Design, Synthesis, In Vitro Biological Activity Evaluation and Stabilized Nanostructured Lipid Carrier Formulation of Newly Synthesized Schiff Bases-Based TMP Moieties
Abstract
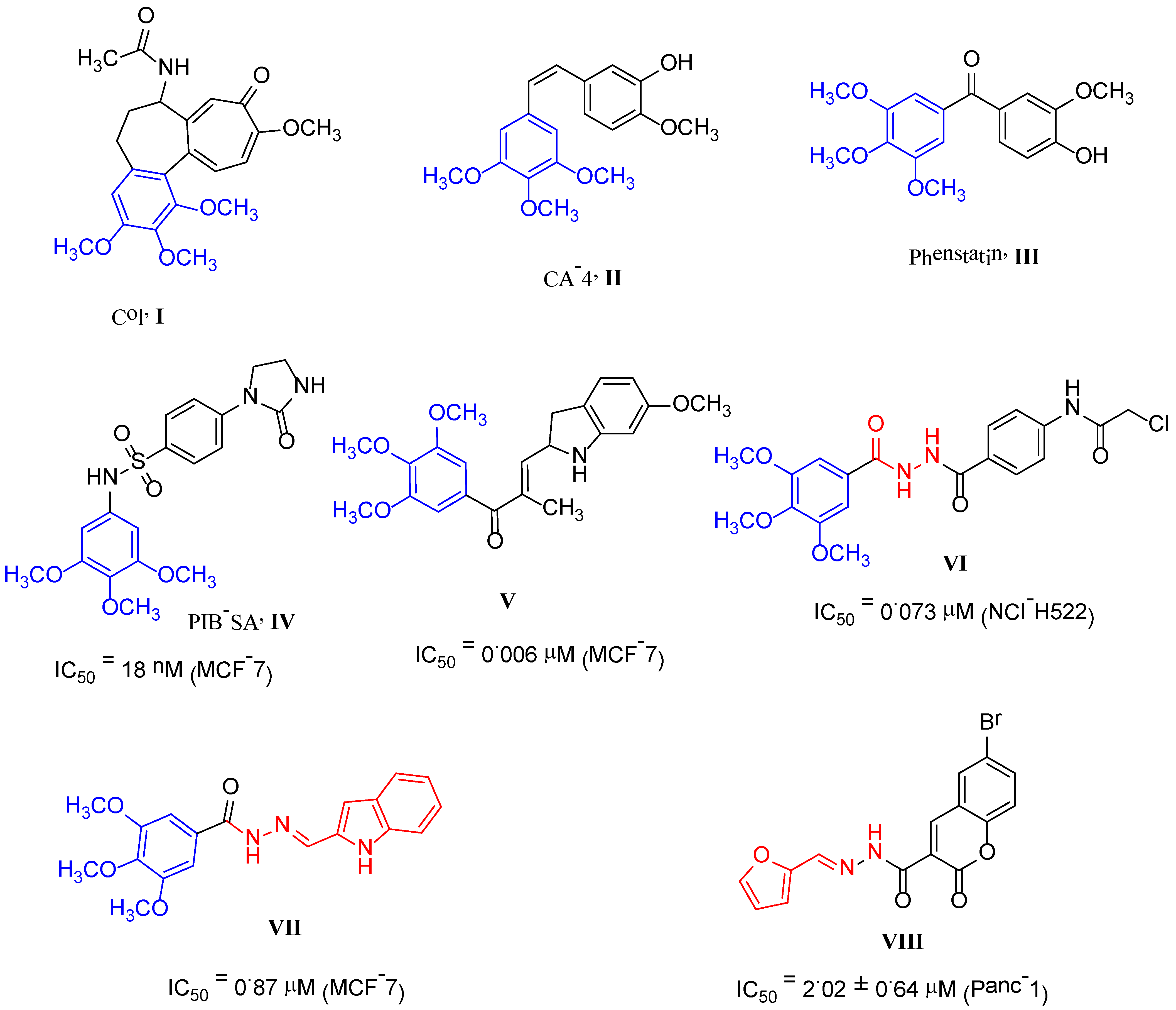
:1. Introduction
2. Results and Discussion
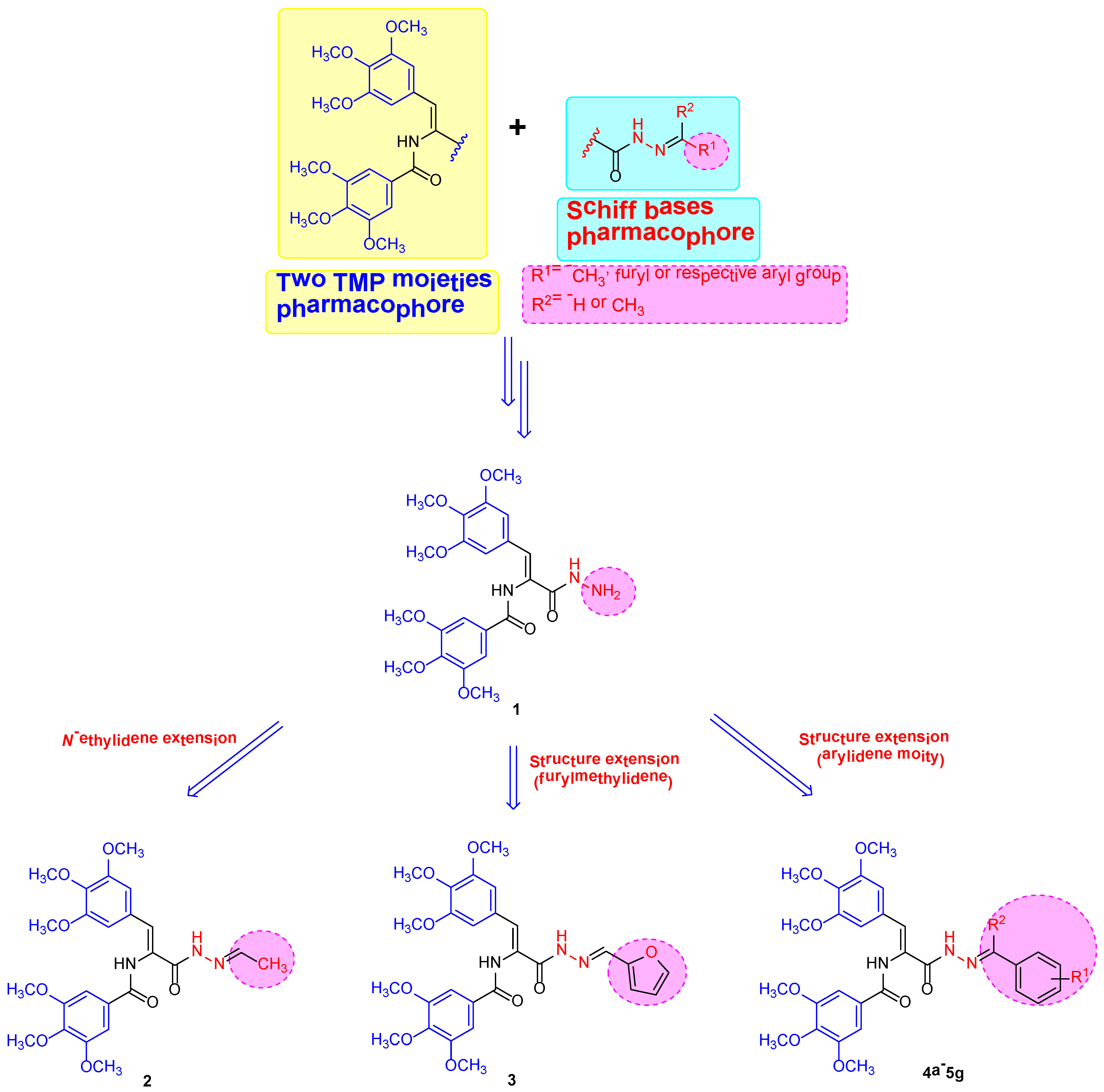
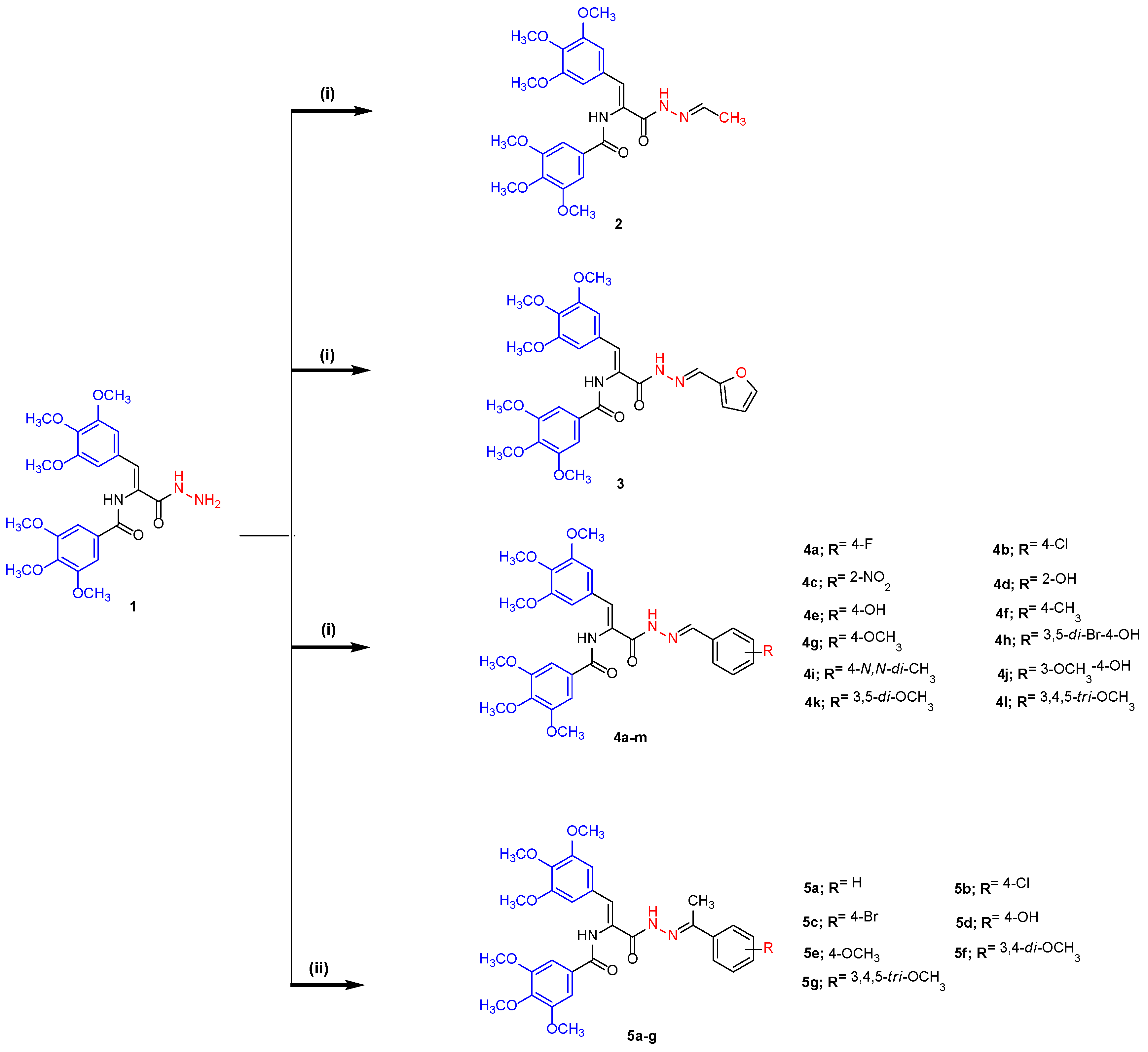
2.1. Chemistry
2.2. Biology
2.2.1. Cytotoxic Assay against MDA-MB-231 Breast Cancer Cell Line
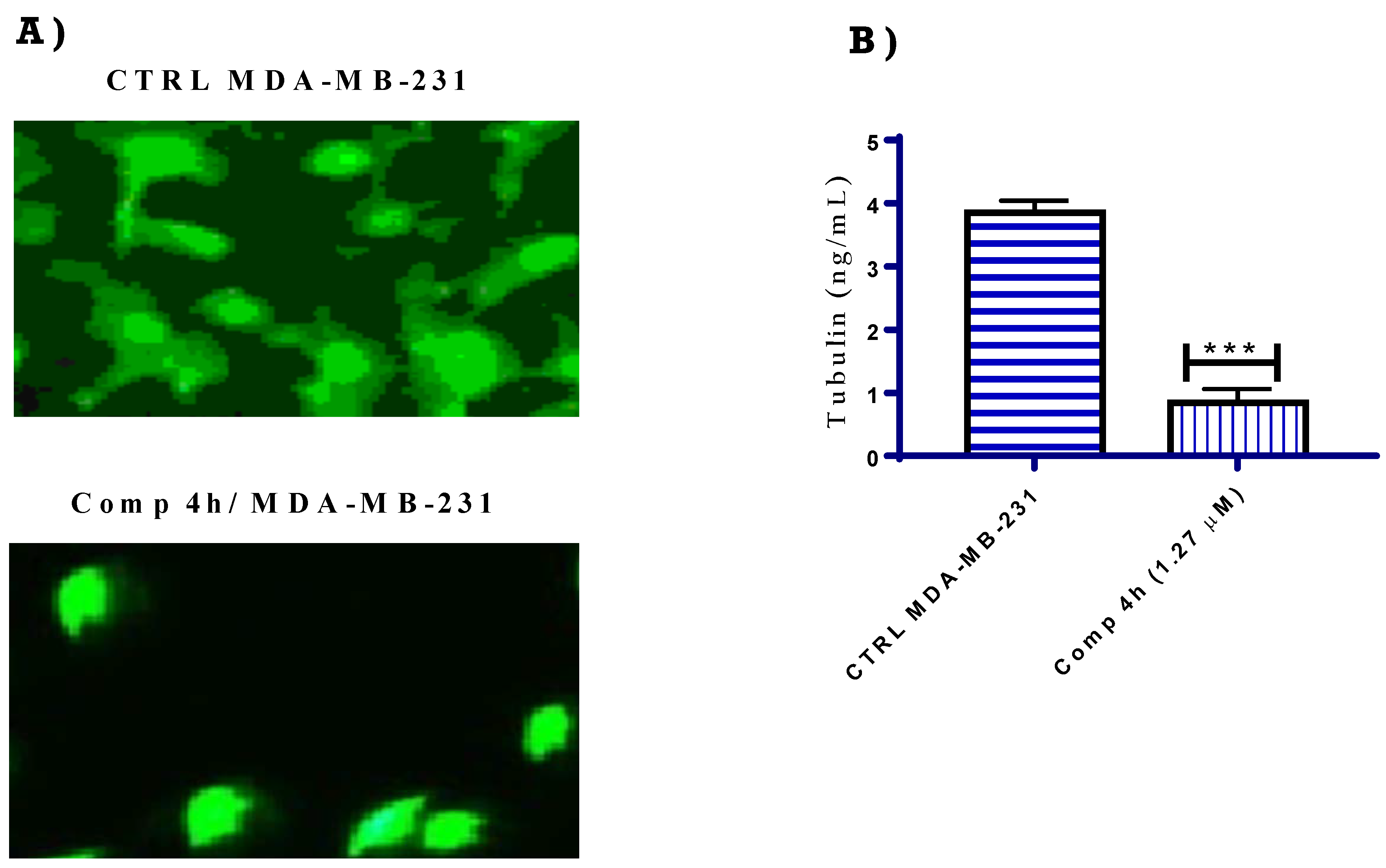
2.2.2. Tubulin Polymerization Inhibition Assays
2.2.3. Cell Cycle Effects
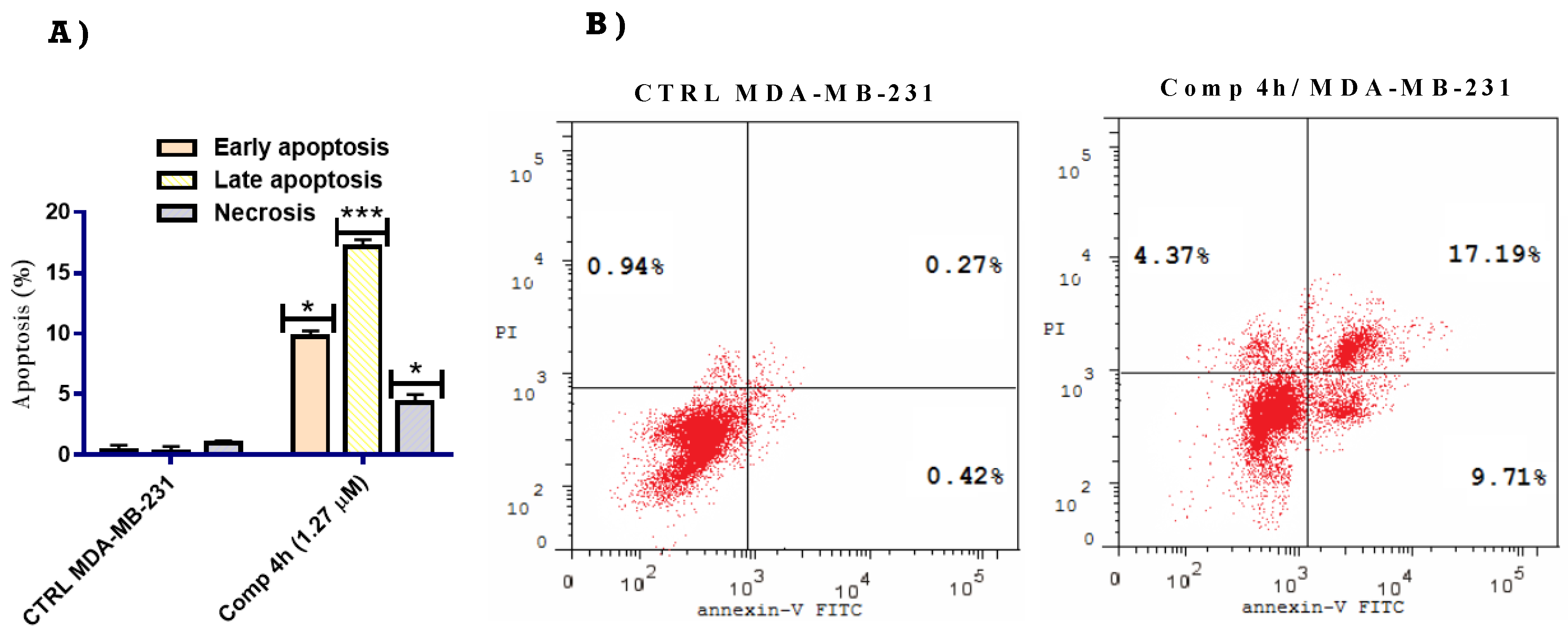
2.2.4. Apoptosis Inducing Effect of Compound 4h
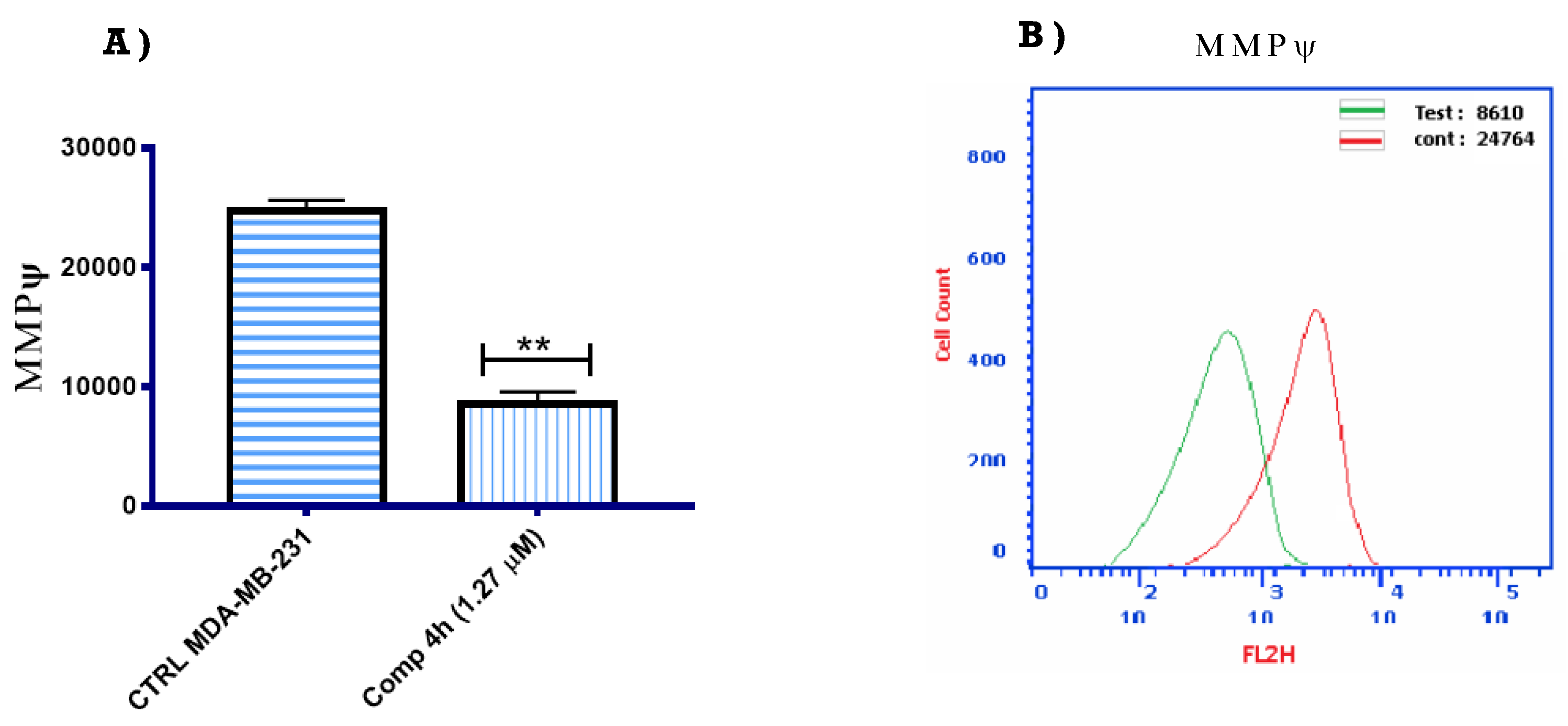
2.2.5. Mitochondrial Membrane Potential
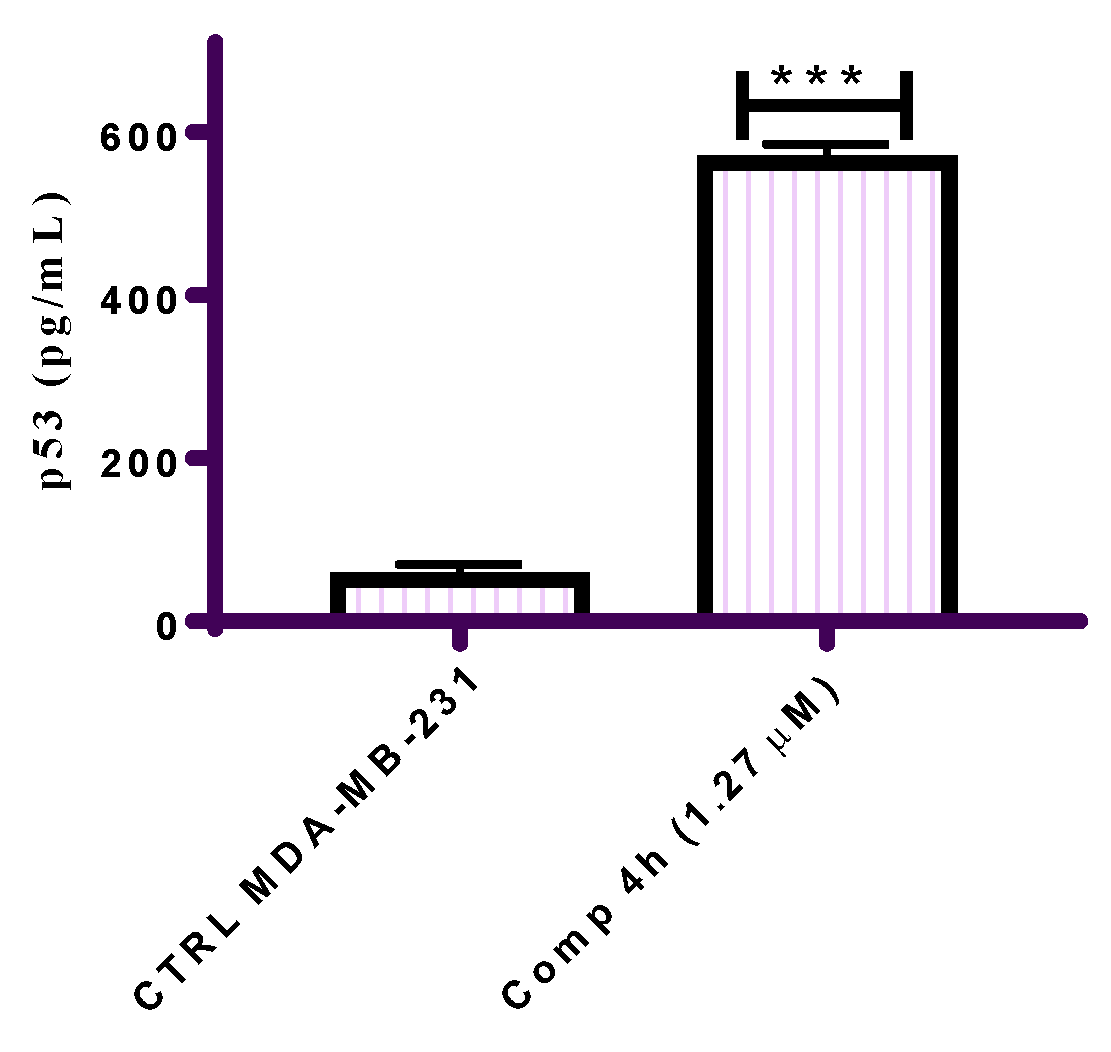
2.2.6. Effect of Compound 4h on the Expression of p53
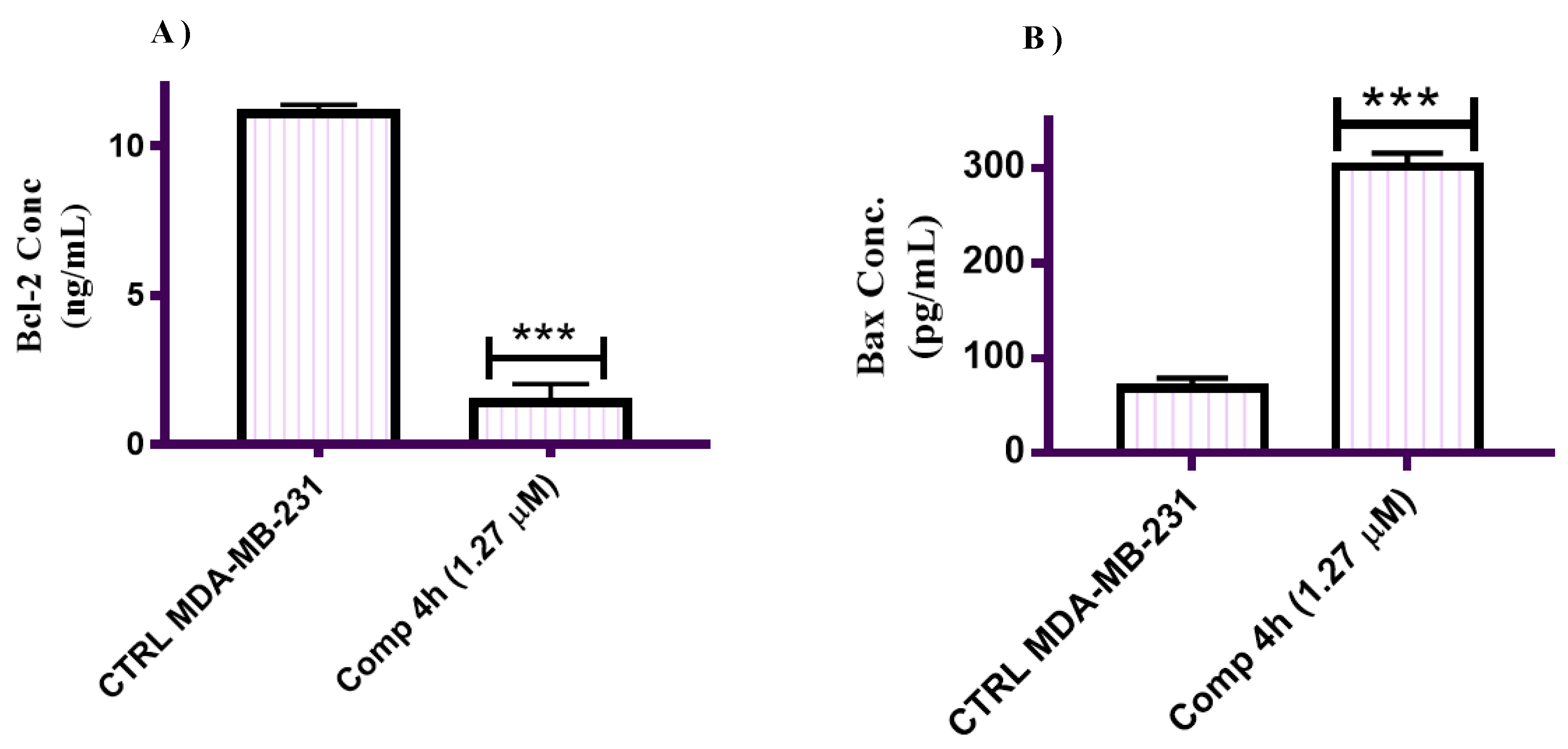
2.2.7. Effect of Compound 4h on the Expression of Bax and Bcl-2
2.2.8. Molecular Docking Study
2.3. Nanostructured Lipid Carrier Fabrication Studies
2.4. Compound 4h Loaded NLCs Design, Preparation and Optimization
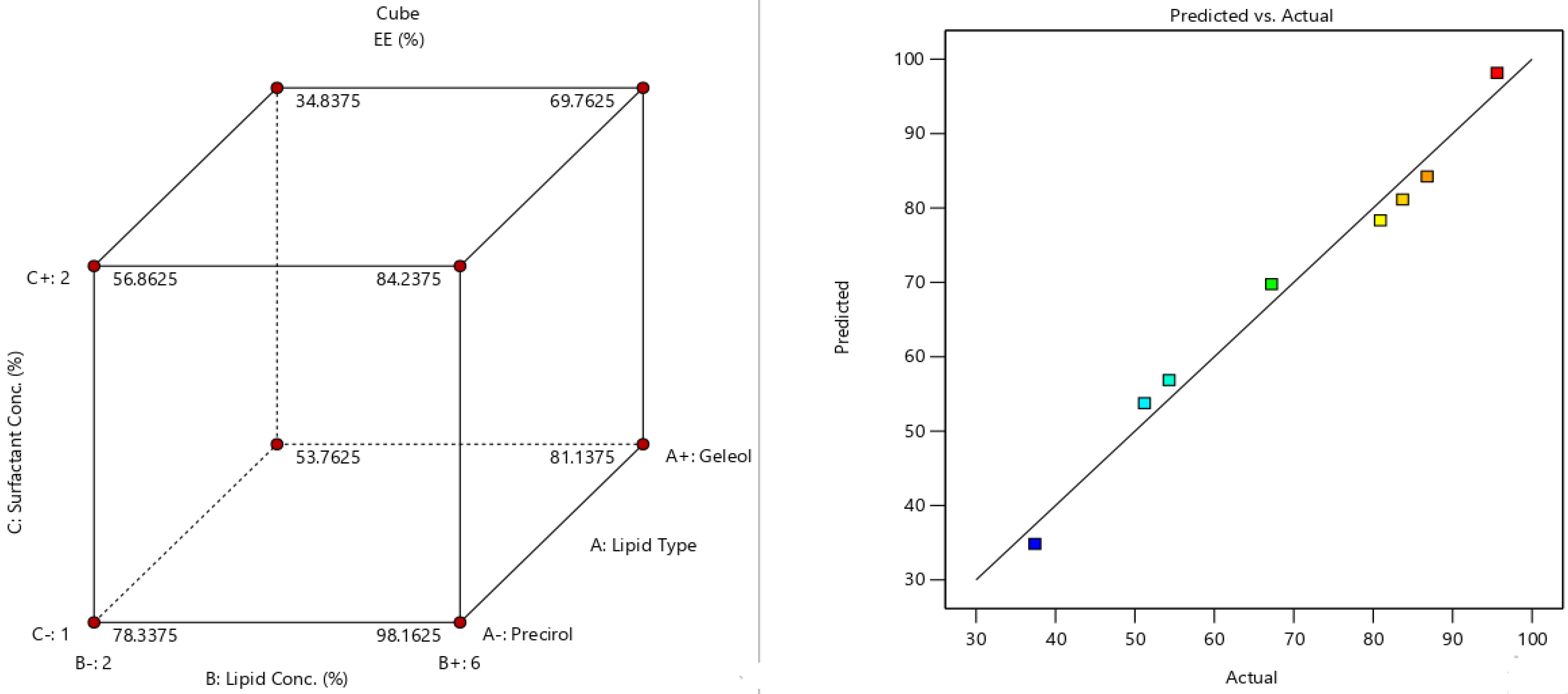
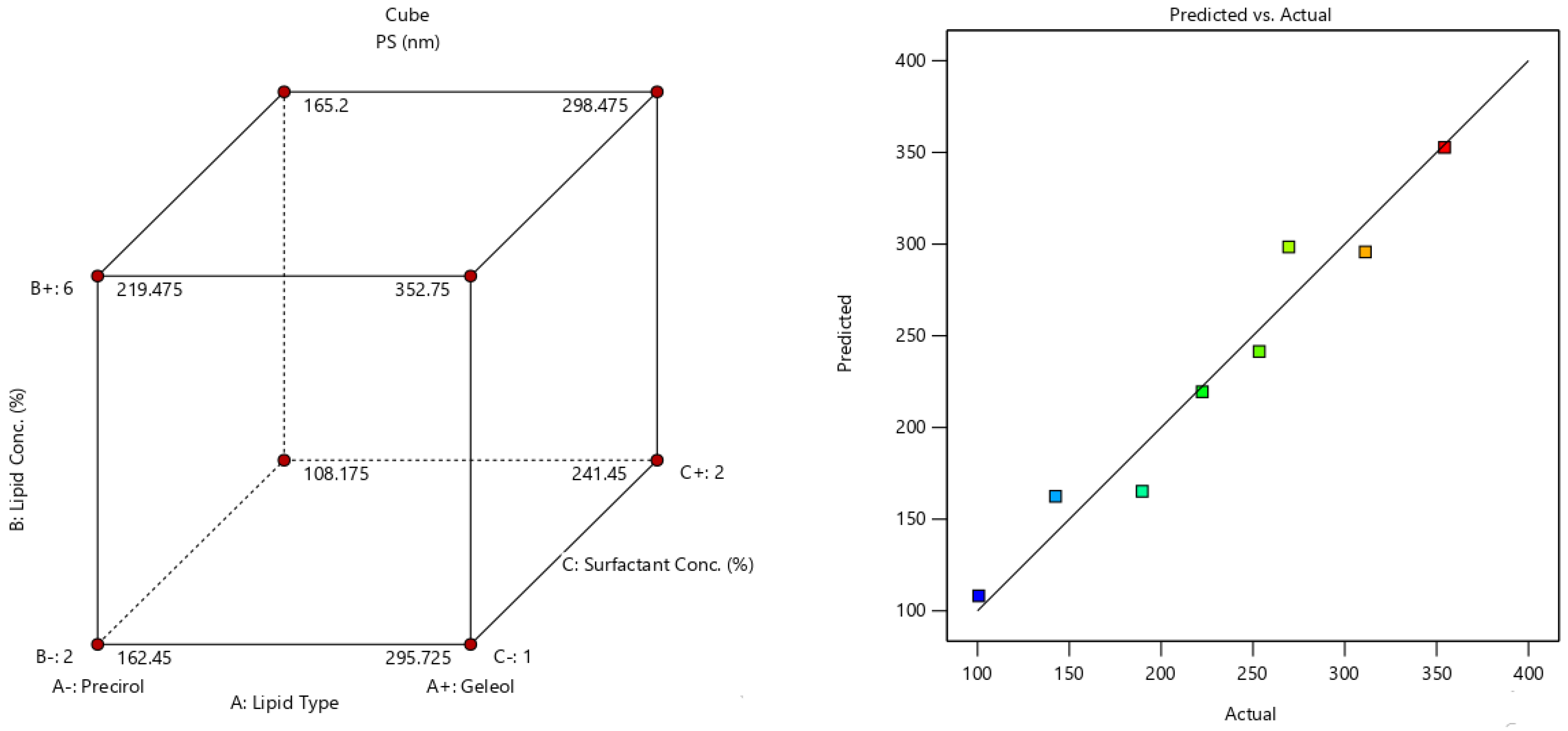
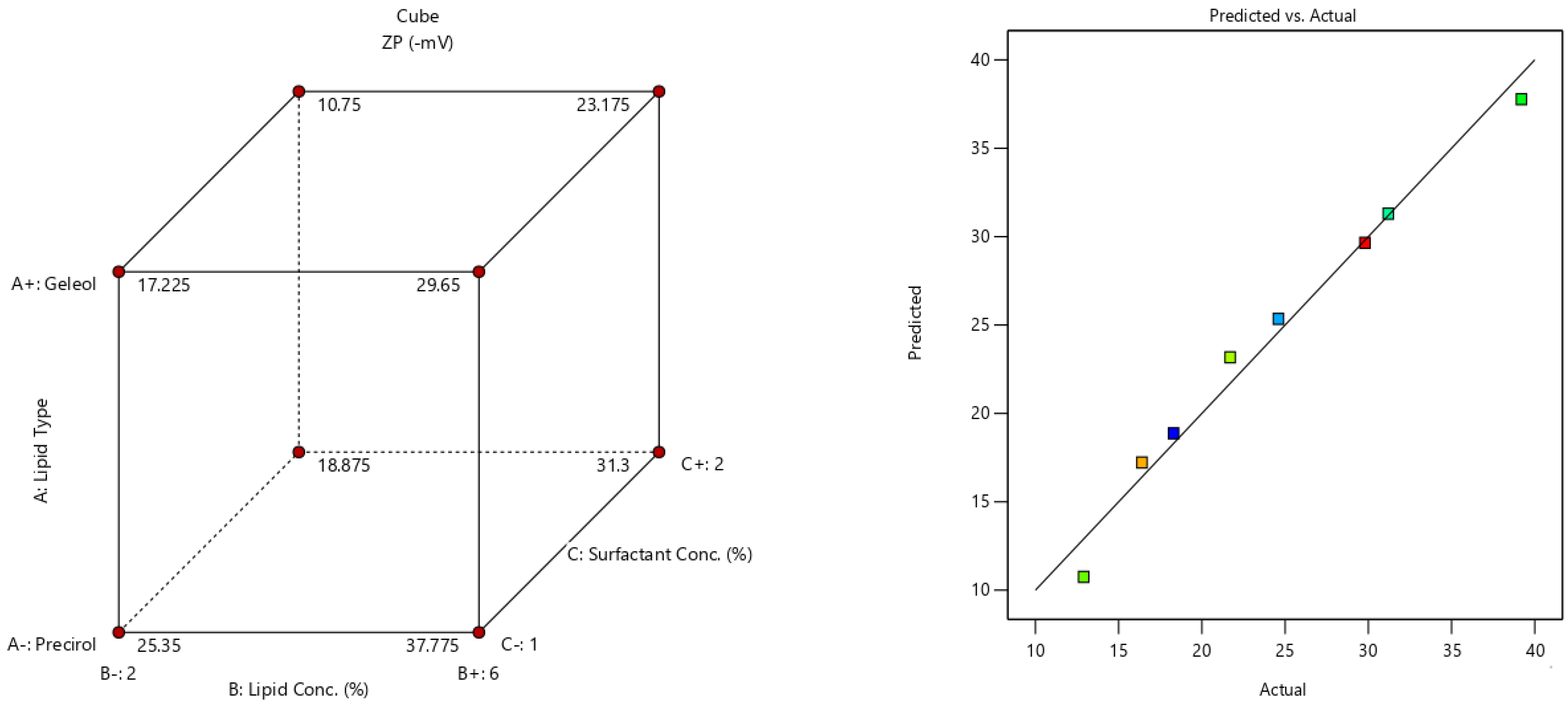
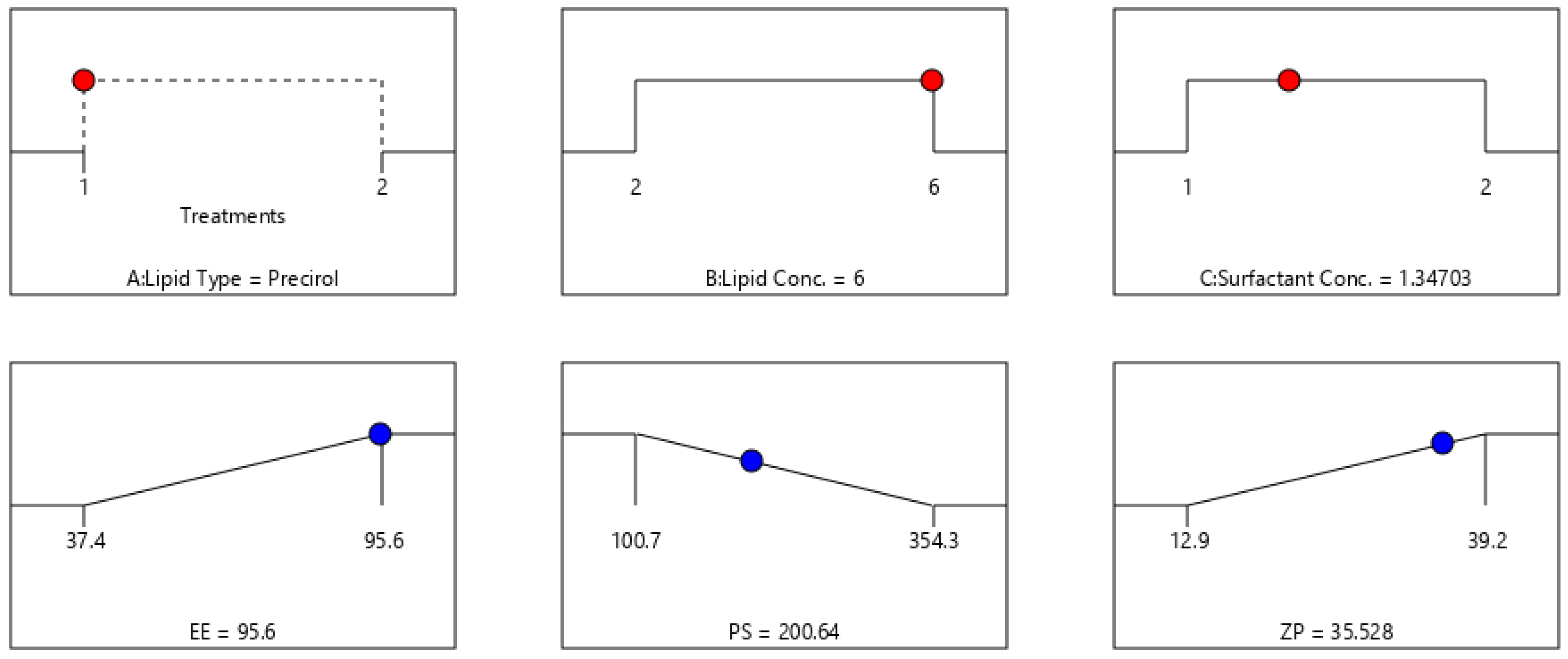
2.5. Fabrication Variables Influence on the Selected Responses
2.6. Statistical Optimization of Fabrication Variables for Election the Optimum Formula
2.7. In Vitro Assessment of the Optimum 4h-Loaded NLC
2.7.1. Particle Morphology Analysis via TEM
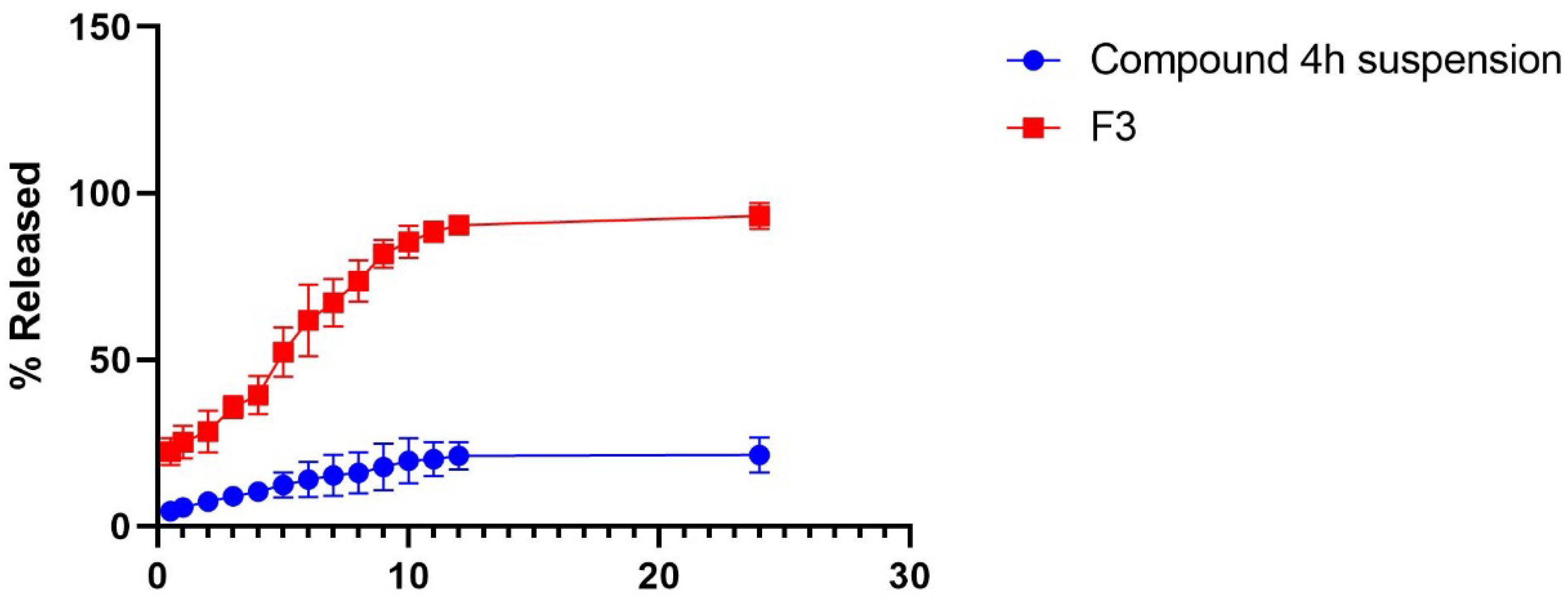
2.7.2. In Vitro Drug Release Experiment
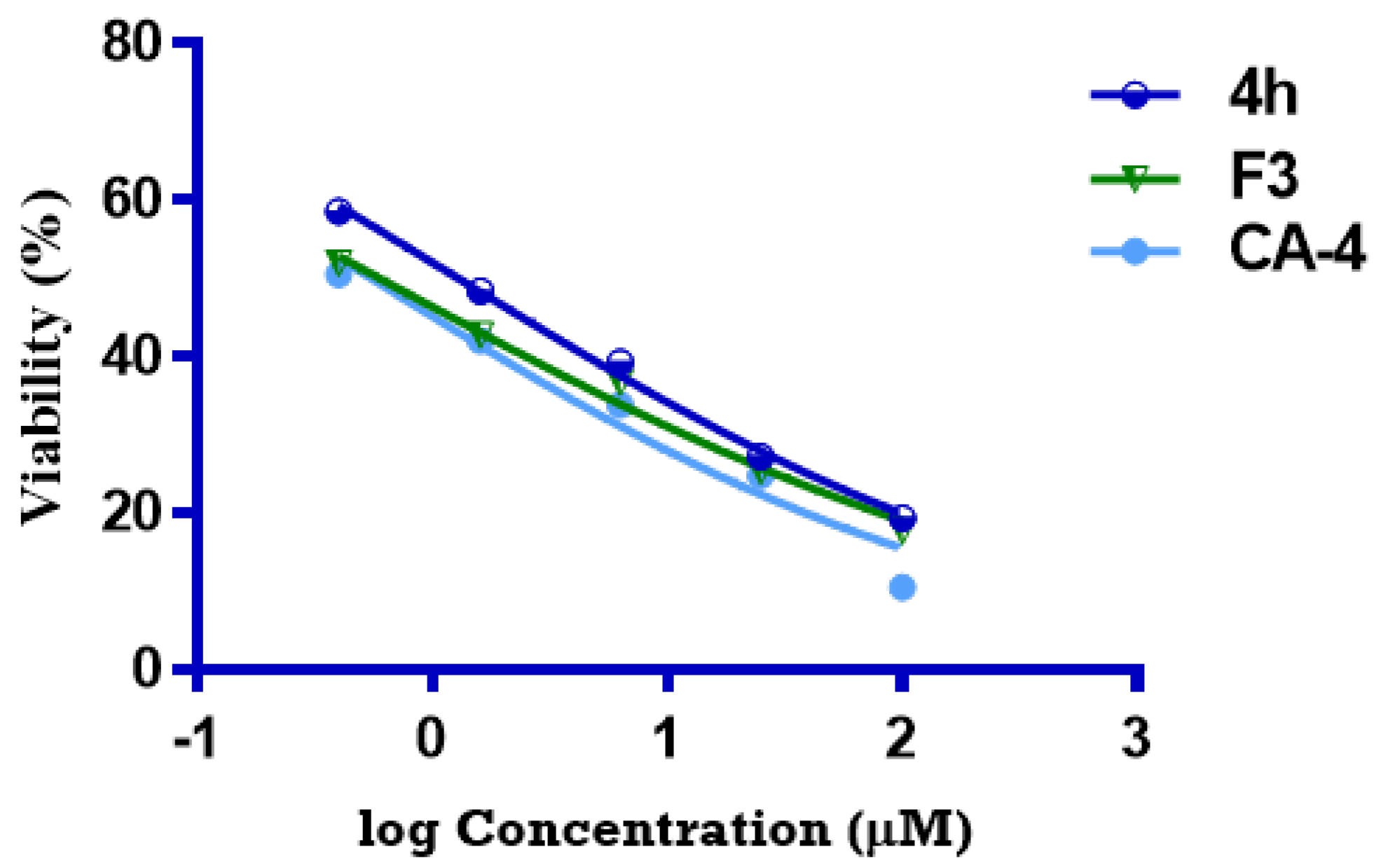
