Cardio- and Neurotoxicity of Selected Anti-COVID-19 Drugs
Abstract
:1. Introduction
2. Results
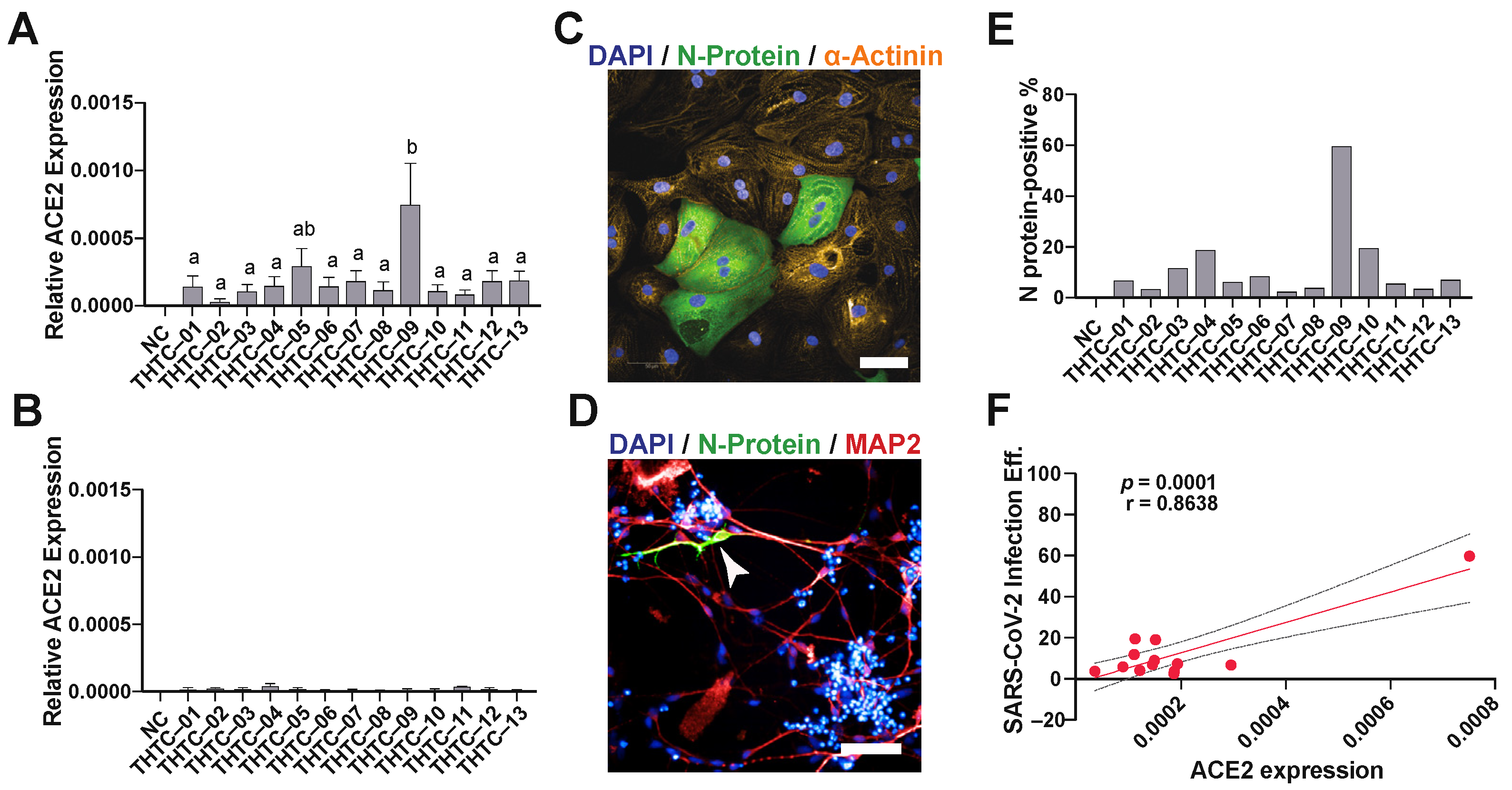
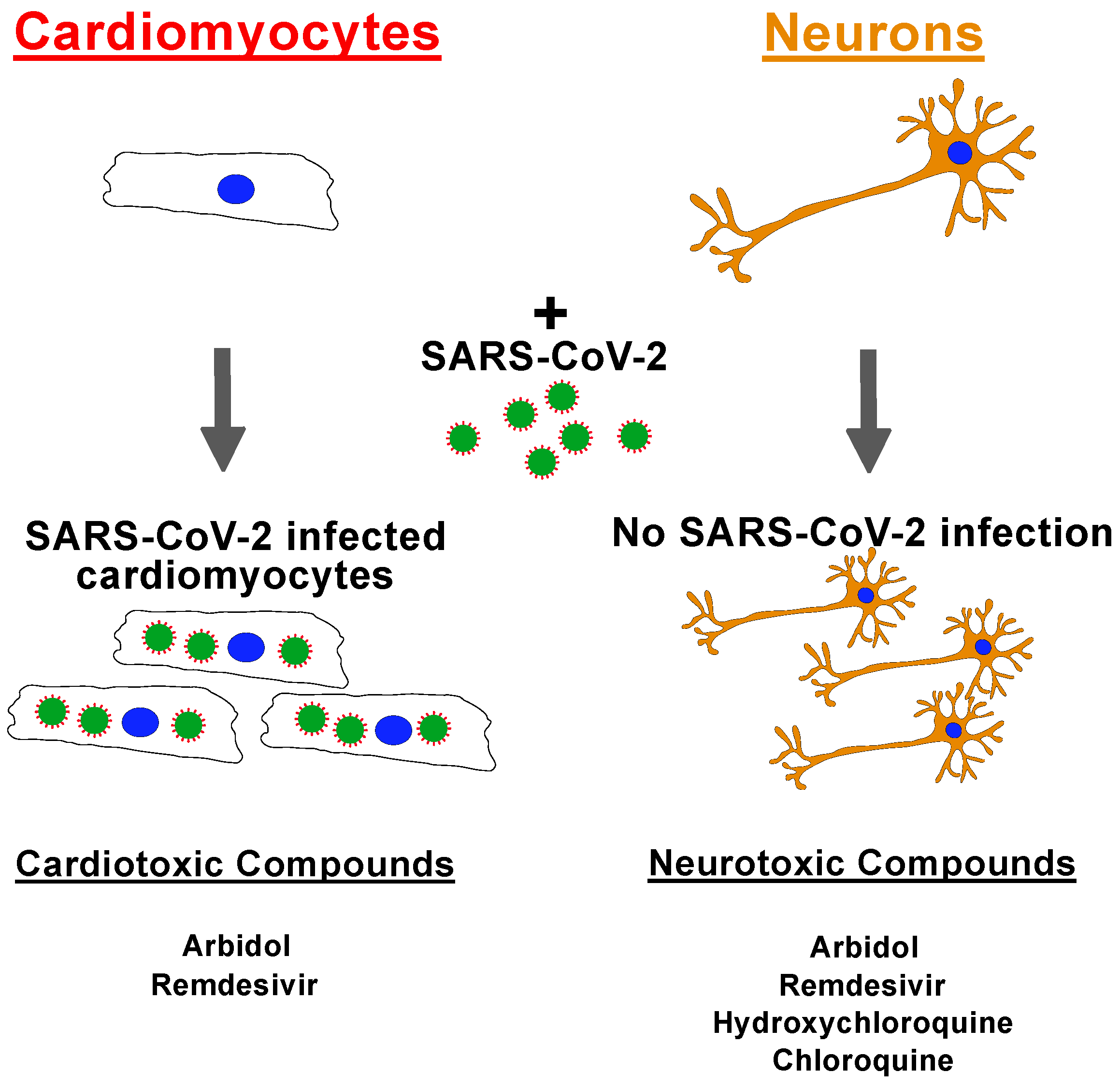
2.1. SARS-CoV-2 Virus Infection of iPSC-Derived Cardiomyocytes and Neurons
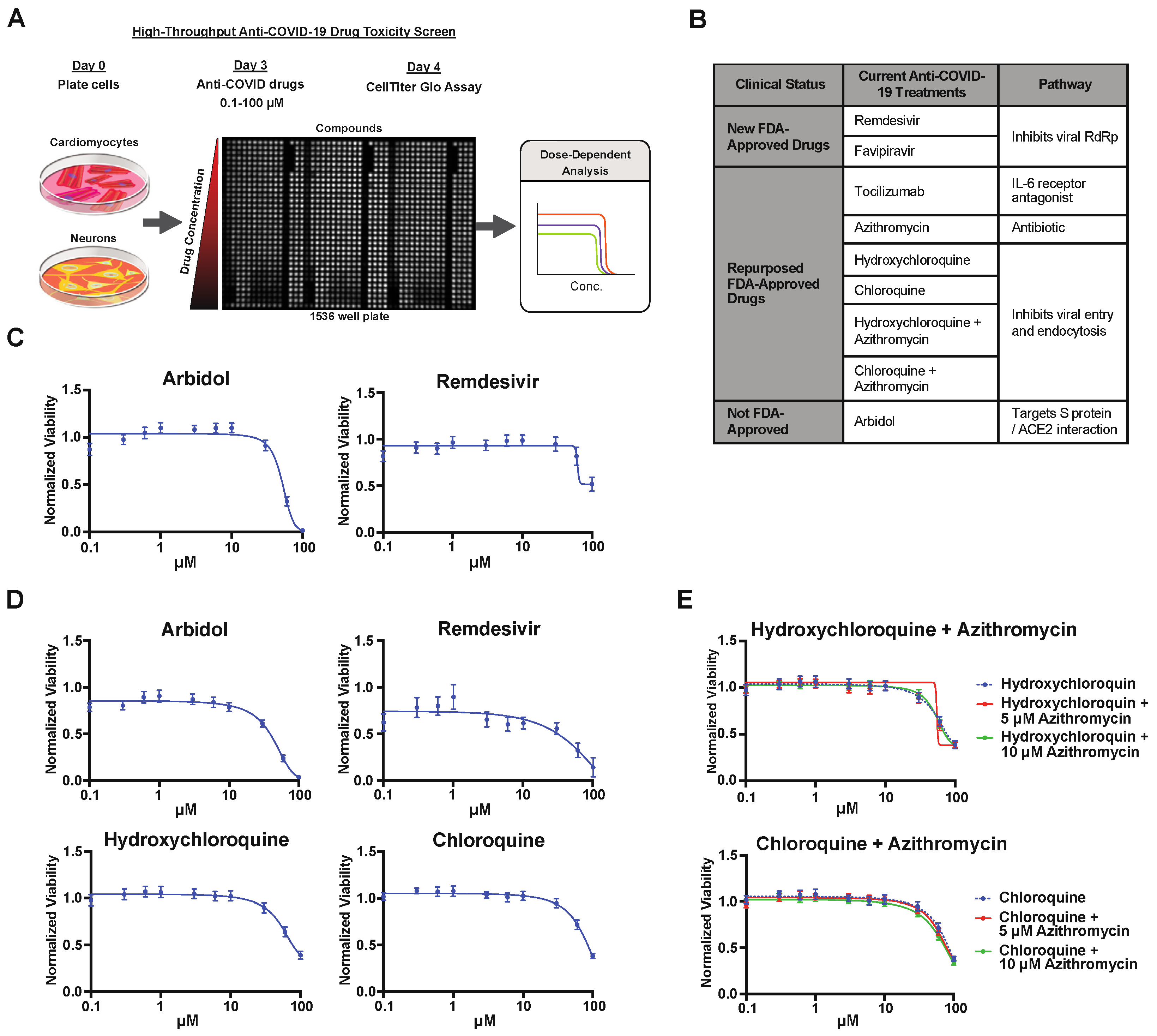
2.2. Drug Screening of Anti-COVID-19 Compounds
3. Discussion
Limitations of Study
4. Materials and Methods
4.1. Immunocytochemistry
4.2. Human iPSCs
4.3. Cardiac Differentiation
4.4. Neuronal Differentiation
4.5. ACE2 Expression
4.6. SARS-CoV-2 Infection
4.7. Toxicity Screening of Anti-COVID-19 Drugs
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehraeen, E.; Behnezhad, F.; Salehi, M.A.; Noori, T.; Harandi, H.; SeyedAlinaghi, S. Olfactory and gustatory dysfunctions due to the coronavirus disease (COVID-19): A review of current evidence. Eur. Arch. Otorhinolaryngol. 2021, 278, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liao, X.; Qian, S.; Yuan, J.; Wang, F.; Liu, Y.; Wang, Z.; Wang, F.S.; Liu, L.; Zhang, Z. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerg. Infect. Dis. 2020, 26, 1320–1323. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gong, X.; Wang, Z.; Chen, R.; Li, T.; Zeng, D.; Li, M. Clinical features of familial clustering in patients infected with 2019 novel coronavirus in Wuhan, China. Virus Res. 2020, 286, 198043. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Nicin, L.; Abplanalp, W.T.; Mellentin, H.; Kattih, B.; Tombor, L.; John, D.; Schmitto, J.D.; Heineke, J.; Emrich, F.; Arsalan, M.; et al. Cell type-specific expression of the putative SARS-CoV-2 receptor ACE2 in human hearts. Eur. Heart J. 2020, 41, 1804–1806. [Google Scholar] [CrossRef] [Green Version]
