Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy
Abstract
1. Introduction
2. Zebrafish In Vitro and In Vivo Screening Systems
2.1. In Vitro Cell Systems Capable of Biomineralization
2.2. Bone Formation in Larval Systems
2.2.1. Opercular Bone Growth
2.2.2. Craniofacial Skeleton
2.2.3. Vertebrae Mineralization
2.3. Ex Vivo Culture of Elasmoid Scales
2.4. Bone Structures Capable of Repair and Regeneration
2.4.1. Regenerating Caudal Fin
2.4.2. Regenerating Elasmoid Scales
2.4.3. Bone Repair
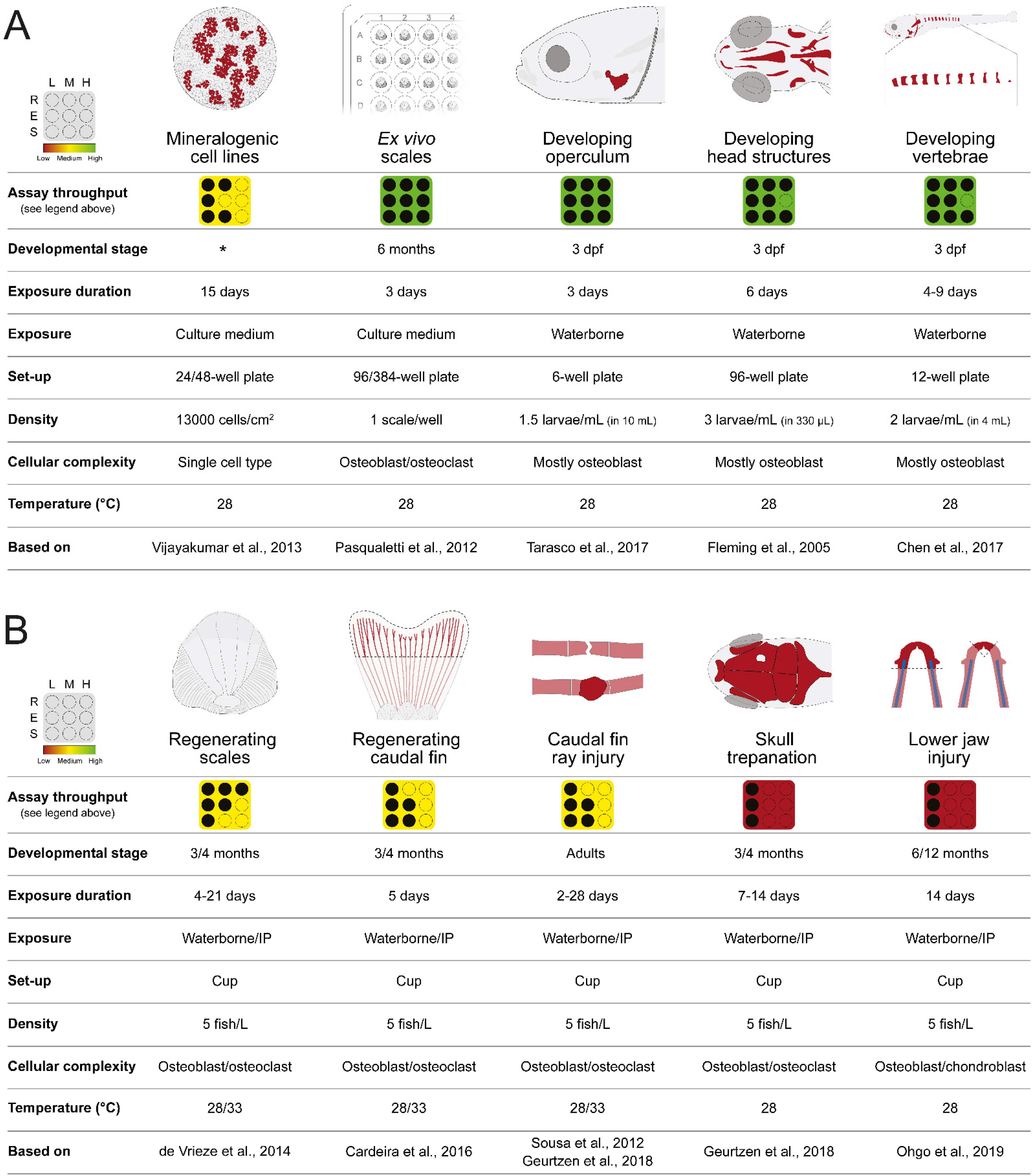
2.4.4. Regenerating Lower Jaw
2.5. Zebrafish Lines for Phenotypic Screening of Bone Anabolic Compounds
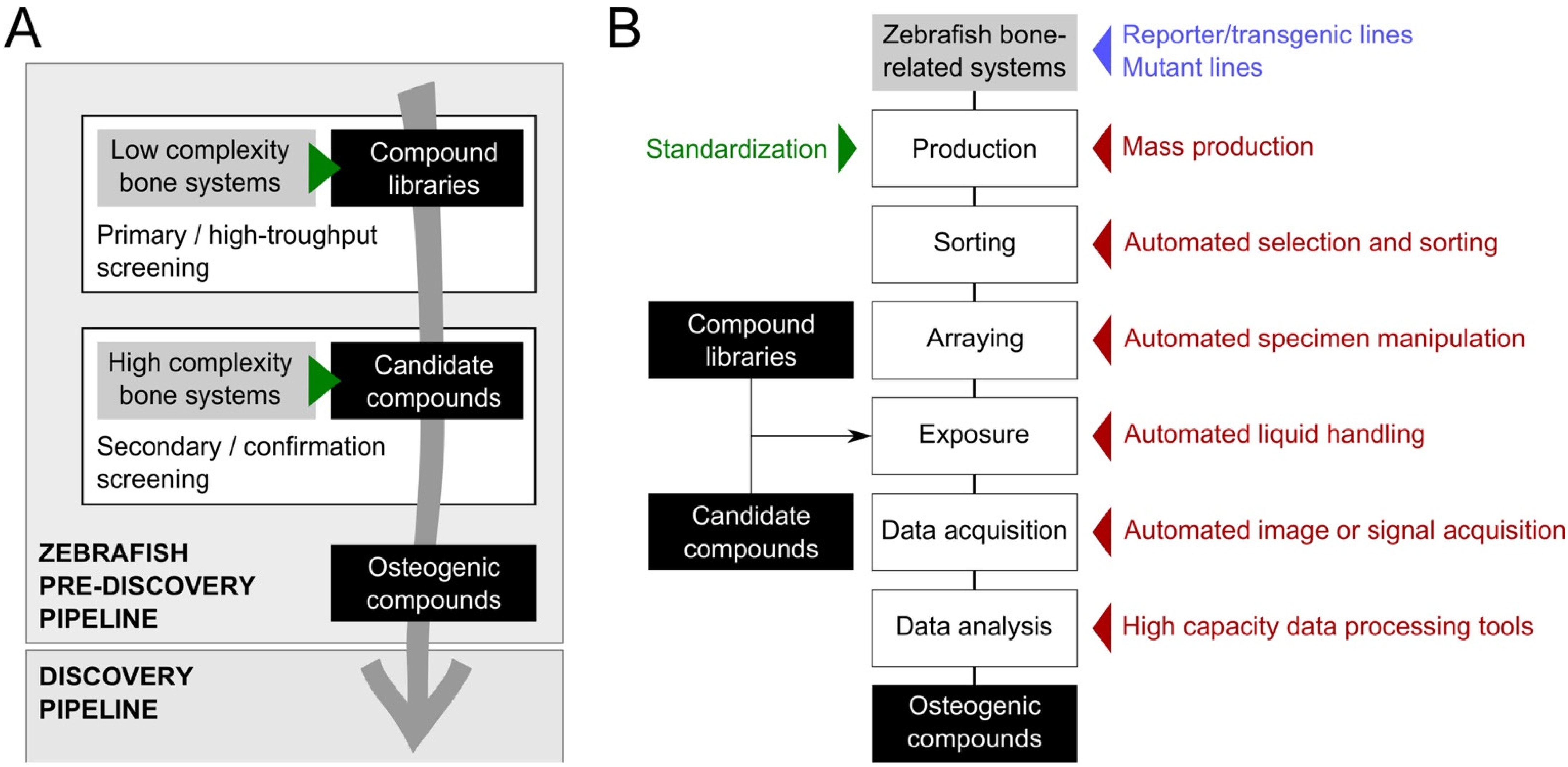
3. Tools to Improve Screening Throughput and Replicability
3.1. Inbred Zebrafish Lines
3.2. Standardized Housing and Feeding
3.3. Mass Production of Synchronized Embryos
3.4. Target Specimen Sorting
3.5. Compound Delivery
3.6. Image Acquisition
3.7. Image Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Compston, J.E.; McClung, M.R.; Leslie, W.D. Osteoporosis. Lancet 2019, 393, 364–376. [Google Scholar] [CrossRef]
- Sözen, T.; Özışık, L.; Başaran, N.Ç. An overview and management of osteoporosis. Eur. J. Rheumatol. 2017, 4, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Cundy, T. Paget’s disease of bone. Metabolism 2018, 80, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Tu, K.N.; Lie, J.D.; Wan, C.K.V.; Cameron, M.; Austel, A.G.; Nguyen, J.K.; Van, K.; Hyun, D. Osteoporosis: A review of treatment options. Pharm. Ther. 2018, 43, 92–104. [Google Scholar]
- Ralston, S.H.; Corral-Gudino, L.; Cooper, C.; Francis, R.M.; Fraser, W.D.; Gennari, L.; Guañabens, N.; Javaid, M.K.; Layfield, R.; O’Neill, T.W.; et al. Diagnosis and management of Paget’s disease of bone in adults: A clinical guideline. J. Bone Miner. Res. 2019, 34, 579–604. [Google Scholar] [CrossRef]
- Denayer, T.; Stöhrn, T.; Van Roy, M. Animal models in translational medicine: Validation and prediction. New Horiz. Transl. Med. 2014, 2, 5–11. [Google Scholar] [CrossRef]
- Strange, K. Drug discovery in fish, flies, and worms. ILAR J. 2016, 57, 133–143. [Google Scholar] [CrossRef]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Giacomotto, J.; Ségalat, L. High-throughput screening and small animal models, where are we? Br. J. Pharmacol. 2010, 160, 204–216. [Google Scholar] [CrossRef]
- Lee, K.Y.; Jang, G.H.; Byun, C.H.; Jeun, M.; Searson, P.C.; Lee, K.H. Zebrafish models for functional and toxicological screening of nanoscale drug delivery systems: Promoting preclinical applications. Biosci. Rep. 2017, 37, BSR20170199. [Google Scholar] [CrossRef]
- Jang, K.; Sato, K.; Igawa, K.; Chung, U.; Kitamori, T. Development of an osteoblast-based 3D continuous-perfusion microfluidic system for drug screening. Anal. Bioanal. Chem. 2008, 390, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, J.P.; Halleen, J.M. Models and screening assays for drug discovery in osteoporosis. Expert Opin. Drug Discov. 2010, 5, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Kolanthai, E.; Veerla, S.C.; Khajuria, D.K.; Mahapatra, D.R. Optical diagnostics of osteoblast cells and osteogenic drug screening. In Proceedings of the Photonic Therapeutics and Diagnostics XII, SPIE, San Francisco, CA, USA, 13–14 February 2016; Volume 9689, p. 96894I. [Google Scholar]
- Czekanska, E.M.; Stoddart, M.J.; Richards, R.G.; Hayes, J.S. In search of an osteoblast cell model for in vitro research. Eur. Cells Mater. 2012, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.-Y.; Wang, M.; Lei, S.; Yang, Q.-X.; Liu, Y.-Q. Rapid screening of active components with an osteoclastic inhibitory effect in Herba epimedii using quantitative pattern-activity relationships based on joint-action models. Molecules 2017, 22, 1767. [Google Scholar] [CrossRef] [PubMed]
- Gibert, Y.; Trengove, M.C.; Ward, A.C. Zebrafish as a genetic model in pre-clinical drug testing and screening. Curr. Med. Chem. 2013, 20, 2458–2466. [Google Scholar] [CrossRef]