2.7.3. Comparative Cytotoxicity Study of Optimized Formula versus Pure 4h
3. Materials and Methods
3.1. General
3.2. Chemistry
3.2.1. General Procedure for the Preparation of N-((Z)-3-((E)-2-(Ethylidene, furo or aryl)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamides (2–4l)
N-((Z)-3-((E)-2-Ethylidenehydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (2)
N-((Z)-3-((E)-2-(Furan-2-ylmethylene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (3)
N-((Z)-3-((E)-2-(4-Fluorobenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4a)
N-((Z)-3-((E)-2-(4-Chlorobenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4b)
3,4,5-Trimethoxy-N-((Z)-3-((E)-2-(2-nitrobenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (4c)
N-((Z)-3-((E)-2-(2-Hydroxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4d)
N-((Z)-3-((E)-2-(4-Hydroxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4e)
3,4,5-Trimethoxy-N-((Z)-3-((E)-2-(4-methylbenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (4f)
3,4,5-Trimethoxy-N-((Z)-3-((E)-2-(4-methoxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (4g)
N-((Z)-3-((E)-2-(3,5-Dibromo-4-hydroxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4h)
N-((Z)-3-((E)-2-(4-(Dimethylamino)benzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4i)
N-((Z)-3-((E)-2-(4-Hydroxy-3-methoxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4j)
N-((Z)-3-((E)-2-(3,5-Dimethoxybenzylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (4k)
3,4,5-Trimethoxy-N-((Z)-3-oxo-3-((E)-2-(3,4,5-trimethoxybenzylidene)hydrazinyl)-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (4l)