- Rice, G.I.; Thomas, D.A.; Grant, P.J.; Turner, A.J.; Hooper, N.M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 2004, 383, 45–51. [Google Scholar] [CrossRef]
- Xiao, L.; Haack, K.K.; Zucker, I.H. Angiotensin II regulates ACE and ACE2 in neurons through p38 mitogen-activated protein kinase and extracellular signal-regulated kinase 1/2 signaling. Am. J. Physiol. Cell Physiol. 2013, 304, C1073–C1079. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Fan, Y.; Chen, M.; Wu, X.; Zhang, L.; He, T.; Wang, H.; Wan, J.; Wang, X.; Lu, Z. Cardiovascular Implications of Fatal Outcomes of Patients With Coronavirus Disease 2019 (COVID-19). JAMA Cardiol. 2020, 5, 811–818. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Qin, M.; Shen, B.; Cai, Y.; Liu, T.; Yang, F.; Gong, W.; Liu, X.; Liang, J.; Zhao, Q.; et al. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiol. 2020, 5, 802–810. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xu, X.; Chen, Z.; Duan, J.; Hashimoto, K.; Yang, L.; Liu, C.; Yang, C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020, 87, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Ramani, S.; Berard, J.A.; Walker, L.A.S. The relationship between neurofilament light chain and cognition in neurological disorders: A scoping review. J. Neurol. Sci. 2021, 420, 117229. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Butowt, R.; von Bartheld, C.S. Anosmia in COVID-19: Underlying Mechanisms and Assessment of an Olfactory Route to Brain Infection. Neuroscientist 2021, 27, 582–603. [Google Scholar] [CrossRef]
- Yanagida, S.; Satsuka, A.; Hayashi, S.; Ono, A.; Kanda, Y. Comprehensive Cardiotoxicity Assessment of COVID-19 Treatments Using Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Toxicol. Sci. 2021, 183, 227–239. