Lancet Reg. Health—Am. 2023, 17, 100388. [Google Scholar] [CrossRef]
- Nathavitharana, R.R.; Garcia-Basteiro, A.L.; Ruhwald, M.; Cobelens, F.; Theron, G. Reimagining the Status Quo: How Close Are We to Rapid Sputum-Free Tuberculosis Diagnostics for All? EBioMedicine 2022, 78, 103939. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Usman, M.; Ahmed, Z. An Efficient Deep Learning-Based Framework for Tuberculosis Detection Using Chest X-Ray Images. Tuberculosis 2022, 136, 102234. [Google Scholar] [CrossRef]
- Karki, M.; Kantipudi, K.; Yang, F.; Yu, H.; Wang, Y.X.J.; Yaniv, Z.; Jaeger, S. Generalization Challenges in Drug-Resistant Tuberculosis Detection from Chest X-Rays. Diagnostics 2022, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Tulo, S.K.; Ramu, P.; Swaminathan, R. Evaluation of Diagnostic Value of Mediastinum for Differentiation of Drug Sensitive, Multi and Extensively Drug Resistant Tuberculosis Using Chest X-Rays. IRBM 2022, 43, 658–669. [Google Scholar] [CrossRef]
- Ureta, J.; Shrestha, A. Identifying Drug-Resistant Tuberculosis from Chest X-Ray Images Using a Simple Convolutional Neural Network. J. Phys. Conf. Ser. 2021, 2071, 012001. [Google Scholar] [CrossRef]
- Tulo, S.K.; Ramu, P.; Swaminathan, R. An Automated Approach to Differentiate Drug Resistant Tuberculosis in Chest X-Ray Images Using Projection Profiling and Mediastinal Features. Public Health Inform. Proc. MIE 2021, 2021, 512–513. [Google Scholar]
- Jaeger, S.; Juarez-Espinosa, O.H.; Candemir, S.; Poostchi, M.; Yang, F.; Kim, L.; Ding, M.; Folio, L.R.; Antani, S.; Gabrielian, A.; et al. Detecting Drug-Resistant Tuberculosis in Chest Radiographs. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, V.; Liauchuk, V.; Kalinovsky, A.; Rosenthal, A.; Gabrielian, A.; Skrahina, A.; Astrauko, A.; Tarasau, A. Utilizing Radiological Images for Predicting Drug Resistance of Lung Tuberculosis. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, S291–S292. [Google Scholar]
- Govindarajan, S.; Swaminathan, R. Analysis of Tuberculosis in Chest Radiographs for Computerized Diagnosis Using Bag of Keypoint Features. J. Med. Syst. 2019, 43, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Liu, X.; Zhang, H.; Zheng, G.; Soomro, N.Q.; Wang, M.; Liu, W. Hybrid Resampling and Multi-Feature Fusion for Automatic Recognition of Cavity Imaging Sign in Lung CT. Future Gener. Comput. Syst. 2019, 99, 558–570. [Google Scholar] [CrossRef]
- Cao, H.; Liu, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Liu, T.; Hung, C.C. A Two-Stage Convolutional Neural Networks for Lung Nodule Detection. IEEE J. Biomed. Health Inform. 2020, 24, 2006–2015. [Google Scholar] [CrossRef]
- Momeny, M.; Neshat, A.A.; Gholizadeh, A.; Jafarnezhad, A.; Rahmanzadeh, E.; Marhamati, M.; Moradi, B.; Ghafoorifar, A.; Zhang, Y.D. Greedy Autoaugment for Classification of Mycobacterium Tuberculosis Image via Generalized Deep CNN Using Mixed Pooling Based on Minimum Square Rough Entropy. Comput. Biol. Med. 2022, 141, 105175. [Google Scholar] [CrossRef]
- Lu, S.Y.; Wang, S.H.; Zhang, X.; Zhang, Y.D. TBNet: A Context-Aware Graph Network for Tuberculosis Diagnosis. Comput. Methods Programs Biomed. 2022, 214, 106587. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Cao, Y.; Sun, X.; Li, B.; Hao, Y. Deep Pre-Trained Networks as a Feature Extractor with XGBoost to Detect Tuberculosis from Chest X-Ray. Comput. Electr. Eng. 2021, 93, 107252. [Google Scholar] [CrossRef]
