Xylan Prebiotics and the Gut Microbiome Promote Health and Wellbeing: Potential Novel Roles for Pentosan Polysulfate
Abstract
1. Introduction
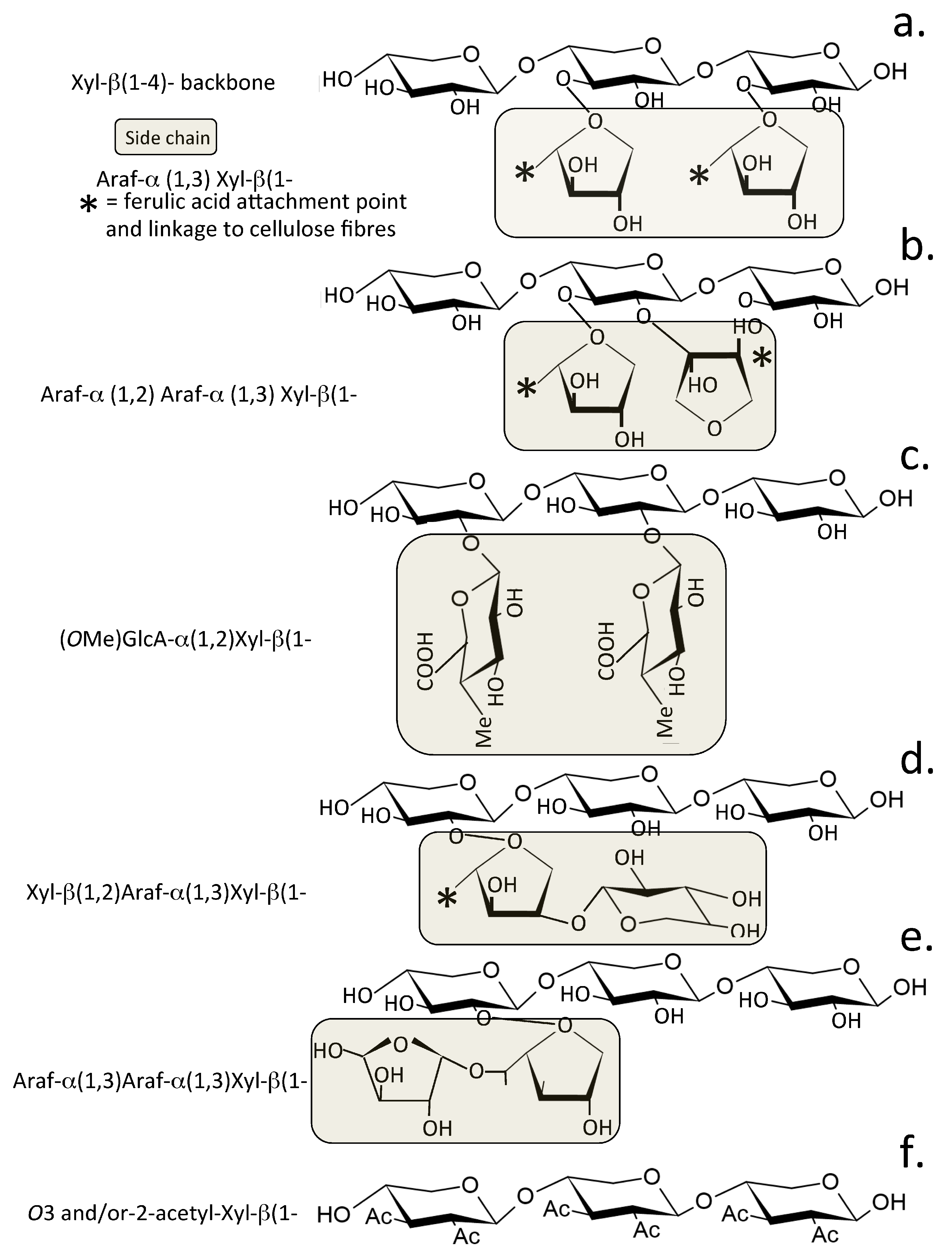
2. Xylans Are Abundant Plant Carbohydrates
2.1. Dietary Xylans
2.2. Pentosan Polysulfate, a Therapeutic Semi-Synthetic Sulfated Xylan
3. Degradation of Xylans in the Gastrointestinal Tract

3.1. The Xylan Regulon and Production of Xylanolytic Enzymes
3.2. Metabolism of GAGs in the Gut Microbiome and the Essential Roles They Play in Gut Homeostasis
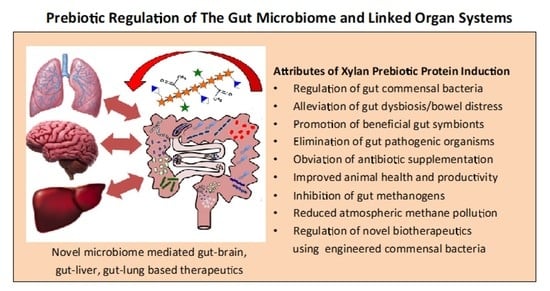
4. Xylans Promote a Healthy Human Microbiome and Linked Organ Systems
5. Manipulation of the Microbiome Increases Farm Animal Productivity
5.1. Use of Recombinant Xylanase as a Food Additive for Monogastric Farm Animals Improves Feedstuff Utilization and Animal Productivity
5.2. Elimination of Antibiotic Supplementation in Animal Foodstuffs
5.3. Manipulation of the Gut Microbiome to Reduce Methane Emissions and Atmospheric Pollution by Ruminant Animals
5.4. The Health Promoting Properties of Xylo-Oligosaccharide Prebiotics in the Treatment of Human Gut Dysbiosis
5.5. The Use of Engineered Commensal Bacteria to Deliver Bioactive Therapeutic Compounds Controlled by a Xylan Protein Induction System
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbio-ta-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, P.; Bienenstock, J.; Kunze, W.A. Vagal pathways for microbiome-brain-gut axis communication. Adv. Exp. Med. Biol. 2014, 817, 115–133. [Google Scholar] [PubMed]
- Fülling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus…. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Ali, M.; Kamal, M.M.; Rahman, M.H.; Siddiqui, M.N.; Haque, M.A.; Saha, K.K.; Rahman, M.A. Functional dairy products as a source of bioactive peptides and probiotics: Current trends and future prospectives. J. Food Sci. Technol. 2022, 59, 1263–1279. [Google Scholar] [CrossRef]
- Singh, S.; Kola, P.; Kaur, D.; Singla, G.; Mishra, V.; Panesar, P.S.; Mallikarjunan, K.; Krishania, M. Therapeutic Potential of Nutraceuticals and Dietary Supplements in the Prevention of Viral Diseases: A Review. Front. Nutr. 2021, 8, 679312. [Google Scholar] [CrossRef]
- Sun, J.; Tian, C.; Diamond, S.; Glass, N.L. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot. Cell 2012, 11, 482–493. [Google Scholar] [CrossRef]
- Tauzin, A.; Laville, E.; Xiao, Y.; Nouaille, S.; Le Bourgeois, P.; Heux, S.; Portais, J.C.; Monsan, P.; Martens, E.C.; Potocki-Veronese, G.; et al. Functional characterization of a gene locus from an uncultured gut Bacteroides conferring xylo-oligosaccharides utilization to Escherichia coli. Mol. Microbiol. 2016, 102, 579–592. [Google Scholar] [CrossRef]
- Zhang, Z.; Raza, M.F.; Zheng, Z.; Zhang, X.; Dong, X.; Zhang, H. Complete genome sequence of Bacillus velezensis ZY-1-1 reveals the genetic basis for its hemicellulosic/cellulosic substrate-inducible xylanase and cellulase activities. 3 Biotech 2018, 8, 465. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, A.; Hundertmark, J.; Guillot, A.; Tacke, F. Molecular and Cellular Mediators of the Gut-Liver Axis in the Progression of Liver Diseases. Front. Med. 2021, 8, 725390. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, B. The Gut-Liver Axis in Health and Disease: The Role of Gut Microbiota-Derived Signals in Liver Injury and Regeneration. Front. Immunol. 2021, 12, 775526. [Google Scholar] [CrossRef] [PubMed]
- Aishwarya, K.G.; Anita-Margret, M. Intermodulation of gut-lung axis microbiome and the implications of biotics to combat COVID-19. J. Biomol. Struct. Dyn. 2021, 26, 1–17. [Google Scholar]
- Gokulan, K.; Joshi, M.; Khare, S.; Bartter, T. Lung microbiome, gut-lung axis and chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2022, 28, 134–138. [Google Scholar] [CrossRef]
- Angoorani, P.; Ejtahed, H.S.; Siadat, S.D.; Sharifi, F.; Larijani, B. Is There Any Link between Cognitive Impairment and Gut Microbiota? A Systematic Review. Gerontology 2022, 9, 1–13. [Google Scholar] [CrossRef]
- Liu, L.; Huh, J.R.; Shah, K. Microbiota and the gut-brain-axis: Implications for new therapeutic design in the CNS. eBioMedicine 2022, 77, 103908. [Google Scholar] [CrossRef]
- Scheller, H.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Hsieh, Y.; Harris, P.J. Xylans of Red and Green Algae: What Is Known about Their Structures and How They Are Synthesised? Polymers 2019, 11, 354. [Google Scholar] [CrossRef]
- Zhang, M.; Chekan, J.R.; Dodd, D.; Hong, P.Y.; Radlinski, L.; Revindran, V.; Nair, S.K.; Mackie, R.I.; Cann, I. Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, E3708–E3717. [Google Scholar] [CrossRef]