- Javidan, Y.; Schilling, T.F. Development of cartilage and bone. Methods Cell Biol. 2004, 76, 415–436. [Google Scholar]
- Hall, B.K. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2015; ISBN 978-0-12-319060-4. [Google Scholar]
- Laizé, V.; Gavaia, P.J.; Cancela, M.L. Fish: A suitable system to model human bone disorders and discover drugs with osteogenic or osteotoxic activities. Drug Discov. Today Dis. Model. 2014, 13, 29–37. [Google Scholar] [CrossRef]
- Tonelli, F.; Bek, J.W.; Besio, R.; De Clercq, A.; Leoni, L.; Salmon, P.; Coucke, P.J.; Willaert, A.; Forlino, A. Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 2020, 11, 489. [Google Scholar] [CrossRef]
- Kwon, R.Y.; Watson, C.J.; Karasik, D. Using zebrafish to study skeletal genomics. Bone 2019, 126, 37–50. [Google Scholar] [CrossRef]
- Fleming, A.; Sato, M.; Goldsmith, P. High-throughput in vivo screening for bone anabolic compounds with zebrafish. J. Biomol. Screen. 2005, 10, 823–831. [Google Scholar] [CrossRef]
- Chen, J.-R.; Lai, Y.-H.; Tsai, J.-J.; Hsiao, C.-D. Live fluorescent staining platform for drug-screening and mechanism-analysis in zebrafish for bone mineralization. Molecules 2017, 22, 2068. [Google Scholar] [CrossRef]
- Huang, H.; Lin, H.; Lan, F.; Wu, Y.; Yang, Z.; Zhang, J. Application of bone transgenic zebrafish in anti-osteoporosis chemical screening. Anim. Model. Exp. Med. 2018, 1, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Rosa, J.T.; Laizé, V.; Gavaia, P.J.; Cancela, M.L. Fish models of induced osteoporosis. Front. Cell Dev. Biol. 2021, 9, 672424. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Cornet, C.; Di Donato, V.; Terriente, J. Combining zebrafish and CRISPR/Cas9: Toward a more efficient drug discovery pipeline. Front. Pharmacol. 2018, 9, 703. [Google Scholar] [CrossRef]
- Bairoch, A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef]
- Laizé, V.; Rosa, J.T.; Tarasco, M.; Cancela, M.L. Status, challenges, and perspectives of fish cell culture—Focus on cell lines capable of in vitro mineralization. In Cellular and Molecular Approaches in Fish Biology; Fernández, I., Fernandes, J., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 381–404. ISBN 9780128222737. [Google Scholar]
- Vijayakumar, P.; Laizé, V.; Cardeira, J.; Trindade, M.; Cancela, M.L. Development of an in vitro cell system from zebrafish suitable to study bone cell differentiation and extracellular matrix mineralization. Zebrafish 2013, 10, 500–509. [Google Scholar] [CrossRef]
- Fernández, I.; Vijayakumar, P.; Marques, C.; Cancela, M.L.; Gavaia, P.J.; Laizé, V. Zebrafish vitamin K epoxide reductases: Expression in vivo, along extracellular matrix mineralization and under phylloquinone and warfarin in vitro exposure. Fish Physiol. Biochem. 2015, 41, 745–759. [Google Scholar] [CrossRef]
- Pombinho, A.R.; Laizé, V.; Molha, D.M.; Marques, S.M.P.; Cancela, M.L. Development of two bone-derived cell lines from the marine teleost Sparus aurata; evidence for extracellular matrix mineralization and cell-type-specific expression of matrix Gla protein and osteocalcin. Cell Tissue Res. 2004, 315, 393–406. [Google Scholar] [CrossRef]
- Surget, G.; Roberto, V.P.; Le Lann, K.; Mira, S.; Guérard, F.; Laizé, V.; Poupart, N.; Cancela, M.L.; Stiger-Pouvreau, V. Marine green macroalgae: A source of natural compounds with mineralogenic and antioxidant activities. J. Appl. Phycol. 2017, 29, 575–584. [Google Scholar] [CrossRef]
- Roberto, V.P.; Surget, G.; Le Lann, K.; Mira, S.; Tarasco, M.; Guérard, F.; Poupart, N.; Laizé, V.; Stiger-Pouvreau, V.; Cancela, M.L. Antioxidant, mineralogenic and osteogenic activities of Spartina alterniflora and Salicornia fragilis extracts rich in polyphenols. Front. Nutr. 2021, 8, 719438. [Google Scholar] [CrossRef] [PubMed]