- Iqbal, A.; Sharif, M. MDA-Net: Multiscale Dual Attention-Based Network for Breast Lesion Segmentation Using Ultrasound Images. J. King Saud Univ. —Comput. Inf. Sci. 2022, 34, 7283–7299. [Google Scholar] [CrossRef]
- Tasci, E.; Uluturk, C.; Ugur, A. A Voting-Based Ensemble Deep Learning Method Focusing on Image Augmentation and Preprocessing Variations for Tuberculosis Detection. Neural. Comput. Appl. 2021, 33, 15541–15555. [Google Scholar] [CrossRef]
- Kukker, A.; Sharma, R. Modified Fuzzy Q Learning Based Classifier for Pneumonia and Tuberculosis. IRBM 2021, 42, 369–377. [Google Scholar] [CrossRef]
- Khatibi, T.; Shahsavari, A.; Farahani, A. Proposing a Novel Multi-Instance Learning Model for Tuberculosis Recognition from Chest X-Ray Images Based on CNNs, Complex Networks and Stacked Ensemble. Phys. Eng. Sci. Med. 2021, 44, 291–311. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z.; Özyurt, F. A Deep Feature Learning Model for Pneumonia Detection Applying a Combination of MRMR Feature Selection and Machine Learning Models. IRBM 2020, 41, 212–222. [Google Scholar] [CrossRef]
- Divya Krishna, K.; Akkala, V.; Bharath, R.; Rajalakshmi, P.; Mohammed, A.M.; Merchant, S.N.; Desai, U.B. Computer Aided Abnormality Detection for Kidney on FPGA Based IoT Enabled Portable Ultrasound Imaging System. IRBM 2016, 37, 189–197. [Google Scholar] [CrossRef]
- Ramaniharan, A.K.; Manoharan, S.C.; Swaminathan, R. Laplace Beltrami Eigen Value Based Classification of Normal and Alzheimer MR Images Using Parametric and Non-Parametric Classifiers. Expert Syst. Appl. 2016, 59, 208–216. [Google Scholar] [CrossRef]
- Caseneuve, G.; Valova, I.; LeBlanc, N.; Thibodeau, M. Chest X-Ray Image Preprocessing for Disease Classification. Procedia Comput. Sci. 2021, 192, 658–665. [Google Scholar] [CrossRef]
- Jun, Z.; Jinglu, H. Image Segmentation Based on 2D Otsu Method with Histogram Analysis. In Proceedings of the Proceedings—International Conference on Computer Science and Software Engineering, CSSE 2008, Wuhan, China, 12–14 December 2008; Volume 6. [Google Scholar]
- Farid, H.; Simoncelli, E.P. Optimally Rotation-Equivariant Directional Derivative Kernels. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics, Berlin, Germany, 26–28 September 2011; Volume 1296. [Google Scholar]
- Scharr, H. Optimal Filters for Extended Optical Flow. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Berlin, Germany, 26–28 September 2011; Volume 3417 LNCS. [Google Scholar]
- Ahamed, K.U.; Islam, M.; Uddin, A.; Akhter, A.; Paul, B.K.; Yousuf, M.A.; Uddin, S.; Quinn, J.M.W.; Moni, M.A. A Deep Learning Approach Using Effective Preprocessing Techniques to Detect COVID-19 from Chest CT-Scan and X-Ray Images. Comput. Biol. Med. 2021, 139, 105014. [Google Scholar] [CrossRef]
- Wang, C.; Peng, J.; Ye, Z. Flattest Histogram Specification with Accurate Brightness Preservation. IET Image Process. 2008, 2, 249–262. [Google Scholar] [CrossRef]
- Bhairannawar, S.S. Efficient Medical Image Enhancement Technique Using Transform HSV Space and Adaptive Histogram Equalization. In Soft Computing Based Medical Image Analysis; Academic Press: New York, NY, USA, 2018. [Google Scholar]
- Noguera, J.M.; Jiménez, J.R.; Ogáyar, C.J.; Segura, R.J. Volume Rendering Strategies on Mobile Devices. In Proceedings of the GRAPP 2012 IVAPP 2012—Proceedings of the International Conference on Computer Graphics Theory and Applications and International Conference on Information Visualization Theory and Applications, Rome, Italy, 24–26 February 2012. [Google Scholar]