- Baker, J.; Duarte, M.E.; Holanda, D.M.; Kim, S.W. Friend or Foe? Impacts of Dietary Xylans, Xylooligosaccharides, and Xylanases on Intestinal Health and Growth Performance of Monogastric Animals. Animals 2021, 11, 609. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.; Takahashi, D.; Wang, Q.; Mikulski, Z.; Chen, A.; Chou, T.F.; Marcovecchio, P.; McArdle, S.; Sethi, A.; Shui, J.W.; et al. Epithelial HVEM maintains intraepithelial T cell survival and contributes to host protection. Sci. Immunol. 2022, 7, eabm6931. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [PubMed]
- Rooks, M.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Cryan, J.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.E.; Lee, N.Y.; Kim, J.H.; Jung, J.H.; Jang, M.K.; Park, S.H.; Lee, M.S.; Kim, D.J.; Kim, H.S.; et al. Role of Microbiota-Derived Metabolites in Alcoholic and Non-Alcoholic Fatty Liver Diseases. Int. J. Mol. Sci. 2021, 23, 426. [Google Scholar] [CrossRef]
- Shi, C.; Yu, C.H.; Yu, W.Y.; Ying, H.Z. Gut-Lung Microbiota in Chronic Pulmonary Diseases: Evolution, Pathogenesis, and Therapeutics. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 9278441. [Google Scholar] [CrossRef]
- Giannos, P.; Prokopidis, K. Gut Dysbiosis and Long COVID-19: Feeling Gutted. J. Med. Virol. 2022, 94, 2917–2918. [Google Scholar] [CrossRef]
- Linares-García, L.; Cárdenas-Barragán, M.E.; Hernández-Ceballos, W.; Pérez-Solano, C.S.; Morales-Guzmán, A.S.; Miller, D.S.; Schmulson, M. Bacterial and Fungal Gut Dysbiosis and Clostridium difficile in COVID-19: A Review. J. Clin. Gastroenterol. 2022, 56, 285. [Google Scholar] [CrossRef]
- Slavin, J. Fiber and prebiotics: Mechanisms and health benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]
- Howarth, N.; Saltzman, E.; Roberts, S.B. Dietary fiber and weight regulation. Nutr. Rev. 2001, 59, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Rosner, B.; Willett, W.W.; Sacks, F.M. Cholesterol-lowering effects of dietary fiber: A meta-analysis. Am. J. Clin. Nutr. 1999, 69, 30–42. [Google Scholar] [CrossRef]
- InterAct-Consortium. Dietary fibre and incidence of type 2 diabetes in eight European countries: The EPIC-InterAct Study and a meta-analysis of prospective studies. Diabetologia 2015, 58, 1394–1408. [Google Scholar] [CrossRef]
- Wu, H.; Chiou, J. Potential Benefits of Probiotics and Prebiotics for Coronary Heart Disease and Stroke. Nutrients 2021, 13, 2878. [Google Scholar] [CrossRef]
- Artemev, A.; Naik, S.; Pougno, A.; Honnavar, P.; Shanbhag, N.M. The Association of Microbiome Dysbiosis with Colorectal Cancer. Cureus 2022, 14, e22156. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Gundamaraju, R.; Jha, N.K.; Gupta, P.K.; Dey, A.; Mandal, C.C.; Ford, B.M. Interplay between Dysbiosis of Gut Microbiome, Lipid Metabolism, and Tumorigenesis: Can Gut Dysbiosis Stand as a Prognostic Marker in Cancer? Dis. Markers 2022, 2022, 2941248. [Google Scholar] [CrossRef]
- Kouzu, K.; Tsujimoto, H.; Kishi, Y.; Ueno, H.; Shinomiya, N. Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines 2022, 10, 380. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, X.; Yang, H.; Tu, T.; Zhang, J.; Luo, H.; Huang, H.; Su, X. PUL-Mediated Plant Cell Wall Polysaccharide Utilization in the Gut Bacteroidetes. Int. J. Mol. Sci. 2021, 22, 3077. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, Y.; Pi, Y.; Gerrits, W.J.J.; de Vries, S.; Shang, L.; Tao, S.; Zhang, S.; Han, D.; Zhu, Z.; et al. Xylan alleviates dietary fiber deprivation-induced dysbiosis by selectively promoting Bifidobacterium pseudocatenulatum in pigs. Microbiome 2021, 9, 227. [Google Scholar] [CrossRef]
- Singh, E.; Banerjee, J.; Arora, T. Prebiotic potential of oligosaccharides: A focus on xylan derived oligosaccharides. Bioact. Carbohydr. Diet. Fibre 2015, 5, 19–30. [Google Scholar] [CrossRef]
- Moya-Pérez, A.; Neef, A.; Sanz, Y. Bifidobacterium pseudocatenulatum CECT 7765 Reduces Obesity-Associated Inflammation by Restoring the Lymphocyte-Macrophage Balance and Gut Microbiota Structure in High-Fat Diet-Fed Mice. PLoS ONE 2015, 10, e0126976. [Google Scholar] [CrossRef] [PubMed]
- Saville, B.A.; Saville, S.H. Functional Attributes and Health Benefits of Novel Prebiotic Oligosaccharides Derived from Xylan, Arabinan, and Mannan; InTech Open: London, UK, 2021. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, Y.; Ajuwon, K.M.; Zhong, R.; Li, T.; Chen, L.; Zhang, H.; Beckers, Y.; Everaert, N. Xylo-Oligosaccharides, Preparation and Application to Human and Animal Health: A Review. Front. Nutr. 2021, 8, 731930. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sood, U.; Gupta, V.; Singh, M.; Scaria, J.; Lal, R. Recent Advancements in the Development of Modern Probiotics for Restoring Human Gut Microbiome Dysbiosis. Indian J. Microbiol. 2020, 60, 12–25. [Google Scholar] [CrossRef]
- Nordberg Karlsson, E.; Schmitz, E.; Linares-Pastén, J.A.; Adlercreutz, P. Endo-xylanases as tools for production of substituted xylooligosaccharides with prebiotic properties. Appl. Microbiol. Biotechnol. 2018, 102, 9081–9088. [Google Scholar] [CrossRef]