3.2.2. General Procedure for the Preparation of N-((Z)-3-((E)-2-(Arylethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamides 5a–g
3,4,5-Trimethoxy-N-((Z)-3-oxo-3-((E)-2-(1-phenylethylidene)hydrazinyl)-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (5a)
N-((Z)-3-((E)-2-(1-(4-Chlorophenyl)ethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (5b)
N-((Z)-3-((E)-2-(1-(4-Bromophenyl)ethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (5c)
N-((Z)-3-((E)-2-(1-(4-Hydroxyphenyl)ethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (5d)
3,4,5-Trimethoxy-N-((Z)-3-((E)-2-(1-(4-methoxyphenyl)ethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)benzamide (5e)
N-((Z)-3-((E)-2-(1-(3,4-Dimethoxyphenyl)ethylidene)hydrazinyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-3,4,5-trimethoxybenzamide (5f)
3,4,5-Trimethoxy-N-((Z)-3-oxo-1-(3,4,5-trimethoxyphenyl)-3-((E)-2-(1-(3,4,5-trimethoxyphenyl)ethylidene)hydrazinyl)prop-1-en-2-yl)benzamide (5g)
3.3. Biological Assays
3.3.1. Cytotoxic Activity against MDA-MB-231 Cell Line
3.3.2. Tubulin Inhibition Assays
3.3.3. Cell Cycle Analysis of Compound 4h
3.3.4. Annexin V FITC/PI Staining Assay for Compound 4h
3.3.5. Mitochondrial Membrane Potential
3.3.6. ELISA Measurements of p53, Bax and Bcl-2
3.3.7. Molecular Docking Study
3.4. Fabrication of Compound 4h Loaded NLCs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mukhtar, E.; Adhami, V.M.; Mukhtar, H. Targeting Microtubules by Natural Agents for Cancer Therapy. Mol. Cancer Ther. 2014, 13, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Eli, S.; Castagna, R.; Mapelli, M.; Parisini, E. Recent Approaches to the Identification of Novel Microtubule-Targeting Agents. Front. Mol. Biosci. 2022, 9, 841777. [Google Scholar] [CrossRef]