- Levoy, M. Display of Surfaces from Volume Data. IEEE Comput. Graph. Appl. 1988, 8, 29–37. [Google Scholar] [CrossRef]
- Virag, I.; Stoicu-Tivadar, L. A Survey of Web Based Medical Imaging Applications. Acta Electroteh. 2015, 56, 365–368. [Google Scholar]
- Congote, J.E.; Novo-Blanco, E.; Kabongo, L.; Ginsburg, D.; Gerhard, S.; Pienaar, R.; Ruiz, O.E. Real-Time Volume Rendering and Tractography Visualization on the Web. J. WSCG 2012, 20, 81–88. [Google Scholar]
- Mobeen, M.M.; Feng, L. High-Performance Volume Rendering on the Ubiquitous WebGL Platform. In Proceedings of the 2012 IEEE 14th International Conference on High Performance Computing and Communication & 2012 IEEE 9th International Conference on Embedded Software and Systems, Liverpool, UK, 25–27 June 2012. [Google Scholar]
- Mahmoudi, S.E.; Akhondi-Asl, A.; Rahmani, R.; Faghih-Roohi, S.; Taimouri, V.; Sabouri, A.; Soltanian-Zadeh, H. Web-Based Interactive 2D/3D Medical Image Processing and Visualization Software. Comput. Methods Programs Biomed. 2010, 98, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Marion, C.; Jomier, J. Real-Time Collaborative Scientific WebGL Visualization with WebSocket. In Proceedings of the Proceedings, Web3D 2012—17th International Conference on 3D Web Technology, Los Angeles, CA, USA, 4–5 August 2012. [Google Scholar]
- Rego, N.; Koes, D. 3Dmol.Js: Molecular Visualization with WebGL. Bioinformatics 2015, 31, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, N.; Iwaniec, M.; Lobur, M. Composite Materials Microlevel Structure Models Visualization Distributed Subsystem Based on WebGL. In Proceedings of the 2016 XII International Conference on Perspective Technologies and Methods in MEMS Design (MEMSTECH), Lviv, Ukraine, 20–24 April 2016. [Google Scholar]
- Sherif, T.; Kassis, N.; Rousseau, M.É.; Adalat, R.; Evans, A.C. Brainbrowser: Distributed, Web-Based Neurological Data Visualization. Front. Neuroinformatics 2015, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Implementing WebGL and HTML5 in Macromolecular Visualization and Modern Computer-Aided Drug Design. Trends Biotechnol. 2017, 35, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Kokelj, Z.; Bohak, C.; Marolt, M. A Web-Based Virtual Reality Environment for Medical Visualization. In Proceedings of the 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 21–25 May 2018. [Google Scholar]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A Dynamic Web Platform for Genome Visualization and Analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef]
- Jiménez, J.; López, A.M.; Cruz, J.; Esteban, F.J.; Navas, J.; Villoslada, P.; Ruiz de Miras, J. A Web Platform for the Interactive Visualization and Analysis of the 3D Fractal Dimension of MRI Data. J. Biomed. Inform. 2014, 51, 176–190. [Google Scholar] [CrossRef]
- Jacinto, H.; Kéchichian, R.; Desvignes, M.; Prost, R.; Valette, S. A Web Interface for 3D Visualization and Interactive Segmentation of Medical Images. In Proceedings of the 17th International Conference on 3D Web Technology, Los Angeles, CA, USA, 4–5 August 2012. [Google Scholar]
- Gonwirat, S.; Surinta, O. DeblurGAN-CNN: Effective Image Denoising and Recognition for Noisy Handwritten Characters. IEEE Access 2022, 10, 90133–90148. [Google Scholar] [CrossRef]