- Ghidoli, M.; Colombo, F.; Sangiorgio, S.; Landoni, M.; Giupponi, L.; Nielsen, E.; Pilu, R. Food Containing Bioactive Flavonoids and Other Phenolic or Sulfur Phytochemicals With Antiviral Effect: Can We Design a Promising Diet Against COVID-19? Front. Nutr. 2021, 8, 661331. [Google Scholar] [CrossRef]
- Subedi, L.; Tchen, S.; Gaire, B.P.; Hu, B.; Hu, K. Adjunctive Nutraceutical Therapies for COVID-19. Int. J. Mol. Sci. 2021, 22, 1963. [Google Scholar] [CrossRef]
- Davison, J.; Wischmeyer, P.E. Probiotic and synbiotic therapy in the critically ill: State of the art. Nutrition 2019, 59, 29–36. [Google Scholar] [CrossRef]
- Manzanares, W.; Lemieux, M.; Langlois, P.L.; Wischmeyer, P.E. Probiotic and synbiotic therapy in critical illness: A systematic review and meta-analysis. Crit. Care 2016, 19, 262. [Google Scholar] [CrossRef]
- Moron, R.; Galvez, J.; Colmenero, M.; Anderson, P.; Cabeza, J.; Rodriguez-Cabezas, M.E. The Importance of the Microbiome in Critically Ill Patients: Role of Nutrition. Nutrients 2019, 11, 3002. [Google Scholar] [CrossRef] [PubMed]
- Wischmeyer, P.; McDonald, D.; Knight, R. Role of the microbiome, probiotics, and ‘dysbiosis therapy’ in critical illness. Curr. Opin. Crit. Care 2016, 22, 347–353. [Google Scholar] [CrossRef]
- Faik, A. Xylan biosynthesis: News from the grass. Plant Physiol. 2010, 153, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Jamme, F.; Rogniaux, H.; Saulnier, L.; Guillon, F. The endosperm cavity of wheat grains contains a highly hydrated gel of arabinoxylan. Plant Sci. 2021, 306, 110845. [Google Scholar]
- Zhang, L.; Gao, C.; Mentink-Vigier, F.; Tang, L.; Zhang, D.; Wang, S.; Cao, S.; Xu, Z.; Liu, X.; Wang, T.; et al. Arabinosyl Deacetylase Modulates the Arabinoxylan Acetylation Profile and Secondary Wall Formation? Plant Cell 2019, 31, 1113–1126. [Google Scholar] [CrossRef]
- Viana, A.; Noseda, M.D.; Gonçalves, A.G.; Duarte, M.E.; Yokoya, N.; Matulewicz, M.C.; Cerezo, A.S. β-D-(1→4), β-D-(1→3) ‘mixed linkage’ xylans from red seaweeds of the order Nemaliales and Palmariales. Carbohydr. Res. 2011, 346, 1023–1028. [Google Scholar] [CrossRef]
- Pawar, P.; Koutaniemi, S.; Tenkanen, M.; Mellerowicz, E.J. Acetylation of woody lignocellulose: Significance and regulation. Front. Plant Sci. 2013, 4, 118. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P. The pathobiology of osteoarthritis and the rationale for the use of pentosan polysulfate for its treatment. Semin. Arthritis Rheum. 1999, 28, 211–267. [Google Scholar] [CrossRef]
- Ghosh, P.; Edelman, J.; March, L.; Smith, M. Effects of pentosan polysulfate in osteoarthritis of the knee: A randomized, double-blind, placebo-controlled pilot study. Curr. Ther. Res. Clin. Exp. 2005, 66, 552–571. [Google Scholar] [CrossRef]
- Kumagai, K.; Shirabe, S.; Miyata, N.; Murata, M.; Yamauchi, A.; Kataoka, Y.; Niwa, M. Sodium pentosan polysulfate resulted in cartilage improvement in knee osteoarthritis—An open clinical trial. BMC Clin. Pharmacol. 2010, 10, 7. [Google Scholar] [CrossRef]
- Anand, R.; Nayyar, S.; Galvin, T.A.; Merril, C.R.; Bigelow, L.B. Sodium pentosan polysulfate (PPS), an anti-HIV agent also exhibits synergism with AZT, lymphoproliferative activity, and virus enhancement. AIDS Res. Hum. Retrovir. 1990, 6, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Herrero, L.; Foo, S.S.; Sheng, K.C.; Chen, W.; Forwood, M.R.; Bucala, R.; Mahalingam, S. Pentosan Polysulfate: A Novel Glycosaminoglycan-Like Molecule for Effective Treatment of Alphavirus-Induced Cartilage Destruction and Inflammatory Disease. J. Virol. 2015, 89, 8063–8076. [Google Scholar] [CrossRef]
- Ma, G.; Yasunaga, J.I.; Ohshima, K.; Matsumoto, T.; Matsuoka, M. Pentosan Polysulfate Demonstrates Anti-human T-Cell Leukemia Virus Type 1 Activities In Vitro and In Vivo. J. Virol. 2019, 93, e00413–e00419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; He, P.; Rodrigues, A.L.; Jeske, W.; Tandon, R.; Bates, J.T.; Bierdeman, M.A.; Fareed, J.; Dordick, J.; Linhardt, R.J. Potential anti SARS CoV-2 activity of pentosan polysulphate and mucopolysaccharide polysulphate. Pharmaceuticals 2022, 15, 258. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Yu, Y.; Zhang, F.; Xia, K.; Zhang, X.; Linhardt, R. Bottom-up and top-down profiling of pentosan polysulfate. Analyst 2019, 144, 4781–4786. [Google Scholar] [CrossRef]
- Rohman, A.; Dijkstra, B.W.; Puspaningsih, N.N.T. β-Xylosidases: Structural Diversity, Catalytic Mechanism, and Inhibition by Monosaccharides. Int. J. Mol. Sci. 2019, 20, 5524. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy). An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Nagy, T.; Nurizzo, D.; Davies, G.J.; Biely, P.; Lakey, J.H.; Bolam, D.N.; Gilbert, H.J. The beta-D-glucuronidase, GlcA67A, of Cellvibrio japonicas utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J. Biol. Chem. 2003, 278, 20286–20292. [Google Scholar] [CrossRef]