- Laizé, V.; Gavaia, P.J.; Tarasco, M.; Viegas, M.N.; Caria, J.; Luis, N.; Cancela, M.L. Osteotoxicity of 3-methylcholanthrene in fish. Ecotoxicol. Environ. Saf. 2018, 161, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Tiago, D.M.; Cancela, M.L.; Aureliano, M.; Laizé, V. Vanadate proliferative and anti-mineralogenic effects are mediated by MAPK and PI-3K/Ras/Erk pathways in a fish chondrocyte cell line. FEBS Lett. 2008, 582, 1381–1385. [Google Scholar] [CrossRef]
- Viegas, M.N.; Dias, J.; Cancela, M.L.; Laizé, V. Polyunsaturated fatty acids regulate cell proliferation, extracellular matrix mineralization and gene expression in a gilthead seabream skeletal cell line. J. Appl. Ichthyol. 2012, 28, 427–432. [Google Scholar] [CrossRef]
- Tiago, D.M.; Laizé, V.; Bargelloni, L.; Ferraresso, S.; Romualdi, C.; Cancela, M.L. Global analysis of gene expression in mineralizing fish vertebra-derived cell lines: New insights into anti-mineralogenic effect of vanadate. BMC Genom. 2011, 12, 310. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef]
- Gavaia, P.J.; Simes, D.C.; Ortiz-Delgado, J.B.; Viegas, C.S.B.; Pinto, J.P.; Kelsh, R.N.; Sarasquete, M.C.; Cancela, M.L. Osteocalcin and matrix Gla protein in zebrafish (Danio rerio) and Senegal sole (Solea senegalensis): Comparative gene and protein expression during larval development through adulthood. Gene Expr. Patterns 2006, 6, 637–652. [Google Scholar] [CrossRef]
- Aceto, J.; Nourizadeh-Lillabadi, R.; Marée, R.; Dardenne, N.; Jeanray, N.; Wehenkel, L.; Aleström, P.; van Loon, J.J.W.A.; Muller, M. Zebrafish bone and general physiology are differently affected by hormones or changes in gravity. PLoS ONE 2015, 10, e0126928. [Google Scholar] [CrossRef]
- Tarasco, M.; Laizé, V.; Cardeira, J.; Cancela, M.L.; Gavaia, P.J. The zebrafish operculum: A powerful system to assess osteogenic bioactivities of molecules with pharmacological and toxicological relevance. Comp. Biochem. Physiol. Part C 2017, 197, 45–52. [Google Scholar] [CrossRef]
- Carson, M.A.; Clarke, S.A. Bioactive compounds from marine organisms: Potential for bone growth and healing. Mar. Drugs 2018, 16, 340. [Google Scholar] [CrossRef]
- Carson, M.A.; Nelson, J.; Cancela, M.L.; Laizé, V.; Gavaia, P.J.; Rae, M.; Heesch, S.; Verzin, E.; Gilmore, B.F.; Clarke, S.A. Screening for osteogenic activity in extracts from Irish marine organisms: The potential of Ceramium pallidum. PLoS ONE 2018, 13, e0207303. [Google Scholar] [CrossRef] [PubMed]
- Carletti, A.; Cardoso, C.; Lobo-Arteaga, J.; Sales, S.; Juliao, D.; Ferreira, I.; Chainho, P.; Dionísio, M.A.; Gaudêncio, M.J.; Afonso, C.; et al. Antioxidant and anti-inflammatory extracts from sea cucumbers and tunicates induce a pro-osteogenic effect in zebrafish larvae. Front. Nutr. 2022, 9, 888360. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Martins, G.; Gavaia, P.J.; Bebianno, M.J.; Cancela, M.L.; Laizé, V. ZEB316: A small stand-alone housing system to study microplastics in small teleosts. Zebrafish 2020, 17, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Cordelières, F.P.; Cancela, M.L.; Laizé, V. ZFBONE: An ImageJ toolset for semi-automatic analysis of zebrafish bone structures. Bone 2020, 138, 115480. [Google Scholar] [CrossRef] [PubMed]
- Mork, L.; Crump, G. Zebrafish craniofacial development: A window into early patterning. In Current Topics in Developmental Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2015; Volume 115, pp. 235–269. ISBN 9780124081413. [Google Scholar]
- Bensimon-Brito, A.; Cardeira, J.; Cancela, M.L.; Huysseune, A.; Witten, P.E. Distinct patterns of notochord mineralization in zebrafish coincide with the localization of Osteocalcin isoform 1 during early vertebral centra formation. BMC Dev. Biol. 2012, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.R.; de Vrieze, E.; Lock, E.-J.; Schulten, I.E.; Flik, G. Elasmoid scales of fishes as model in biomedical bone research. J. Appl. Ichthyol. 2012, 28, 382–387. [Google Scholar] [CrossRef]