- Gonwirat, S.; Surinta, O. Optimal Weighted Parameters of Ensemble Convolutional Neural Networks Based on a Differential Evolution Algorithm for Enhancing Pornographic Image Classification. Eng. Appl. Sci. Res. 2021, 48, 560–569. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the The 3rd International Conference for Learning Representations, San Diego, CA, USA, 7 May 2015; Bengio, Y., LeCun, Y., Eds.; pp. 1–15. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Wang, F.; Jiang, M.; Qian, C.; Yang, S.; Li, C.; Zhang, H.; Wang, X.; Tang, X. Residual Attention Network for Image Classification. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, ICML 2019, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 1997. [Google Scholar]
- Abdar, M.; Salari, S.; Qahremani, S.; Lam, H.K.; Karray, F.; Hussain, S.; Khosravi, A.; Acharya, U.R.; Makarenkov, V.; Nahavandi, S. UncertaintyFuseNet: Robust Uncertainty-Aware Hierarchical Feature Fusion Model with Ensemble Monte Carlo Dropout for COVID-19 Detection. Inf. Fusion. 2023, 90, 364–381. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Wang, S.; Wang, J.; Liu, J.; Jin, Q.; Sun, L. Multiscale Attention Guided Network for COVID-19 Diagnosis Using Chest X-Ray Images. IEEE J. Biomed. Health Inform. 2021, 25, 1336–1346. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A Deep Neural Network for Detection and Diagnosis of COVID-19 from Chest x-Ray Images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.I.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Munich, Germany, 5–9 October 2015; Volume 9351. [Google Scholar]
- Liu, W.; Luo, J.; Yang, Y.; Wang, W.; Deng, J.; Yu, L. Automatic Lung Segmentation in Chest X-Ray Images Using Improved U-Net. Sci. Rep. 2022, 12, 8649. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans Pattern. Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Jain, A.; Kumar, A.; Susan, S. Evaluating Deep Neural Network Ensembles by Majority Voting Cum Meta-Learning Scheme. arXiv 2022, arXiv:2105.03819. [Google Scholar]
- Prasitpuriprecha, C.; Pitakaso, R.; Gonwirat, S.; Enkvetchakul, P.; Preeprem, T.; Jantama, S.S.; Kaewta, C.; Weerayuth, N.; Srichok, T.; Khonjun, S.; et al. Embedded AMIS-Deep Learning with Dialog-Based Object Query System for Multi-Class Tuberculosis Drug Response Classification. Diagnostics 2022, 12, 2980. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; Volume 2017-October. [Google Scholar]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Saunders: Philadelphia, PA, USA, 2014. [Google Scholar]
- Yang, F.; Yu, H.; Kantipudi, K.; Karki, M.; Kassim, Y.M.; Rosenthal, A.; Hurt, D.E.; Yaniv, Z.; Jaeger, S. Differentiating between Drug-Sensitive and Drug-Resistant Tuberculosis with Machine Learning for Clinical and Radiological Features. Quant. Imaging. Med. Surg. 2022, 12, 67587–67687. [Google Scholar] [CrossRef]
- Shi, M.; Gao, J.; Zhang, M.Q. Web3DMol: Interactive Protein Structure Visualization Based on WebGL. Nucleic. Acids Res. 2017, 45, W523–W527. [Google Scholar] [CrossRef][Green Version]
- Rosenthal, A.; Gabrielian, A.; Engle, E.; Hurt, D.E.; Alexandru, S.; Crudu, V.; Sergueev, E.; Kirichenko, V.; Lapitskii, V.; Snezhko, E.; et al. The TB Portals: An Open-Access, Web-Based Platform for Global Drug-Resistant- Tuberculosis Data Sharing and Analysis. J. Clin. Microbiol. 2017, 55, 3267–3282. [Google Scholar] [CrossRef]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two Public Chest X-Ray Datasets for Computer-Aided Screening of Pulmonary Diseases. Quant. Imaging. Med. Surg. 2014, 4, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Belarus Public Health Belarus Tuberculosis Portal. Available online: http://tuberculosis.by/ (accessed on 9 July 2022).