- Nurizzo, D.; Nagy, T.; Gilbert, H.J.; Davies, G.J. The structural basis for catalysis and specificity of the Pseudomonas cellulosa glucuronidase, GlcA67A. Structure 2002, 10, 547–556. [Google Scholar] [CrossRef]
- Koupenova, M.; Corkrey, H.A.; Vitseva, O.; Tanriverdi, K.; Somasundaran, M.; Liu, P.; Soofi, S.; Bhandari, R.; Godwin, M.; Parsi, K.M.; et al. SARS-CoV-2 Initiates Programmed Cell Death in Platelets. Circ. Res. 2021, 129, 631–646. [Google Scholar] [CrossRef]
- Mendis, M.; Leclerc, E.; Simsek, S. Arabinoxylans, gut microbiota and immunity. Carbohydr. Polym. 2016, 139, 159–166. [Google Scholar] [CrossRef]
- Pereira, G.; Abdel-Hamid, A.M.; Dutta, S.; D’Alessandro-Gabazza, C.N.; Wefers, D.; Farris, J.A.; Bajaj, S.; Wawrzak, Z.; Atomi, H.; Mackie, R.I.; et al. Degradation of complex arabinoxylans by human colonic Bacteroidetes. Nat. Commun. 2021, 12, 459. [Google Scholar] [CrossRef]
- Turnbaugh, P.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Childs, C.; Röytiö, H.; Alhoniemi, E.; Fekete, A.A.; Forssten, S.D.; Hudjec, N.; Lim, Y.N.; Steger, C.J.; Yaqoob, P.; Tuohy, K.M.; et al. Xylo-oligosaccharides alone or in synbiotic combination with Bifidobacterium animalis subsp. lactis induce bifidogenesis and modulate markers of immune function in healthy adults: A double-blind, placebo-controlled, randomised, factorial cross-over study. Br. J. Nutr. 2014, 111, 1945–1956. [Google Scholar] [CrossRef]
- Christensen, E.; Licht, T.R.; Leser, T.D.; Bahl, M.I. Dietary xylo-oligosaccharide stimulates intestinal bifidobacteria and lactobacilli but has limited effect on intestinal integrity in rats. BMC Res. Notes 2014, 7, 660. [Google Scholar] [CrossRef]
- Majumdar, S.; Bhattacharyya, D.K.; Bhowal, J. Evaluation of nutraceutical application of xylooligosaccharide enzymatically produced from cauliflower stalk for its value addition through a sustainable approach. Food Funct. 2021, 12, 5501–5523. [Google Scholar] [CrossRef]
- Pang, J.; Wang, S.; Wang, Z.; Wu, Y.; Zhang, X.; Pi, Y.; Han, D.; Zhang, S.; Wang, J. Xylo-oligosaccharide alleviates Salmonella induced inflammation by stimulating Bifidobacterium animalis and inhibiting Salmonella colonization. FASEB J. 2021, 35, e21977. [Google Scholar] [CrossRef]
- Chow, V.; Nong, G.; St John, F.J.; Sawhney, N.; Rice, J.D.; Preston, J.F. Bacterial xylan utilization regulons: Systems for coupling depolymerization of methylglucuronoxylans with assimilation and metabolism. J. Ind. Microbiol. Biotechnol. 2021, 49, kuab080. [Google Scholar] [CrossRef]
- Kawai, K.; Kamochi, R.; Oiki, S.; Murata, K.; Hashimoto, W. Probiotics in human gut microbiota can degrade host glycosaminoglycans. Sci. Rep. 2018, 8, 10674. [Google Scholar] [CrossRef]
- Cartmell, A.; Lowe, E.C.; Baslé, A.; Firbank, S.J.; Ndeh, D.A.; Murray, H.; Terrapon, N.; Lombard, V.; Henrissat, B.; Turnbull, J.E.; et al. How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl. Acad. Sci. USA 2017, 114, 7037–7042. [Google Scholar] [CrossRef]
- Egan, M.; Jiang, H.; O’Connell Motherway, M.; Oscarson, S.; van Sinderen, D. Glycosulfatase-Encoding Gene Cluster in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2016, 82, 6611–6623. [Google Scholar] [CrossRef] [PubMed]
- Ervin, S.; Simpson, J.B.; Gibbs, M.E.; Creekmore, B.C.; Lim, L.; Walton, W.G.; Gharaibeh, R.Z.; Redinbo, M.R. Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases. Biochemistry 2020, 59, 3939–3950. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.; Seyed Hameed, A.S.; Meng, X.; Liu, W. Utilization of glycosaminoglycans by the human gut microbiota: Participating bacteria and their enzymatic machineries. Gut Microbes 2022, 14, 2068367. [Google Scholar] [CrossRef] [PubMed]
- Coker, J.; Moyne, O.; Rodionov, D.A.; Zengler, K. Carbohydrates great and small, from dietary fiber to sialic acids: How glycans influence the gut microbiome and affect human health. Gut Microbes 2021, 13, 1869502. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, N.; Li, Z.; Wang, X.; Shi, H.; Xue, C.; Li, R.W.; Tang, Q. Chondroitin sulfate disaccharides modified the structure and function of the murine gut microbiome under healthy and stressed conditions. Sci. Rep. 2017, 7, 6783. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Ai, X.; Fu, T.; Ren, L.; Shang, Q.; Li, G.; Yu, G. In vitro fermentation of hyaluronan by human gut microbiota: Changes in microbiota community and potential degradation mechanism. Carbohydr. Polym. 2021, 269, 118313. [Google Scholar] [CrossRef]
- Chen, S.; He, Y.; Hu, Z.; Lu, S.; Yin, X.; Ma, X.; Lv, C.; Jin, G. Heparanase Mediates Intestinal Inflammation and Injury in a Mouse Model of Sepsis. J. Histochem. Cytochem. 2017, 65, 241–249. [Google Scholar] [CrossRef]