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Cancer 2004, 4, 253–265. [Google Scholar]
- Chatterjee, S.; Som, S.; Varshney, N.; Satyadev, P.V.S.; Sanyal, K.; Paul, R. Mechanics of microtubule organizing center clustering and spindle positioning in budding yeast Cryptococcus neoformans. Phys. Rev. E 2021, 104, 034402. [Google Scholar] [CrossRef]
- Li, G.; Hu, X.; Wu, X.; Zhang, Y. Microtubule-Targeted Self-Assembly Triggers Prometaphase–Metaphase Oscillations Suppressing Tumor Growth. Nano Lett. 2021, 21, 3052–3059. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar]
- Barreca, M.; Stathis, A.; Barraja, P.; Bertoni, F. An overview on anti-tubulin agents for the treatment of lymphoma patients. Pharmacol. Ther. 2020, 211, 107552. [Google Scholar]
- Qi, F.; Zhou, J.; Liu, M. Microtubule-interfering agents, spindle defects, and interkinetochore tension. J. Cell. Physiol. 2020, 235, 26–30. [Google Scholar] [CrossRef]
- Arnst, K.E.; Banerjee, S.; Chen, H.; Deng, S.; Hwang, D.-J.; Li, W.; Miller, D.D. Current advances of tubulin inhibitors as dual acting small molecules for cancer therapy. Med. Res. Rev. 2019, 39, 1398–1426. [Google Scholar] [CrossRef]
- Li, L.; Jiang, S.; Li, X.; Liu, Y.; Su, J.; Chen, J. Recent advances in trimethoxyphenyl (TMP) based tubulin inhibitors targeting the colchicine binding site. Eur. J. Med. Chem. 2018, 151, 482–494. [Google Scholar] [CrossRef]