- Pasqualetti, S.; Banfi, G.; Mariotti, M. The zebrafish scale as model to study the bone mineralization process. J. Mol. Histol. 2012, 43, 589–595. [Google Scholar] [CrossRef]
- Pasqualetti, S.; Banfi, G.; Mariotti, M. Osteoblast and osteoclast behavior in zebrafish cultured scales. Cell Tissue Res. 2012, 350, 69–75. [Google Scholar] [CrossRef]
- Torvanger, I.; Metz, J.R.; Olsvik, P.A.; Søfteland, L.; Lie, K.K. Benzo(a)pyrene reduces osteoclast and osteoblast activity in ex-vivo scales of zebrafish (Danio rerio (Hamilton-Buchanan, 1822)) and goldfish (Carassius auratus (Linnaeus, 1758)). J. Appl. Ichthyol. 2018, 34, 431–439. [Google Scholar] [CrossRef]
- de Vrieze, E.; Zethof, J.; Schulte-merker, S.; Flik, G.; Metz, J.R. Identification of novel osteogenic compounds by an ex-vivo sp7:luciferase zebrafish scale assay. Bone 2015, 74, 106–113. [Google Scholar] [CrossRef]
- de Vrieze, E.; van Kessel, M.A.H.J.; Peters, H.M.; Spanings, F.A.T.; Flik, G.; Metz, J.R. Prednisolone induces osteoporosis-like phenotype in regenerating zebrafish scales. Osteoporos. Int. 2014, 25, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Sire, J.-Y.; Girondot, M.; Babiar, O. Marking zebrafish, Danio rerio (Cyprinidae), using scale regeneration. J. Exp. Zool. 2000, 286, 297–304. [Google Scholar] [CrossRef]
- de Vrieze, E.; Sharif, F.; Metz, J.R.; Flik, G.; Richardson, M.K. Matrix metalloproteinases in osteoclasts of ontogenetic and regenerating zebrafish scales. Bone 2011, 48, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Cardeira, J.; Gavaia, P.J.; Fernández, I.; Cengiz, I.F.; Moreira-Silva, J.; Oliveira, J.M.; Reis, R.L.; Cancela, M.L.; Laizé, V. Quantitative assessment of the regenerative and mineralogenic performances of the zebrafish caudal fin. Sci. Rep. 2016, 6, 39191. [Google Scholar] [CrossRef]
- Sehring, I.M.; Weidinger, G. Recent advancements in understanding fin regeneration in zebrafish. WIREs Dev. Biol. 2020, 9, e367. [Google Scholar] [CrossRef]
- Pfefferli, C.; Jaźwińska, A. The art of fin regeneration in zebrafish. Regeneration 2015, 2, 72–83. [Google Scholar] [CrossRef]
- Hou, Y.; Lee, H.J.; Chen, Y.; Ge, J.; Osman, F.O.I.; McAdow, A.R.; Mokalled, M.H.; Johnson, S.L.; Zhao, G.; Wang, T. Cellular diversity of the regenerating caudal fin. Sci. Adv. 2020, 6, eaba2084. [Google Scholar] [CrossRef]
- Lebedeva, L.; Zhumabayeva, B.; Gebauer, T.; Kisselev, I.; Aitasheva, Z. Zebrafish (Danio rerio) as a model for understanding the process of caudal fin regeneration. Zebrafish 2020, 17, 359–372. [Google Scholar] [CrossRef]
- Iwasaki, M.; Kuroda, J.; Kawakami, K.; Wada, H. Epidermal regulation of bone morphogenesis through the development and regeneration of osteoblasts in the zebrafish scale. Dev. Biol. 2018, 437, 105–119. [Google Scholar] [CrossRef]
- Bergen, D.J.M.; Tong, Q.; Shukla, A.; Newham, E.; Zethof, J.; Lundberg, M.; Ryan, R.; Youlten, S.E.; Frysz, M.; Croucher, P.I.; et al. Regenerating zebrafish scales express a subset of evolutionary conserved genes involved in human skeletal disease. BMC Biol. 2022, 20, 21. [Google Scholar] [CrossRef]
- Kinkel, M.D.; Eames, S.C.; Philipson, L.H.; Prince, V.E. Intraperitoneal injection into adult zebrafish. J. Vis. Exp. 2010, e2126. [Google Scholar] [CrossRef] [PubMed]
- Monstad-Rios, A.T.; Watson, C.J.; Kwon, R.Y. ScreenCube: A 3D printed system for rapid and cost-effective chemical screening in adult zebrafish. Zebrafish 2018, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Akimenko, M.A.; Marí-Beffa, M.; Becerra, J.; Géraudie, J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 2003, 226, 190–201. [Google Scholar] [CrossRef]
- Poss, K.D.; Keating, M.T.; Nechiporuk, A. Tales of regeneration in zebrafish. Dev. Dyn. 2003, 226, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Cardeira, J.; Viegas, M.N.; Caria, J.; Martins, G.; Gavaia, P.J.; Cancela, M.L.; Laizé, V. Anti-osteogenic activity of cadmium in zebrafish. Fishes 2019, 4, 11. [Google Scholar] [CrossRef]