- Yanagibashi, T.; Hosono, A.; Oyama, A.; Tsuda, M.; Suzuki, A.; Hachimura, S.; Takahashi, Y.; Momose, Y.; Itoh, K.; Hirayama, K.; et al. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA + B cells. Immunobiology 2013, 218, 645–651. [Google Scholar]
- Duan, R.; Chen, X.; Wang, F.; Zhang, T.; Ling, P. Oral administration of heparin or heparosan increases the Lactobacillus population in gut microbiota of rats. Carbohydr. Polym. 2013, 94, 100–105. [Google Scholar] [CrossRef]
- Shang, Q.; Li, Q.; Zhang, M.; Song, G.; Shi, J.; Jiang, H.; Cai, C.; Hao, J.; Li, G.; Yu, G. Dietary keratan sulfate from shark cartilage modulates gut microbiota and increases the abundance of lactobacillus spp. Mar. Drugs 2016, 14, 224. [Google Scholar] [CrossRef]
- Floer, M.; Götte, M.; Wild, M.K.; Heidemann, J.; Gassar, E.S.; Domschke, W.; Kiesel, L.; Luegering, A.; Kucharzik, T. Enoxaparin improves the course of dextran sodium sulfate-induced colitis in syndecan-1-deficient mice. Am. J. Pathol. 2010, 176, 146–157. [Google Scholar] [CrossRef]
- Barsotti, G.; Cupisti, A.; Gervasi, G.; Bartoli, C.; Barsotti, M.; Pasquariello, A.; Moriconi, L.; Giovannetti, S. Effects of oral administration of heparan sulphate in the rat remnant kidney model. Nephron 1999, 81, 310–316. [Google Scholar] [CrossRef]
- Kim, Y.; Kessler, S.P.; Obery, D.R.; Homer, C.R.; McDonald, C.; de la Motte, C.A. Hyaluronan 35kDa treatment protects mice from Citrobacter rodentium infection and induces epithelial tight junction protein ZO-1 in vivo. Matrix Biol. J. Int. Soc. Matrix Biol. 2017, 62, 28–39. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Salyers, A.; Vercellotti, J.R.; West, S.E.; Wilkins, T.D. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 1977, 33, 319–322. [Google Scholar] [CrossRef]
- Sonnenburg, J.; Xu, J.; Leip, D.D.; Chen, C.-H.; Westover, B.P.; Weatherford, J.; Buhler, J.D.; Gordon, J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005, 307, 1955–1959. [Google Scholar] [CrossRef]
- Hamai, A.; Hashimoto, N.; Mochizuki, H.; Kato, F.; Makiguchi, Y.; Horie, K.; Suzuki, S. Two distinct chondroitin sulfate ABC lyases: An endoeliminase yielding tetrasaccharides and an exoeliminase preferentially acting on oligosaccharides. J. Biol. Chem. 1997, 272, 9123–9130. [Google Scholar] [CrossRef]
- Drzewiecka, D. Significance and roles of Proteus spp. bacteria in natural environments. Microb. Ecol. 2016, 72, 741–758. [Google Scholar] [CrossRef]
- Prabhakar, V.; Capila, I.; Bosques, C.J.; Pojasek, K.; Sasisekharan, R. Chondroitinase ABC I from Proteus vulgaris: Cloning, recombinant expression and active site identification. Biochem. J. 2005, 386, 103–112. [Google Scholar] [CrossRef]
- Prabhakar, V.; Capila, I.; Soundararajan, V.; Raman, R.; Sasisekharan, R. Recombinant expression, purification, and biochemical characterization of chondroitinase ABC II from Proteus vulgaris. J. Biol. Chem. 2009, 284, 974–982. [Google Scholar] [CrossRef]
- Salyers, A.; West, S.E.; Vercellotti, J.R.; Wilkins, T.D. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl. Environ. Microbiol. 1977, 34, 529. [Google Scholar] [CrossRef] [PubMed]
- Crociani, F.; Alessandrini, A.; Mucci, M.M.; Biavati, B. Degradation of complex carbohydrates by Bifidobacterium spp. Int. J. Food Microbiol. 1994, 24, 199–210. [Google Scholar] [CrossRef]
- Kato, K.; Ishiwa, A. The role of carbohydrates in infection strategies of enteric pathogens. Trop. Med. Health 2015, 43, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Henry-Stanley, M.; Hess, D.J.; Erlandsen, S.L.; Wells, C.L. Ability of the heparan sulfate proteoglycan syndecan-1 to participate in bacterial translocation across the intestinal epithelial barrier. Shock 2005, 24, 571–576. [Google Scholar] [CrossRef]
- Simon, M.; McClanahan, R.H.; Shah, J.F.; Repko, T.; Modi, N.B. Metabolism of [3H]pentosan polysulfate sodium (PPS) in healthy human volunteers. Xenobiotica 2005, 35, 775–784. [Google Scholar] [CrossRef]
- Taneja, R. Current status of oral pentosan polysulphate in bladder pain syndrome/interstitial cystitis. Int. Urogynecol. J. 2020, 32, 1107–1115. [Google Scholar] [CrossRef]
- Tseng, C.; Chang, S.J.; Meng, E.; Chang, H.C.; Lee, Y.J. The efficacy of pentosan polysulfate monotherapy for preventing recurrent urinary tract infections in women: A multicenter open-label randomized controlled trial. J. Formos. Med. Assoc. 2020, 119, 1314–1319. [Google Scholar] [CrossRef]
- Li, M.; Li, G.; Shang, Q.; Chen, X.; Liu, W.; Pi, X.; Zhu, L.; Yin, Y.; Yu, G.; Wang, X. In vitro fermentation of alginate and its derivatives by human gut microbiota. Anaerobe 2016, 39, 19–25. [Google Scholar] [CrossRef]