- Chest X-ray (Covid-19 & Pneumonia) | Kaggle. Available online: https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia (accessed on 14 December 2022).
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern. Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Berlin, Germany, 8 September 2018; Volume 11211 LNCS. [Google Scholar]
- Zhang, J.; Xie, Y.; Xia, Y.; Shen, C. Attention Residual Learning for Skin Lesion Classification. IEEE Trans Med. Imaging. 2019, 38, 2092–2103. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Liu, Y.; Wang, X.; Wang, L.; Liu, D. A Modified Leaky ReLU Scheme (MLRS) for Topology Optimization with Multiple Materials. Appl. Math. Comput. 2019, 352, 188–204. [Google Scholar] [CrossRef]
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.; Chen, J.; Liu, X.; Pietikäinen, M. Deep Learning for Generic Object Detection: A Survey. Int. J. Comput. Vis. 2020, 128, 261–318. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. In IEEE Transactions on Pattern Analysis and Machine Intelligence; IEEE: Toulouse, France, 2022; Volume 44. [Google Scholar] [CrossRef]
- Algan, G.; Ulusoy, I. Image Classification with Deep Learning in the Presence of Noisy Labels: A Survey. Knowl. Based Syst. 2021, 215, 106771. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR, Honolulu, HI, USA, 26 July 2017. [Google Scholar]
- Howard, E. Gardner Intelligence Reframed: Multiple Intelligences for the 21st Century; Basic Books: New York, NY, USA, 2000. [Google Scholar]
- Pitakaso, R.; Nanthasamroeng, N.; Srichok, T.; Khonjun, S.; Weerayuth, N.; Kotmongkol, T.; Pornprasert, P.; Pranet, K. A Novel Artificial Multiple Intelligence System (AMIS) for Agricultural Product Transborder Logistics Network Design in the Greater Mekong Subregion (GMS). Computation 2022, 10, 126. [Google Scholar] [CrossRef]
- Xia, Y.; Chen, K.; Yang, Y. Multi-Label Classification with Weighted Classifier Selection and Stacked Ensemble. Inf. Sci. 2021, 557, 421–442. [Google Scholar] [CrossRef]
| Methods | 5-cv | Test Set | ||||
|---|---|---|---|---|---|---|
| AUC | F-Measure | Accuracy | AUC | F-Measure | Accuracy | |
| N-1 | 69.2 ± 0.29 | 61.9 ± 0.17 | 65.3 ± 0.15 | 68.5 | 61.3 | 64.5 |
| N-2 | 72.8 ± 0.21 | 66.2 ± 0.27 | 68.7 ± 0.07 | 72.4 | 65.1 | 67.9 |
| N-3 | 72.9 ± 0.07 | 67.8 ± 0.25 | 70.2 ± 0.18 | 72.5 | 66.3 | 69.4 |
| N-4 | 76.1 ± 0.17 | 67.4 ± 0.14 | 72.4 ± 0.13 | 75.4 | 68.9 | 71.8 |
| N-5 | 77.6 ± 0.19 | 72.8 ± 0.09 | 73.9 ± 0.21 | 76.3 | 71.7 | 73.6 |
| N-6 | 79.8 ± 0.25 | 74.2 ± 0.27 | 76.7 ± 0.09 | 79.1 | 73.5 | 75.4 |
| N-7 | 88.2 ± 0.29 | 82.6 ± 0.25 | 85.3 ± 0.26 | 87.8 | 81.4 | 84.1 |
| N-8 | 90.3 ± 0.04 | 84.8 ± 0.18 | 87.9 ± 0.17 | 89.2 | 85.5 | 87.3 |
| N-9 | 91.9 ± 0.07 | 87.5 ± 0.08 | 89.8 ± 0.05 | 91.6 | 85.8 | 89.3 |
| N-10 | 91.5 ± 0.24 | 87.8 ± 0.09 | 90.1 ± 0.19 | 90.7 | 86.1 | 88.5 |
| N-11 | 94.8 ± 0.18 | 91.2 ± 0.28 | 91.5 ± 0.24 | 94.1 | 90.5 | 91.1 |
| N-12 | 95.3 ± 0.07 | 91.7 ± 0.10 | 93.9 ± 0.08 | 94.7 | 90.9 | 92.6 |
| KPI | Image Segmentation | Data Augmentation | Decision Fusion Strategy | ||||
|---|---|---|---|---|---|---|---|
| No Image Segmentation | Use Image Segmentation | No Data Augmentation | Use Data Augmentation | UWA | AMIS-WCE | AMIS-WLCE | |
| AUC | 74.0 | 91.4 | 80.3 | 85.1 | 80.6 | 83.0 | 84.5 |
| F-measure | 72.6 | 90.2 | 78.9 | 83.9 | 79.2 | 81.7 | 83.4 |
| Accuracy | 70.4 | 88.8 | 77.1 | 82.2 | 77.2 | 80.0 | 81.7 |
| Methods | Classes | Features | Region in CXR | Accuracy | ||