- Sousa, S.; Valerio, F.; Jacinto, A. A new zebrafish bone crush injury model. Biol. Open 2012, 1, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Tomecka, M.J.; Ethiraj, L.P.; Sánchez, L.M.; Roehl, H.H.; Carney, T.J. Clinical pathologies of bone fracture modelled in zebrafish. Dis. Model. Mech. 2019, 12, dmm037630. [Google Scholar] [CrossRef]
- Geurtzen, K.; Knopf, F.; Wehner, D.; Huitema, L.F.A.; Schulte-Merker, S.; Weidinger, G. Mature osteoblasts dedifferentiate in response to traumatic bone injury in the zebrafish fin and skull. Development 2014, 141, 2225–2234. [Google Scholar] [CrossRef]
- Ohgo, S.; Ichinose, S.; Yokota, H.; Sato-Maeda, M.; Shoji, W.; Wada, N. Tissue regeneration during lower jaw restoration in zebrafish shows some features of epimorphic regeneration. Dev. Growth Differ. 2019, 61, 419–430. [Google Scholar] [CrossRef]
- Witten, P.E.; Harris, M.P.; Huysseune, A.; Winkler, C. Small teleost fish provide new insights into human skeletal diseases. In Methods in Cell Biology; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; Volume 138, pp. 321–346. [Google Scholar]
- Carnovali, M.; Banfi, G.; Mariotti, M. Zebrafish models of human skeletal disorders: Embryo and adult swimming together. BioMed Res. Int. 2019, 2019, 1253710. [Google Scholar] [CrossRef]
- Valenti, M.T.; Marchetto, G.; Mottes, M.; Dalle Carbonare, L. Zebrafish: A suitable tool for the study of cell signaling in bone. Cells 2020, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Mullins, M.C.; Hammerschmidt, M.; Haffter, P.; Nüsslein-Volhard, C. Large-scale mutagenesis in the zebrafish: In search of genes controlling development in a vertebrate. Curr. Biol. 1994, 4, 189–202. [Google Scholar] [CrossRef]
- Fisher, S.; Jagadeeswaran, P.; Halpern, M.E. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 2003, 264, 64–76. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Liu, B.; Du, H.; Zhao, S.; Li, Y.; Cheng, X.; Wang, S.; Lin, J.; Zhou, J.; Qiu, G.; et al. The progress of CRISPR/Cas9-mediated gene editing in generating mouse/zebrafish models of human skeletal diseases. Comput. Struct. Biotechnol. J. 2019, 17, 954–962. [Google Scholar] [CrossRef]
- Kemp, J.P.; Morris, J.A.; Medina-Gomez, C.; Forgetta, V.; Warrington, N.M.; Youlten, S.E.; Zheng, J.; Gregson, C.L.; Grundberg, E.; Trajanoska, K.; et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017, 49, 1468–1475. [Google Scholar] [CrossRef]
- Gregson, C.L.; Newell, F.; Leo, P.J.; Clark, G.R.; Paternoster, L.; Marshall, M.; Forgetta, V.; Morris, J.A.; Ge, B.; Bao, X.; et al. Genome-wide association study of extreme high bone mass: Contribution of common genetic variation to extreme BMD phenotypes and potential novel BMD-associated genes. Bone 2018, 114, 62–71. [Google Scholar] [CrossRef]
- Kim, S.K. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE 2018, 13, e0200785. [Google Scholar] [CrossRef]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
- Meyer, A.; Van De Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [CrossRef] [PubMed]
- Bergen, D.J.M.; Kague, E.; Hammond, C.L. Zebrafish as an emerging model for osteoporosis: A primary testing platform for screening new osteo-active compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef]
- Luderman, L.N.; Unlu, G.; Knapik, E.W. Zebrafish developmental models of skeletal diseases. In Current Topics in Developmental Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 124, pp. 81–124. ISBN 0070-2153. [Google Scholar]
- Marí-Beffa, M.; Mesa-Román, A.B.; Duran, I. Zebrafish models for human skeletal disorders. Front. Genet. 2021, 12, 675331. [Google Scholar] [CrossRef]
- Dietrich, K.; Fiedler, I.A.K.; Kurzyukova, A.; López-Delgado, A.C.; McGowan, L.M.; Geurtzen, K.; Hammond, C.L.; Busse, B.; Knopf, F. Skeletal biology and disease modeling in zebrafish. J. Bone Miner. Res. 2021, 36, 436–458. [Google Scholar] [CrossRef] [PubMed]