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Wong, A.Y.; MacKenna, B.; Morton, C.E.; Schultze, A.; Walker, A.J.; Bhaskaran, K.; Brown, J.P.; Rentsch, C.T.; Williamson, E.; Drysdale, H.; et al. Use of non-steroidal anti-inflammatory drugs and risk of death from COVID-19: An OpenSAFELY cohort analysis based on two cohorts. Ann. Rheum. Dis. 2021, 80, 943–951. [Google Scholar] [CrossRef]
- Kalra, R.S.; Tomar, D.; Meena, A.S.; Kandimalla, R. SARS-CoV-2, ACE2, and Hydroxychloroquine: Cardiovascular Complications, Therapeutics, and Clinical Readouts in the Current Settings. Pathogens 2020, 9, 546. [Google Scholar] [CrossRef]
- Huang, C.Y.; Nicholson, M.W.; Wang, J.Y.; Ting, C.Y.; Tsai, M.H.; Cheng, Y.C.; Liu, C.L.; Chan, D.Z.H.; Lee, Y.C.; Hsu, C.C.; et al. Population-based high-throughput toxicity screen of human iPSC-derived cardiomyocytes and neurons. Cell Rep. 2022, 39, 110643. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kryshtal, D.O.; Feaster, T.K.; Sanchez-Freire, V.; Zhang, J.; Kamp, T.J.; Hong, C.C.; Wu, J.C.; Knollmann, B.C. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. J. Mol. Cell Cardiol. 2015, 85, 79–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrini, L.; Albecka, A.; Mallery, D.L.; Kellner, M.J.; Paul, D.; Carter, A.P.; James, L.C.; Lancaster, M.A. SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27, 951–961.e955. [Google Scholar] [CrossRef] [PubMed]
- Simoneau, C.R.; Ott, M. Modeling Multi-organ Infection by SARS-CoV-2 Using Stem Cell Technology. Cell Stem Cell 2020, 27, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffre, F.; et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef]
- Kim, K.; Johnson, J.A.; Derendorf, H. Differences in drug pharmacokinetics between East Asians and Caucasians and the role of genetic polymorphisms. J. Clin. Pharmacol. 2004, 44, 1083–1105. [Google Scholar] [CrossRef]