- Sun, K.; Sun, Z.; Zhao, F.; Shan, G.; Meng, Q. Recent advances in research of colchicine binding site inhibitors and their interaction modes with tubulin. Future Med. Chem. 2021, 13, 839–858. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, W.; Tian, L.; Mao, Y.; Song, C. Targeting tubulin-colchicine site for cancer therapy: Inhibitors, antibody-drug conjugates and degradation agents. Curr. Top. Med. Chem. 2019, 19, 1289–1304. [Google Scholar]
- Kerr, D.J.; Hamel, E.; Jung, M.K.; Flynn, B.L. The concise synthesis of chalcone, indanone and indenone analogues of combretastatin A4. Bioorg. Med. Chem. 2007, 15, 3290–3298. [Google Scholar] [CrossRef]
- Shawky, A.M.; Ibrahim, N.A.; Abdalla, A.N.; Abourehab, M.A.S.; Gouda, A.M. Novel pyrrolizines bearing 3,4,5-trimethoxyphenyl moiety: Design, synthesis, molecular docking, and biological evaluation as potential multi-target cytotoxic agents. J. Enzym. Inhib. Med. Chem. 2021, 36, 1312–1332. [Google Scholar] [CrossRef]
- Ansari, M.; Shokrzadeh, M.; Karima, S.; Rajaei, S.; Fallah, M.; Ghassemi-Barghi, N.; Ghasemian, M.; Emami, S. New thiazole-2(3H)-thiones containing 4-(3,4,5-trimethoxyphenyl) moiety as anticancer agents. Eur. J. Med. Chem. 2020, 185, 111784. [Google Scholar] [CrossRef]
- Hamze, A.; Alami, M.; Provot, O. Developments of isoCombretastatin A-4 derivatives as highly cytotoxic agents. Eur. J. Med. Chem. 2020, 190, 112110. [Google Scholar] [CrossRef]
- Naaz, F.; Haider, M.R.; Shafi, S.; Yar, M.S. Anti-tubulin agents of natural origin: Targeting taxol, vinca, and colchicine binding domains. Eur. J. Med. Chem. 2019, 171, 310–331. [Google Scholar]