|---|---|---|---|---|---|---|
| AUC | F-Measure | Accuracy | ||||
| Ureta and Shrestha [11] | DS vs. DR | CNN | Whole | 67.0 | - | - |
| Tulo et al. [12] | DS vs. DR | Shape | Mediastinum + Lung | 93.6 | - | |
| Kovalev et al. [14] | DS vs. DR | Texture and Shape | Lung | - | - | 61.7 |
| Karki et al. [9] | DS vs. DR | CNN | Lung excluded | 79.0 | - | 72.0 |
| Proposed method | DS vs. DR | Ensemble CNN | Whole | 97.2 | 94.9 | 95.2 |
| Jaeger et al. [13] | DS vs. MDR | Texture, Shape and Edge | Lung | 66 | 61 | 62 |
| Tulo et al. [10] | DS vs. MDR | Shape | Mediastinum + Lungs | 87.3 | 82.4 | 82.5 |
| Proposed method | DS vs.MDR | Ensemble CNN | Whole | 92.5 | 90.1 | 90.6 |
| Tulo et al. [10] | DS vs. XDR | Shape | Mediastinum + Lungs | 93.5 | 87.0 | 87.0 |
| Proposed method | DS vs. XDR | Ensemble CNN | Whole | 95.7 | 92.1 | 92.5 |
| Tulo et al. [10] | MDR vs. XDR | Shape | Mediastinum + Lungs | 86.6 | 81.0 | 81.0 |
| Proposed method | MDR vs. XDR | Ensemble CNN | Whole | 90.8 | 86.6 | 88.6 |
| Pairwise Comparison | AUC | F-Measure | Accuracy |
|---|---|---|---|
| DS vs. DR | 34.1 | 3.3 | 43.3 |
| DS vs. MDR | 23.1 | 30.2 | 28.1 |
| DS vs. XDR | 2.4 | 6.4 | 6.2 |
| MDR vs. XDR | 4.8 | 10.5 | 9.4 |
| Average | 16.1 | 12.6 | 21.7 |
| Classes | Features | Region in CXR | Accuracy | |
|---|---|---|---|---|
| Simonyan and Zisserman [53] | TB vs. non-TB | VGG16 | Whole | 83.8 |
| Wang et al. [54] | TB vs. non-TB | ResNet50 | Whole | 82.2 |
| Tan and Le [55] | TB vs. non-TB | EfficientNet | Whole | 83.3 |
| Sandler et al. [56] | TB vs. non-TB | MobileNetv2 | Whole | 80.2 |
| Abdar et al. [57] | TB vs. non-TB | FuseNet | Whole | 91.6 |
| Li et al. [58] | TB vs. non-TB | MAG-SD | Whole | 96.1 |
| Khan et al. [59] | TB vs. non-TB | CoroNet | Whole | 93.6 |
| Ahmed et al. [8] | TB vs. non-TB | TBXNet | Whole | 98.9 |
| Proposed method | TB vs. non-TB | Ensemble CNN | Whole | 99.4 |
| Classes | Features | Region in CXR | Accuracy | |
|---|---|---|---|---|
| Simonyan and Zisserman [53] | TB vs. non-TB | VGG16 | Whole | 73.3 |
| Wang et al. [54] | TB vs. non-TB | ResNet50 | Whole | 65.8 |
| Tan and Le [55] | TB vs. non-TB | EfficientNet | Whole | 87.5 |
| Sandler et al. [56] | TB vs. non-TB | MobileNetv2 | Whole | 58.3 |
| Abdar et al. [57] | TB vs. non-TB | FuseNet | Whole | 94.6 |
| Li et al. [58] | TB vs. non-TB | MAG-SD | Whole | 96.1 |
| Khan et al. [59] | TB vs. non-TB | CoroNet | Whole | 93.7 |
| Ahmed et al. [8] | TB vs. non-TB | TBXNet | Whole | 99.2 |
| Proposed method | TB vs. non-TB | Ensemble CNN | Whole | 99.3 |
| Classes | Features | Region in CXR | Accuracy | |
|---|---|---|---|---|
| Simonyan and Zisserman [53] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | VGG16 | Whole | 64.4 |
| Wang et al. [54] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | ResNet50 | Whole | 62.7 |
| Tan and Le [55] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | EfficientNet | Whole | 45.4 |
| Sandler et al. [56] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | MobileNetv2 | Whole | 81.7 |
| Abdar et al. [57] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | FuseNet | Whole | 90.6 |
| Li et al. [58] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | MAG-SD | Whole | 94.1 |
| Khan et al. [59] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | CoroNet | Whole | 89.6 |
| Ahmed et al. [8] | TB vs. non-TB vs. COVID-19 vs. Pneumonia | TBXNet | Whole | 95.1 |
| Proposed method | TB vs. non-TB vs. COVID-19 vs. Pneumonia | Ensemble CNN | Whole | 97.4 |