- Lin, K.M.; Lau, J.K.; Smith, R.; Phillips, P.; Antal, E.; Poland, R.E. Comparison of alprazolam plasma levels in normal Asian and Caucasian male volunteers. Psychopharmacology 1988, 96, 365–369. [Google Scholar] [CrossRef]
- Dang, M.-T.N.; Hambleton, J.; Kayser, S.R. The Influence of Ethnicity on Warfarin Dosage Requirement. Ann. Pharmacother. 2005, 39, 1008–1012. [Google Scholar] [CrossRef]
- Liao, J.K. Safety and efficacy of statins in Asians. Am. J. Cardiol. 2007, 99, 410–414. [Google Scholar] [CrossRef] [Green Version]
- Mercuro, N.J.; Yen, C.F.; Shim, D.J.; Maher, T.R.; McCoy, C.M.; Zimetbaum, P.J.; Gold, H.S. Risk of QT Interval Prolongation Associated With Use of Hydroxychloroquine With or Without Concomitant Azithromycin Among Hospitalized Patients Testing Positive for Coronavirus Disease 2019 (COVID-19). JAMA Cardiol. 2020, 5, 1036–1041. [Google Scholar] [CrossRef]
- Zhou, X.; Hou, H.; Yang, L.; Ding, G.; Wei, T.; Li, C.; Heng, Y.; Liu, R.; Ma, M.; Hu, Z.; et al. Arbidol is associated with increased in-hospital mortality among 109 patients with severe COVID-19: A multicenter, retrospective study. J. Glob. Health 2021, 11, 05017. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; He, X.; Qiu, F.; Zhu, X.; Zhao, M.; Li-Ling, J.; Su, X.; Zhao, L. Pharmacokinetics of single and multiple oral doses of arbidol in healthy Chinese volunteers. Int. J. Clin. Pharmacol. Ther. 2013, 51, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Nabati, M.; Parsaee, H. Potential Cardiotoxic Effects of Remdesivir on Cardiovascular System: A Literature Review. Cardiovasc. Toxicol. 2022, 22, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Humeniuk, R.; Mathias, A.; Kirby, B.J.; Lutz, J.D.; Cao, H.; Osinusi, A.; Babusis, D.; Porter, D.; Wei, X.; Ling, J.; et al. Pharmacokinetic, Pharmacodynamic, and Drug-Interaction Profile of Remdesivir, a SARS-CoV-2 Replication Inhibitor. Clin. Pharmacokinet. 