- Gaspari, R.; Prota, A.E.; Bargsten, K.; Cavalli, A.; Steinmetz, M.O. Structural Basis of cis- and trans-Combretastatin Binding to Tubulin. Chem 2017, 2, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Pettit, G.R.; Toki, B.; Herald, D.L.; Verdier-Pinard, P.; Boyd, M.R.; Hamel, E.; Pettit, R.K. Antineoplastic Agents. 379. Synthesis of Phenstatin Phosphate1a. J. Med. Chem. 1998, 41, 1688–1695. [Google Scholar] [CrossRef]
- Fortin, S.; Wei, L.; Moreau, E.; Lacroix, J.; Côté, M.-F.; Petitclerc, É.; Kotra, L.P.; Gaudreault, R.C. Substituted phenyl 4-(2-oxoimidazolidin-1-yl)benzenesulfonamides as antimitotics. Antiproliferative, antiangiogenic and antitumoral activity, and quantitative structure-activity relationships. Eur. J. Med. Chem. 2011, 46, 5327–5342. [Google Scholar] [CrossRef]
- Yan, J.; Chen, J.; Zhang, S.; Hu, J.; Huang, L.; Li, X. Synthesis, Evaluation, and Mechanism Study of Novel Indole-Chalcone Derivatives Exerting Effective Antitumor Activity through Microtubule Destabilization In Vitro and In Vivo. J. Med. Chem. 2016, 59, 5264–5283. [Google Scholar] [CrossRef]
- Maklad, R.M.; AbdelHafez, E.-S.M.N.; Abdelhamid, D.; Aly, O.M. Tubulin inhibitors: Discovery of a new scaffold targeting extra-binding residues within the colchicine site through anchoring substituents properly adapted to their pocket by a semi-flexible linker. Bioorg. Chem. 2020, 99, 103767. [Google Scholar] [CrossRef]
- Uddin, N.; Rashid, F.; Ali, S.; Tirmizi, S.A.; Ahmad, I.; Zaib, S.; Zubair, M.; Diaconescu, P.L.; Tahir, M.N.; Iqbal, J.; et al. Synthesis, characterization, and anticancer activity of Schiff bases. J. Biomol. Struct. Dyn. 2020, 38, 3246–3259. [Google Scholar] [CrossRef]