| Number of Input Images | Number of Correct Classifications | %Correct Classification | Number of Wrong Classification | %Wrong Classification | |
|---|---|---|---|---|---|
| Non-TB | 81 | 78 | 96.3 | 3 | 3.7 |
| DS-TB | 78 | 71 | 91.0 | 7 | 9.0 |
| DR-TB | 74 | 68 | 91.9 | 6 | 8.1 |
| MDR-TB | 102 | 94 | 92.2 | 8 | 7.8 |
| XDR-TB | 93 | 86 | 92.5 | 7 | 7.5 |
| Total | 428 | 397 | 463.8 | 31 | 36.2 |
| Average | 86 | 79 | 92.8 | 6 | 7.2 |
| Questionnaire to Pictures | Level of Agreement (Strongly Agree Level is 10) |
|---|---|
| TB/DR-TDDS provides quick results. | 9.42 |
| TB/DR-TDDS provides the correct results. | 9.45 |
| TB-DRC-DSS is beneficial to your work. | 9.32 |
| You will choose the TB/DR-TDDS to assist in your diagnosis. | 9.39 |
| What rating will you give the TB/DR-TDDS about your reliability? | 9.48 |
| How would you rate the TB/DR-TDDS in relation to your preferences? | 9.52 |
| Dataset A [71] | Dataset B [72] | Dataset C [73] | Portal Dataset [70] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-TB | TB | Non-TB | TB | Non-TB | TB | Pneumonia | COVID | DS-TB | DR-TB | MDR-TB | XDR-TB | |
| Train dataset | 324 | 315 | 324 | 245 | 1266 | 315 | 3879 | 460 | 1299 | 375 | 1653 | 688 |
| Test dataset | 82 | 79 | 82 | 61 | 317 | 79 | 970 | 116 | 308 | 93 | 434 | 169 |
| Total dataset | 406 | 394 | 406 | 306 | 1583 | 394 | 4,849 | 576 | 1607 | 468 | 2087 | 857 |
| Aggregate Dataset | |||||
|---|---|---|---|---|---|
| Non-TB | DS-TB | DR-TB | MDR-TB | XDR-TB | |
| Train dataset | 1591 | 1299 | 375 | 1653 | 688 |
| Test dataset | 398 | 308 | 93 | 434 | 169 |
| Total dataset | 1989 | 1607 | 468 | 2087 | 857 |
| Studies | Classes | Features | Region in CXR | AUC | Accuracy F-Measure | Accuracy |
|---|---|---|---|---|---|---|
| Ureta and Shrestha [11] | DS vs. DR | CNN | Whole | 67.0 | - | - |
| Tulo et al. [12] | DS vs. DR | Shape | Mediastinum + Lung | 93.6 | ||
| Jaeger et al. [13] | DS vs. MDR | Texture, Shape, and Edge | Lung | 66 | 61 | 62 |
| Kovalev et al. [14] | DS vs. DR | Texture and Shape | Lung | - | - | 61.7 |
| Tulo et al. [10] | DS vs. MDR | Shape | Mediastinum + Lungs | 87.3 | 82.4 | 82.5 |
| Tulo et al. [10] | MDR vs. XDR | Shape | Mediastinum + Lungs | 86.6 | 81.0 | 81.0 |
| Tulo et al. [10] | DS vs. XDR | Shape | Mediastinum + Lungs | 93.5 | 87.0 | 87.0 |
| Karki et al. [9] | DS vs. DR | CNN | Lung excluded | 79.0 | - | 72.0 |
| Simonyan and Zisserman [53] | TB vs. non-TB | VGG16 | Lungs | - | - | 83.8 (A), 73.3(B), |
| Wang et al. [54] | TB vs. non-TB | ResNet50 | Lungs | - | - | 82.2 (A), 65.8(B), 64.4(C) |
| Tan and Le [55] | TB vs. non-TB | EfficientNet | Lungs | - | - | 83.3 (A), 87.5 (B), 62.7 (C) |
| Sandler et al. [56] | TB vs. non-TB | MobileNetv2 | Lungs | - | - | 80.2 (A), 58.3 (B), 45.4 (C) |
| Abdar et al. [57] | TB vs. non-TB | FuseNet | Lungs | - | - | 91.6 (A), 94.6 (B), 81.7 (C) |
| Li et al. [58] | TB vs. non-TB | MAG-SD | Lungs | - | - | 96.1 (A), 96.1 (B), 90.6 (C) |
| Khan et al. [59] | TB vs. non-TB | CoroNet | Lungs | - | - | 93.6 (A), 93.7 (B), 94.1 (C) |
| Ahmed et al. [8] | TB vs. non-TB | TBXNet | Lungs | - | - | 98.98 (A), 99.2 (B), 95.1 (C) |
| Proposed Method (set A) | TB vs. non-TB | Ensemble CNN | Lungs | - | - | - |
| Proposed Method (set B) | TB vs. non-TB | Ensemble CNN | Lungs | - | - | - |
| Proposed Method (set C) | TB vs. non-TB vs. COVID-19 vs. Pneumonia | Ensemble CNN | Lungs | - | - | - |
| Proposed method | DR vs. DS | Ensemble CNN | Lungs | - | - | - |