2021, 60, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Perez-Bermejo, J.A.; Kang, S.; Rockwood, S.J.; Simoneau, C.R.; Joy, D.A.; Silva, A.C.; Ramadoss, G.N.; Flanigan, W.R.; Fozouni, P.; Li, H.; et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci. Transl. Med. 2021, 13. [Google Scholar] [CrossRef]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950. [Google Scholar] [CrossRef]
- Eftekhar, S.P.; Kazemi, S.; Barary, M.; Javanian, M.; Ebrahimpour, S.; Ziaei, N. Effect of Hydroxychloroquine and Azithromycin on QT Interval Prolongation and Other Cardiac Arrhythmias in COVID-19 Confirmed Patients. Cardiovasc. Ther. 2021, 2021, 6683098. [Google Scholar] [CrossRef]
- Varatharaj, A.; Thomas, N.; Ellul, M.A.; Davies, N.W.S.; Pollak, T.A.; Tenorio, E.L.; Sultan, M.; Easton, A.; Breen, G.; Zandi, M.; et al. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: A UK-wide surveillance study. Lancet Psychiatry 2020, 7, 875–882. [Google Scholar] [CrossRef]
- Isacson, O. The Consequences of Coronavirus-Induced Cytokine Storm Are Associated With Neurological Diseases, Which May Be Preventable. Front. Neurol. 2020, 11, 745. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Q.; Xia, X.; Liu, K.; Yu, Z.; Tao, W.; Gong, W.; Han, J.J. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell 2020, 19. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, e00510-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zhang, W.; Zhang, J.; He, J.; Zhu, F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19). Hla 2020, 96, 194–196. [Google Scholar] [CrossRef] [PubMed]