- Mortier, G.R.; Cohn, D.H.; Cormier-Daire, V.; Hall, C.; Krakow, D.; Mundlos, S.; Nishimura, G.; Robertson, S.; Sangiorgi, L.; Savarirayan, R.; et al. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. Part A 2019, 179, 2393–2419. [Google Scholar] [CrossRef] [PubMed]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Antinucci, P.; Hindges, R. A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 2016, 6, 29490. [Google Scholar] [CrossRef]
- Guryev, V.; Koudijs, M.J.; Berezikov, E.; Johnson, S.L.; Plasterk, R.H.A.; Van Eeden, F.J.M.; Cuppen, E. Genetic variation in the zebrafish. Genome Res. 2006, 16, 491–497. [Google Scholar] [CrossRef]
- Lawrence, C.; Mason, T. Zebrafish housing systems: A review of basic operating principles and considerations for design and functionality. ILAR J. 2012, 53, 179–191. [Google Scholar] [CrossRef]
- Lawrence, C. The husbandry of zebrafish (Danio rerio): A review. Aquaculture 2007, 269, 1–20. [Google Scholar] [CrossRef]
- Ribas, L.; Valdivieso, A.; Díaz, N.; Piferrer, F. Appropriate rearing density in domesticated zebrafish to avoid masculinization: Links with the stress response. J. Exp. Biol. 2017, 220, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.; Jennings, M. Guidance on the Housing and Care of Zebrafish Danio rerio; eBook; Royal Society for the Prevention of Cruelty to Animals (RSPCA): Horsham, UK, 2011. [Google Scholar]
- Lawrence, C. New frontiers for zebrafish management. Methods Cell Biol. 2016, 135, 483–508. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Lee, C.J.; Paull, G.C.; Tyler, C.R. Improving zebrafish laboratory welfare and scientific research through understanding their natural history. Biol. Rev. 2022, 97, 1038–1056. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, L.; de Girolamo, P. Laboratory Fish in Biomedical Research; Academic Press: Cambridge, MA, USA, 2021; ISBN 9780128212455. [Google Scholar]
- Barnard, D.E.; Lewis, S.M.; Teter, B.B.; Thigpen, J.E. Open- and closed-formula laboratory animal diets and their importance to research. J. Am. Assoc. Lab. Anim. Sci. 2009, 48, 709–713. [Google Scholar] [PubMed]
- Ulloa, P.E.; Medrano, J.F.; Feijoo, C.G. Zebrafish as animal model for aquaculture nutrition research. Front. Genet. 2014, 5, 313. [Google Scholar] [CrossRef]
- Carvalho, A.P.; Araújo, L.; Santos, M.M. Rearing zebrafish (Danio rerio) larvae without live food: Evaluation of a commercial, a practical and a purified starter diet on larval performance. Aquac. Res. 2006, 37, 1107–1111. [Google Scholar] [CrossRef]
- Lawrence, C.; James, A.; Mobley, S. Successful replacement of Artemia salina nauplii with marine rotifers (Brachionus plicatilis) in the diet of preadult zebrafish (Danio rerio). Zebrafish 2015, 12, 366–371. [Google Scholar] [CrossRef]
- Lawrence, C.; Best, J.; James, A.; Maloney, K. The effects of feeding frequency on growth and reproduction in zebrafish (Danio rerio). Aquaculture 2012, 368–369, 103–108. [Google Scholar] [CrossRef]
- Monteiro, J.F.; Martins, S.; Farias, M.; Costa, T.; Certal, A.C. The impact of two different cold-extruded feeds and feeding regimens on zebrafish survival, growth and reproductive performance. J. Dev. Biol. 2018, 6, 15. [Google Scholar] [CrossRef]
- Best, J.; Adatto, I.; Cockington, J.; James, A.; Lawrence, C. A novel method for rearing first-feeding larval zebrafish: Polyculture with type L saltwater rotifers (Brachionus plicatilis). Zebrafish 2010, 7, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.; Diogo, P.; Pinto, W.; Gavaia, P.J. Early transition to microdiets improves growth, reproductive performance and reduces skeletal anomalies in zebrafish (Danio rerio). Zebrafish 2019, 16, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Diogo, P.; Martins, G.; Gavaia, P.; Pinto, W.; Dias, J.; Cancela, L.; Martínez-Páramo, S. Assessment of nutritional supplementation in phospholipids on the reproductive performance of zebrafish, Danio rerio (Hamilton, 1822). J. Appl. Ichthyol. 2015, 31, 3–9. [Google Scholar] [CrossRef]