| Proposed method | DS vs. MDR | Ensemble CNN | Lungs | - | - | - |
| Proposed method | DS vs. XDR | Ensemble CNN | Lungs | - | - | - |
| Proposed method | Non-TB vs. DR vs. VS vs. MDR vs. XDR | Ensemble CNN | Lungs | - | - | - |
| Variables | Update Method | |
|---|---|---|
| Sum of number of work packages (WP) that have selected IB b from iteration 1 through iteration t | ||
| is objective value of WP e in iteration t. | ||
| (6) | ||
| When | ||
| Current global best WP is updated | ||
| Current IB’s best WP is updated | ||
| Random number is iteratively updated | ||
| Name of IB | IB-Equation | Equation Number |
|---|---|---|
| IB-1 | (7) | |
| IB-2 | (8) | |
| IB-3 | (9) | |
| IB-4 | (10) | |
| IB-5 | (11) | |
| IB-6 | (12) | |
| IB-7 | (13) | |
| IB-8 | (14) |
| Details of W and Y Values | UAM | AMIS-WCE | AMIS-WLCE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CNN (i) | Weighted (Wi) | Classes (j) | Weighted (Wij) | Yij | Prediction Class | Prediction Class | Prediction Class | |||
| M-1 | 0.5 | C-1 | 0.10 | 0.6 | C-1 = 0.90 C-2 = 0.37 C-3 = 0.33 | C-2 | C-1 = 0.37 C-2 = 0.34 C-3 = 0.28 | C-1 | C-1 = 0.12 C-2 = 0.06 C-3 = 0.13 | C-3 |
| C-2 | 0.20 | 0.3 | ||||||||
| C-3 | 0.20 | 0.1 | ||||||||
| M-2 | 0.2 | C-1 | 0.02 | 0.1 | ||||||
| C-2 | 0.03 | 0.5 | ||||||||
| C-3 | 0.15 | 0.4 | ||||||||
| M-3 | 0.3 | C-1 | 0.03 | 0.2 | ||||||
| C-2 | 0.07 | 0.3 | ||||||||
| C-3 | 0.20 | 0.5 | ||||||||
| #No | No. Seg | Seg. | No. Aug | Aug. | UWA | AMIS-WCE | AMIS-WLCE |
|---|---|---|---|---|---|---|---|
| N-1 | √ | √ | √ | ||||
| N-2 | √ | √ | √ | ||||
| N-3 | √ | √ | √ | ||||
| N-4 | √ | √ | √ | ||||
| N-5 | √ | √ | √ | ||||
| N-6 | √ | √ | √ | ||||
| N-7 | √ | √ | √ | ||||
| N-8 | √ | √ | √ | ||||
| N-9 | √ | √ | √ | ||||
| N-10 | √ | √ | √ | ||||
| N-11 | √ | √ | √ | ||||
| N-12 | √ | √ | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasitpuriprecha, C.; Jantama, S.S.; Preeprem, T.; Pitakaso, R.; Srichok, T.; Khonjun, S.; Weerayuth, N.; Gonwirat, S.; Enkvetchakul, P.; Kaewta, C.; et al. Drug-Resistant Tuberculosis Treatment Recommendation, and Multi-Class Tuberculosis Detection and Classification Using Ensemble Deep Learning-Based System. Pharmaceuticals 2023, 16, 13. https://doi.org/10.3390/ph16010013
Prasitpuriprecha C, Jantama SS, Preeprem T, Pitakaso R, Srichok T, Khonjun S, Weerayuth N, Gonwirat S, Enkvetchakul P, Kaewta C, et al. Drug-Resistant Tuberculosis Treatment Recommendation, and Multi-Class Tuberculosis Detection and Classification Using Ensemble Deep Learning-Based System. Pharmaceuticals. 2023; 16(1):13. https://doi.org/10.3390/ph16010013
Chicago/Turabian StylePrasitpuriprecha, Chutinun, Sirima Suvarnakuta Jantama, Thanawadee Preeprem, Rapeepan Pitakaso, Thanatkij Srichok, Surajet Khonjun, Nantawatana Weerayuth, Sarayut Gonwirat, Prem Enkvetchakul, Chutchai Kaewta, and et al. 2023. "Drug-Resistant Tuberculosis Treatment Recommendation, and Multi-Class Tuberculosis Detection and Classification Using Ensemble Deep Learning-Based System" Pharmaceuticals 16, no. 1: 13. https://doi.org/10.3390/ph16010013
APA StylePrasitpuriprecha, C., Jantama, S. S., Preeprem, T., Pitakaso, R., Srichok, T., Khonjun, S., Weerayuth, N., Gonwirat, S., Enkvetchakul, P., Kaewta, C., & Nanthasamroeng, N. (2023). Drug-Resistant Tuberculosis Treatment Recommendation, and Multi-Class Tuberculosis Detection and Classification Using Ensemble Deep Learning-Based System. Pharmaceuticals, 16(1), 13. https://doi.org/10.3390/ph16010013