- Pisanti, S.; Deelen, J.; Gallina, A.M.; Caputo, M.; Citro, M.; Abate, M.; Sacchi, N.; Vecchione, C.; Martinelli, R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J. Translational Med. 2020, 18, 352. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Li, L.H.; Hsu, W.T.; Cheng, Y.C.; Nicholson, M.W.; Liu, C.L.; Ting, C.Y.; Ko, H.W.; Syu, S.H.; Wen, C.H.; et al. Copy number variant hotspots in Han Taiwanese population induced pluripotent stem cell lines—Lessons from establishing the Taiwan human disease iPSC Consortium Bank. J. Biomed. Sci. 2020, 27, 92. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicholson, M.W.; Huang, C.-Y.; Wang, J.-Y.; Ting, C.-Y.; Cheng, Y.-C.; Chan, D.Z.H.; Lee, Y.-C.; Hsu, C.-C.; Hsu, Y.-H.; Chang, C.M.C.; et al. Cardio- and Neurotoxicity of Selected Anti-COVID-19 Drugs. Pharmaceuticals 2022, 15, 765. https://doi.org/10.3390/ph15060765
Nicholson MW, Huang C-Y, Wang J-Y, Ting C-Y, Cheng Y-C, Chan DZH, Lee Y-C, Hsu C-C, Hsu Y-H, Chang CMC, et al. Cardio- and Neurotoxicity of Selected Anti-COVID-19 Drugs. Pharmaceuticals. 2022; 15(6):765. https://doi.org/10.3390/ph15060765
Chicago/Turabian StyleNicholson, Martin W., Ching-Ying Huang, Jyun-Yuan Wang, Chien-Yu Ting, Yu-Che Cheng, Darien Z. H. Chan, Yi-Chan Lee, Ching-Chuan Hsu, Yu-Hung Hsu, Cindy M. C. Chang, and et al. 2022. "Cardio- and Neurotoxicity of Selected Anti-COVID-19 Drugs" Pharmaceuticals 15, no. 6: 765. https://doi.org/10.3390/ph15060765
APA StyleNicholson, M. W., Huang, C.-Y., Wang, J.-Y., Ting, C.-Y., Cheng, Y.-C., Chan, D. Z. H., Lee, Y.-C., Hsu, C.-C., Hsu, Y.-H., Chang, C. M. C., Hsieh, M. L., Cheng, Y.-Y., Lin, Y.-L., Chen, C.-H., Wu, Y.-T., Hacker, T. A., Wu, J. C., Kamp, T. J., & Hsieh, P. C. H. (2022). Cardio- and Neurotoxicity of Selected Anti-COVID-19 Drugs. Pharmaceuticals, 15(6), 765. https://doi.org/10.3390/ph15060765