- Graf, S.F.; Hötzel, S.; Liebel, U.; Stemmer, A.; Knapp, H.F. Image-based fluidic sorting system for automated zebrafish egg sorting into multiwell plates. J. Lab. Autom. 2011, 16, 105–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- White, D.T.; Eroglu, A.U.; Wang, G.; Zhang, L.; Sengupta, S.; Ding, D.; Rajpurohit, S.K.; Walker, S.L.; Ji, H.; Qian, J.; et al. ARQiv-HTS, a versatile whole-organism screening platform enabling in vivo drug discovery at high-throughput rates. Nat. Protoc. 2016, 11, 2432–2453. [Google Scholar] [CrossRef]
- Reischl, M.; Bartschat, A.; Liebel, U.; Gehrig, J.; Müller, F.; Mikut, R. ZebrafishMiner: An open source software for interactive evaluation of domain-specific fluorescence in zebrafish. Curr. Dir. Biomed. Eng. 2017, 3, 199–202. [Google Scholar] [CrossRef]
- Liu, T.; Nie, J.; Li, G.; Guo, L.; Wong, S.T.C. ZFIQ: A software package for zebrafish biology. Bioinformatics 2008, 24, 438–439. [Google Scholar] [CrossRef][Green Version]
- Adatto, I.; Lawrence, C.; Thompson, M.; Zon, L.I. A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS ONE 2011, 6, e21715. [Google Scholar] [CrossRef]
- Mandrell, D.; Truong, L.; Jephson, C.; Sarker, M.R.; Moore, A.; Lang, C.; Simonich, M.T.; Tanguay, R.L. Automated zebrafish chorion removal and single embryo placement: Optimizing throughput of zebrafish developmental toxicity screens. J. Lab. Autom. 2012, 17, 66–74. [Google Scholar] [CrossRef]
- Letamendia, A.; Quevedo, C.; Ibarbia, I.; Virto, J.M.; Holgado, O.; Diez, M.; Izpisua Belmonte, J.C.; Callol-Massot, C. Development and validation of an automated high-throughput system for zebrafish in vivo screenings. PLoS ONE 2012, 7, e36690. [Google Scholar] [CrossRef]
- Cubbage, C.C.; Mabee, P.M. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J. Morphol. 1996, 229, 121–160. [Google Scholar] [CrossRef]
- Spaink, H.P.; Cui, C.; Wiweger, M.I.; Jansen, H.J.; Veneman, W.J.; Marín-Juez, R.; De Sonneville, J.; Ordas, A.; Torraca, V.; van der Ent, W.; et al. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 2013, 62, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, X.; Gelinas, D.; Ciruna, B.; Sun, Y. A fully automated robotic system for microinjection of zebrafish embryos. PLoS ONE 2007, 2, e862. [Google Scholar] [CrossRef]
- Chi, Z.; Xu, Q.; Ai, N.; Ge, W. Design and development of an automatic microinjection system for high-throughput injection of zebrafish larvae. IEEE Robot. Autom. Lett. 2022, 7, 1848–1855. [Google Scholar] [CrossRef]
- Early, J.J.; Cole, K.L.H.; Williamson, J.M.; Swire, M.; Kamadurai, H.; Muskavitch, M.; Lyons, D.A. An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. eLife 2018, 7, e35136. [Google Scholar] [CrossRef] [PubMed]
- Høgset, H.; Horgan, C.C.; Armstrong, J.P.K.; Bergholt, M.S.; Torraca, V.; Chen, Q.; Keane, T.J.; Bugeon, L.; Dallman, M.J.; Mostowy, S.; et al. In vivo biomolecular imaging of zebrafish embryos using confocal Raman spectroscopy. Nat. Commun. 2020, 11, 6172. [Google Scholar] [CrossRef]
- Logan, S.L.; Dudley, C.; Baker, R.P.; Taormina, M.J.; Hay, E.A.; Parthasarathy, R. Automated high-throughput light-sheet fluorescence microscopy of larval zebrafish. PLoS ONE 2018, 13, e0198705. [Google Scholar] [CrossRef]
- Hur, M.; Gistelinck, C.A.; Huber, P.; Lee, J.; Thompson, M.H.; Monstad-Rios, A.T.; Watson, C.J.; McMenamin, S.K.; Willaert, A.; Parichy, D.M.; et al. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. eLife 2017, 6, e26014. [Google Scholar] [CrossRef]
- Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F.A.; Kausler, B.X.; Ledesma-Carbayo, M.J.; Marée, R.; Mikula, K.; Pantazis, P.; et al. Automated processing of zebrafish imaging data: A survey. Zebrafish 2013, 10, 401–421. [Google Scholar] [CrossRef]
- Fernández, I.; Gavaia, P.J.; Laizé, V.; Cancela, M.L. Fish as a model to assess chemical toxicity in bone. Aquat. Toxicol. 2018, 194, 208–226. [Google Scholar] [CrossRef]