- Barbeyron, T.; Brillet-Guéguen, L.; Carré, W.; Carrière, C.; Caron, C.; Czjzek, M.; Hoebeke, M.; Michel, G. Matching the Diversity of Sulfated Biomolecules: Creation of a Classification Database for Sulfatases Reflecting Their Substrate Specificity. PLoS ONE 2016, 11, e0164846. [Google Scholar] [CrossRef]
- Hansen, E.; Lozupone, C.A.; Rey, F.E.; Wu, M.; Guruge, J.L.; Narra, A.; Goodfellow, J.; Zaneveld, J.R.; McDonald, D.T.; Goodrich, J.A.; et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4599–4606. [Google Scholar] [CrossRef]
- Nava, G.; Carbonero, F.; Croix, J.A.; Greenberg, E.; Gaskins, H.R. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012, 6, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Rey, F.; Gonzalez, M.D.; Cheng, J.; Wu, M.; Ahern, P.P.; Gordon, J.I. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. USA 2013, 110, 13582–13587. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.; Chadwick, V.S.; Murray, A. Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett. Appl. Microbiol. 2006, 43, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Annunziata, I.; Roma, G.; Ballabio, A. Sulfatases and sulfatase modifying factors: An exclusive and promiscuous relationship. Hum. Mol. Genet. 2005, 14, 3203–3217. [Google Scholar] [CrossRef]
- Leth, M.; Ejby, M.; Workman, C.; Ewald, D.A.; Pedersen, S.S.; Sternberg, C.; Bahl, M.I.; Licht, T.R.; Aachmann, F.L.; Westereng, B.; et al. Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat. Microbiol. 2018, 3, 570–580. [Google Scholar] [CrossRef]
- Di Costanzo, M.; De Paulis, N.; Biasucci, G. Butyrate: A Link between Early Life Nutrition and Gut Microbiome in the Development of Food Allergy. Life 2021, 11, 384. [Google Scholar] [CrossRef]
- Méndez, C.; Bueno, S.M.; Kalergis, A.M. Contribution of Gut Microbiota to Immune Tolerance in Infants. J. Immunol. Res. 2021, 2021, 7823316. [Google Scholar] [CrossRef]
- Riazi-Rad, F.; Behrouzi, A.; Mazaheri, H.; Katebi, A.; Ajdary, S. Impact of gut microbiota on immune system. Acta Microbiol. Immunol. Hung. 2021, 68, 135–144. [Google Scholar] [CrossRef]
- Wagner, C.; Torow, N.; Hornef, M.W.; Lelouard, H. Spatial and temporal key steps in early-life intestinal immune system development and education. FEBS J. 2021, 289, 4731–4751. [Google Scholar] [CrossRef]
- Yokanovich, L.; Newberry, R.D.; Knoop, K.A. Regulation of oral antigen delivery early in life: Implications for oral tolerance and food allergy. Clin. Exp. Allergy 2021, 51, 518–526. [Google Scholar] [CrossRef]
- Jandhyala, S.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Li, A.; Jiang, J.; Yuan, X.; Zhang, P.; Xi, C.; Wu, L.; Wang, Z.; Chen, J.; Lu, J.; et al. Metagenomic analysis reveals gut bacterial signatures for diagnosis and treatment outcome prediction in bipolar depression. Psychiatry Res. 2022, 307, 114326. [Google Scholar] [CrossRef]
- Levi Mortera, S.; Vernocchi, P.; Basadonne, I.; Zandonà, A.; Chierici, M.; Durighello, M.; Marzano, V.; Gardini, S.; Gasbarrini, A.; Urbani, A.; et al. A metaproteomic-based gut microbiota profiling in children affected by autism spectrum disorders. J. Proteom. 2022, 251, 104407. [Google Scholar] [CrossRef]
- Sorboni, S.; Moghaddam, H.S.; Jafarzadeh-Esfehani, R.; Soleimanpour, S. A Comprehensive Review on the Role of the Gut Microbiome in Human Neurological Disorders. Clin. Microbiol. Rev. 2022, 35, e00338-20. [Google Scholar] [CrossRef]
- Curry, K.; Nute, M.G.; Treangen, T.J. It takes guts to learn: Machine learning techniques for disease detection from the gut microbiome. Emerg. Top. Life Sci. 2021, 5, 815–827. [Google Scholar] [CrossRef]
- Jain, N. The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut Microbes 2020, 12, 1824564. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef] [PubMed]
- Kreft, L.; Hoffmann, C.; Ohnmacht, C. Therapeutic Potential of the Intestinal Microbiota for Immunomodulation of Food Allergies. Front. Immunol. 2020, 11, 1853. [Google Scholar] [CrossRef]
- Pero-Gascon, R.; Hemeryck, L.Y.; Poma, G.; Falony, G.; Nawrot, T.S.; Raes, J.; Vanhaecke, L.; De Boevre, M.; Covaci, A.; De Saeger, S. FLEXiGUT: Rationale for exposomics associations with chronic low-grade gut inflammation. Environ. Int. 2022, 158, 106906. [Google Scholar] [CrossRef]
- Berding, K.; Cryan, J.F. Microbiota-targeted interventions for mental health. Curr. Opin. Psychiatry 2022, 35, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.; Porrelli, A.; Vacca, M.; Comte, B.; Nimptsch, K.; Pinart, M.; Pischon, T.; Pujos-Guillot, E.; De Angelis, M. Metaproteomics Approach and Pathway Modulation in Obesity and Diabetes: A Narrative Review. Nutrients 2021, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre-Arbogast, S.; Thomas, A.; Samieri, C. Dietary factors and brain health. Curr. Opin. Lipidol. 2022, 33, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Sihag, J.; Di Marzo, V. (Wh)olistic (E)ndocannabinoidome-Microbiome-Axis Modulation through (N)utrition (WHEN) to Curb Obesity and Related Disorders. Lipids Health Dis. 2022, 21, 9. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.; Vlamakis, H.; Lee, J.W.J.; Besse, L.A.; Xanthakis, V.; Vasan, R.S.; Shaw, S.Y.; Xavier, R.J. Population study of the gut microbiome: Associations with diet, lifestyle, and cardiometabolic disease. Genome Med. 2021, 13, 188. [Google Scholar] [CrossRef] [PubMed]