- Cordeiro, R.; Kachroo, M. Synthesis and biological evaluation of anti-tubercular activity of Schiff bases of 2-Amino thiazoles. Bioorg. Med. Chem. Lett. 2020, 30, 127655. [Google Scholar] [CrossRef]
- Singh, G.; Kalra, P.; Singh, A.; Sharma, G.; Espinosa-Ruíz, C.; Esteban, M.A. A quick microwave preparation of isatin hydrazone schiff base conjugated organosilicon compounds: Exploration of their antibacterial, antifungal, and antioxidative potentials. J. Organomet. Chem. 2021, 953, 122051. [Google Scholar] [CrossRef]
- Sundaree, S.; Vaddula, B.R.; Tantak, M.P.; Khandagale, S.B.; Shi, C.; Shah, K.; Kumar, D. Synthesis and anticancer activity study of indolyl hydrazide–hydrazones. Med. Chem. Res. 2016, 25, 941–950. [Google Scholar] [CrossRef]
- Acharya, S.; Maji, M.; Chakraborty, M.P.; Bhattacharya, I.; Das, R.; Gupta, A.; Mukherjee, A. Disruption of the Microtubule Network and Inhibition of VEGFR2 Phosphorylation by Cytotoxic N,O-Coordinated Pt(II) and Ru(II) Complexes of Trimethoxy Aniline-Based Schiff Bases. Inorg. Chem. 2021, 60, 3418–3430. [Google Scholar] [CrossRef]
- Nasr, T.; Bondock, S.; Youns, M. Anticancer activity of new coumarin substituted hydrazide–hydrazone derivatives. Eur. J. Med. Chem. 2014, 76, 539–548. [Google Scholar] [CrossRef]
- Zaki, I.; El-ata, S.A.A.; Fayad, E.; Ali, O.A.A.; Almaaty, A.H.A.; Saad, A.S. Evaluation of Synthetic 2,4-Disubstituted-benzo[g]quinoxaline Derivatives as Potential Anticancer Agents. Pharmaceuticals 2021, 14, 853. [Google Scholar] [CrossRef]