| Parameters | Description | |
|---|---|---|
| Filters | Mechanical | Filter pads; cleaned daily and changed monthly |
| Chemical | Activated charcoal; changed every 6 months | |
| Biological | Bio-balls or ceramic rings hosting nitrifying bacteria (Nitrosomonas and Nitrobacter) | |
| Germicidal light | Ultraviolet light at 254 nm; bulbs changed after 6000 h of use | |
| Temperature | 24–29 °C (ideally 28.5 ± 0.5 °C) | |
| Photoperiod | 14 h of light|10 h of dark (automated light system to be checked regularly) | |
| Water | Type | Dechlorinated water (ideally filtered reverse osmosis water) |
| pH | 6.5–8.0 adjusted with sodium bicarbonate | |
| Conductivity | 150 to 1700 µS adjusted with commercial salts | |
| Hardness | 3–8 d (ideally 4–5 d) | |
| Ammonia | < 0.1 mg/L (as close to 0 mg/L as possible) | |
| Nitrites | < 0.3 mg/L (as close to 0 mg/L as possible) | |
| Nitrates | <25 mg/L | |
| Renewal | 5–10% in a daily basis (occasionally up to 15%) | |
| Fish density | 5 adults/L, 25 juveniles/L and 100 larvae/L | |
| Tool (Company) | ZF Standardized Production | ZF Mass Production | ZF Sorting | Compound Handling | ZF Exposure | ZF Handling | Signal Acquisition | Imaging | Data Analysis | URL/Reference * |
|---|---|---|---|---|---|---|---|---|---|---|
| ZEBRAFEED (Sparos Lda.) | X | www.sparos.pt | ||||||||
| GemmaMicro (Skretting) | X | www.skretting.com | ||||||||
| MEPS—Mass embryo production systems (Aquatic Habitats) | X | www.mbki.com | ||||||||
| iSPAWN (Tecniplast) | X | www.tecniplast.it | ||||||||
| COPAS FP-1000/2000 (Union Biometrica) | X | www.unionbio.com | ||||||||
| ZebraFactor (Swiss Center for Electronics and Microtechnology) | X | [113] | ||||||||
| Dispensing/sorting robot for small aquatic organisms | X | X | www.lifesciencemethods.com | |||||||
| ARQiv—Automated reporter quantification system in vivo | X | X | X | X | [114] | |||||
| ScreenCube | X | [66] | ||||||||
| Microinjection robot | X | www.lifesciencemethods.com | ||||||||
| VAST BioImager (Union Biometrica) | X | X | X | X | www.unionbio.com | |||||
| Imaging robot for small aquatic organisms | X | X | X | www.lifesciencemethods.com | ||||||
| HCS LCI (Leica) | X | X | X | www.leica-microsystems.com | ||||||
| Imaging Machine (Acquifer) | X | X | X | www.acquifer.de | ||||||
| ImageXpress (Molecular Devices) | X | X | X | www.moleculardevices.com | ||||||
| EnSight multimode plate reader (PerkinElmer) | X | X | X | www.perkinelmer.com | ||||||
| IN Cell Analyzer (GE Healthcare) | X | X | X | www.gehealthcare.com | ||||||
| COPAS Vision (Union Biometrica) | X | X | X | X | www.unionbio.com | |||||
| Micro computed tomography (Brucker) | X | X | www.bruker.com | |||||||
| ZebrafishMiner | X | [115] | ||||||||
| ZFIQ zebrafish image quantitator | X | [116] | ||||||||
| ZFBONE toolset | X | [47] | ||||||||
| ImageJ | X | imagej.nih.gov | ||||||||
| Image-Pro (Media Cybernetics) | X | www.mediacy.com |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, J.T.; Tarasco, M.; Gavaia, P.J.; Cancela, M.L.; Laizé, V. Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy. Pharmaceuticals 2022, 15, 983. https://doi.org/10.3390/ph15080983
Rosa JT, Tarasco M, Gavaia PJ, Cancela ML, Laizé V. Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy. Pharmaceuticals. 2022; 15(8):983. https://doi.org/10.3390/ph15080983
Chicago/Turabian StyleRosa, Joana T., Marco Tarasco, Paulo J. Gavaia, M. Leonor Cancela, and Vincent Laizé. 2022. "Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy" Pharmaceuticals 15, no. 8: 983. https://doi.org/10.3390/ph15080983
APA StyleRosa, J. T., Tarasco, M., Gavaia, P. J., Cancela, M. L., & Laizé, V. (2022). Screening of Mineralogenic and Osteogenic Compounds in Zebrafish—Tools to Improve Assay Throughput and Data Accuracy. Pharmaceuticals, 15(8), 983. https://doi.org/10.3390/ph15080983