- Wali, J.; Milner, A.J.; Luk, A.W.S.; Pulpitel, T.J.; Dodgson, T.; Facey, H.J.W.; Wahl, D.; Kebede, M.A.; Senior, A.M.; Sullivan, M.A.; et al. Impact of dietary carbohydrate type and protein-carbohydrate interaction on metabolic health. Nat. Metab. 2021, 3, 810–828. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.L.; Christensen, H.; Dusemund, B.; Kouba, M.; Kos Durjava, M.; López-Alonso, M.; López Puente, S.; et al. Safety and efficacy of RONOZYME®WX CT/L (endo-1,4-β-xylanase) as a feed additive for sows for reproduction. EFSA J. 2019, 17, e05790. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.L.; Christensen, H.; Dusemund, B.; Kouba, M.; Kos Durjava, M.; López-Alonso, M.; López Puente, S.; et al. Efficacy of RONOZYME®WX (endo-1,4-β-xylanase) as a feed additive for laying hens. EFSA J. 2019, 17, e05919. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; et al. Safety and efficacy of RONOZYME®WX (endo-1,4-β-xylanase) as a feed additive for laying hens. EFSA J. 2017, 15, e05020. [Google Scholar]
- Woyengo, T.; Bogota, K.J.; Noll, S.L.; Wilson, J. Enhancing nutrient utilization of broiler chickens through supplemental enzymes. Poult. Sci. 2019, 98, 1302–1309. [Google Scholar] [CrossRef]
- Seidavi, A.; Tavakoli, M.; Asroosh, F.; Scanes, C.G.; Abd El-Hack, M.E.; Naiel, M.A.E.; Taha, A.E.; Aleya, L.; El-Tarabily, K.A.; Swelum, A.A. Antioxidant and antimicrobial activities of phytonutrients as antibiotic substitutes in poultry feed. Environ. Sci. Pollut. Res. Int. 2021, 29, 5006–5031. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Lin, Z.; Yu, C.; Qiu, M.; Peng, H.; Jiang, X.; Du, H.; Li, Q.; Liu, Y.; Zhang, Z.; et al. Effects of Lactobacillus plantarum on growth traits, slaughter performance, serum markers and intestinal bacterial community of Daheng broilers. J. Anim. Physiol. Anim. Nutr. 2021, 106, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mishra, B.; Bedford, M.R.; Jha, R. Effects of supplemental xylanase and xylooligosaccharides on production performance and gut health variables of broiler chickens. J. Anim. Sci. Biotechnol. 2021, 12, 98. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wu, S.; Li, B.; Tan, J.; Yan, J.; Wang, Y.; Tang, Z.; Liu, M.; Fu, C.; Zhang, H.; et al. Dietary ferulic acid and vanillic acid on inflammation, gut barrier function and growth performance in lipopolysaccharide-challenged piglets. Anim. Nutr. 2022, 8, 144–152. [Google Scholar] [CrossRef]
- Segura-Wang, M.; Grabner, N.; Koestelbauer, A.; Klose, V.; Ghanbari, M. Genome-Resolved Metagenomics of the Chicken Gut Microbiome. Front. Microbiol. 2021, 12, 726923. [Google Scholar] [CrossRef]
- Bohra, V.; Dafale, N.A.; Purohit, H.J. Understanding the alteration in rumen microbiome and CAZymes profile with diet and host through comparative metagenomic approach. Arch. Microbiol. 2019, 201, 1385–1397. [Google Scholar] [CrossRef]
- Duarte, M.; Kim, S.W. Intestinal microbiota and its interaction to intestinal health in nursery pigs. Anim. Nutr. 2022, 8, 169–184. [Google Scholar] [CrossRef]
- Charoensiddhi, S.; Abraham, R.E.; Su, P.; Zhang, W. Seaweed and seaweed-derived metabolites as prebiotics. Adv. Food Nutr. Res. 2020, 91, 97–156. [Google Scholar] [PubMed]
- Costa, M.; Cardoso, C.; Afonso, C.; Bandarra, N.M.; Prates, J.A.M. Current knowledge and future perspectives of the use of seaweeds for livestock production and meat quality: A systematic review. J. Anim. Physiol. Anim. Nutr. 2021, 105, 1075–1102. [Google Scholar] [CrossRef]
- Morais, T.; Inacio, A.; Coutinho, T.; Ministro, M.; Cotas, J.; Periera, L.; Bahcevandziev, K. Seaweed Potential in the Animal Feed: A Review. J. Mar. Sci. Eng. 2020, 8, 559. [Google Scholar] [CrossRef]
- Min, B.; Parker, D.; Brauer, D.; Waldrip, H.; Lockard, C.; Hales, K.; Akbay, A.; Augyte, S.T. he role of seaweed as a potential dietary supplementation for enteric methane mitigation in ruminants: Challenges and opportunities. Anim. Nutr. 2021, 7, 1371–1387. [Google Scholar] [CrossRef] [PubMed]
- Reisinger, A.; Clark, H.; Cowie, A.L.; Emmet-Booth, J.; Gonzalez Fischer, C.; Herrero, M.; Howden, M.; Leahy, S. How necessary and feasible are reductions of methane emissions from livestock to support stringent temperature goals? Philos. Trans. R. Soc. A 2021, 379, 20200452. [Google Scholar] [CrossRef] [PubMed]
- FutureFeed. 2021. Available online: https://www.csiro.au/en/research/animals/livestock/futurefeed (accessed on 7 January 2021).
- Aachary, A.; Prapulla, S.G. Xylooligosaccharides (XOS) as an emerging prebiotic: Microbial synthesis, utilization, structural characterization, bioactive properties, and applications. Compr. Rev. Food Sci. Food Saf. 2011, 10, 2–16. [Google Scholar] [CrossRef]
- Servin, A. Antagonistic activities of Lactobacilli and Bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef]
- Sarkar, S.; Mazumder, P.M.; Banerjee, S. Oligosaccharide and Flavanoid Mediated Prebiotic Interventions to Treat Gut Dysbiosis Associated Cognitive Decline. J. Neuroimmune Pharmacol. 2022. [Google Scholar] [CrossRef]
- Savignac, H.; Tramullas, M.; Kiely, B.; Dinan, T.G.; Cryan, J.F. Bifobacteria modulate cognitive processes in anxious mouse strain. Behav. Brain Res. 2015, 287, 59–72. [Google Scholar] [CrossRef]
- Steidler, L.; Robinson, K.; Chamberlain, L.; Schofield, K.M.; Remaut, E.; Le Page, R.W.; Wells, J.M. Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect. Immun. 1998, 66, 3183–3189. [Google Scholar] [CrossRef]
- Schotte, L.; Steidler, L.; Vandekerckhove, J.; Remaut, E. Secretion of biologically active murine interleukin-10 by Lactococcus lactis. Enzyme Microb. Technol. 2000, 27, 761–765. [Google Scholar] [CrossRef]
- Steidler, L.; Hans, W.; Schotte, L.; Neirynck, S.; Obermeier, F.; Falk, W.; Fiers, W.; Remaut, E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. [Google Scholar] [CrossRef]
- Sturm, A.; Dignass, A.U. Epithelial restitution and wound healing in inflammatory bowel disease. World J. Gastroenterol. 2008, 14, 348–353. [Google Scholar] [CrossRef]
- Farrar, M.; Whitehead, T.R.; Lan, J.; Dilger, P.; Thorpe, R.; Holland, K.T.; Carding, S.R. Engineering of the gut commensal bacterium Bacteroides ovatus to produce and secrete biologically active murine interleukin-2 in response to xylan. J. Appl. Microbiol. 2005, 98, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Hamady, Z.; Farrar, M.D.; Whitehead, T.R.; Holland, K.T.; Lodge, J.P.A.; Carding, S.R. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology 2008, 154, 3165–3174. [Google Scholar] [CrossRef] [PubMed]
- Hamady, Z.; Scott, N.; Farrar, M.D.; Lodge, J.P.; Holland, K.T.; Whitehead, T.; Carding, S.R. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut 2010, 59, 461–469. [Google Scholar] [CrossRef]
- Hamady, Z.; Scott, N.; Farrar, M.D.; Wadhwa, M.; Dilger, P.; Whitehead, T.R.; Thorpe, R.; Holland, K.T.; Lodge, J.P.; Carding, S.R. Treatment of colitis with a commensal gut bacterium engineered to secrete human TGF-β1 under the control of dietary xylan 1. Inflamm. Bowel. Dis. 2011, 17, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Hamady, Z. Novel xylan-controlled delivery of therapeutic proteins to inflamed colon by the human anaerobic commensal bacterium. Ann. R. Coll. Surg. Engl. 2013, 95, 235–240. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, M.M.; Melrose, J. Xylan Prebiotics and the Gut Microbiome Promote Health and Wellbeing: Potential Novel Roles for Pentosan Polysulfate. Pharmaceuticals 2022, 15, 1151. https://doi.org/10.3390/ph15091151
Smith MM, Melrose J. Xylan Prebiotics and the Gut Microbiome Promote Health and Wellbeing: Potential Novel Roles for Pentosan Polysulfate. Pharmaceuticals. 2022; 15(9):1151. https://doi.org/10.3390/ph15091151
Chicago/Turabian StyleSmith, Margaret M., and James Melrose. 2022. "Xylan Prebiotics and the Gut Microbiome Promote Health and Wellbeing: Potential Novel Roles for Pentosan Polysulfate" Pharmaceuticals 15, no. 9: 1151. https://doi.org/10.3390/ph15091151
APA StyleSmith, M. M., & Melrose, J. (2022). Xylan Prebiotics and the Gut Microbiome Promote Health and Wellbeing: Potential Novel Roles for Pentosan Polysulfate. Pharmaceuticals, 15(9), 1151. https://doi.org/10.3390/ph15091151