- Almaaty, A.H.A.; Toson, E.E.M.; El-Sayed, E.-S.H.; Tantawy, M.A.M.; Fayad, E.; Ali, O.A.A.; Zaki, I. 5-Aryl-1-Arylideneamino-1H-Imidazole-2(3H)-Thiones: Synthesis and In Vitro Anticancer Evaluation. Molecules 2021, 26, 1706. [Google Scholar] [CrossRef]
- Zaki, I.; Abdelhameid, M.K.; El-Deen, I.M.; Wahab, A.H.A.A.; Ashmawy, A.M.; Mohamed, K.O. Design, synthesis and screening of 1, 2, 4-triazinone derivatives as potential antitumor agents with apoptosis inducing activity on MCF-7 breast cancer cell line. Eur. J. Med. Chem. 2018, 156, 563–579. [Google Scholar] [CrossRef]
- Zaki, I.; Masoud, R.E.; Hamoud, M.M.S.; Ali, O.A.A.; Abualnaja, M.; Fayad, E.; Almaaty, A.H.A.; Elnaghia, L.K. Design, synthesis and cytotoxicity screening of new synthesized pyrimidine-5-carbonitrile derivatives showing marked apoptotic effect. J. Mol. Struct. 2022, 1259, 132749. [Google Scholar] [CrossRef]
- Tang, S.; Zhou, Z.; Jiang, Z.; Zhu, W.; Qiao, D. Indole-Based Tubulin Inhibitors: Binding Modes and SARs Investigations. Molecules 2022, 27, 1587. [Google Scholar] [CrossRef]
- Liao, W.-L.; Lin, J.-Y.; Shieh, J.-C.; Yeh, H.-F.; Hsieh, Y.-H.; Cheng, Y.-C.; Lee, H.-J.; Shen, C.-Y.; Cheng, C.-W. Induction of G2/M Phase Arrest by Diosgenin via Activation of Chk1 Kinase and Cdc25C Regulatory Pathways to Promote Apoptosis in Human Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 172. [Google Scholar] [CrossRef] [Green Version]
- Li, M. The role of P53 up-regulated modulator of apoptosis (PUMA) in ovarian development, cardiovascular and neurodegenerative diseases. Apoptosis 2021, 26, 235–247. [Google Scholar] [CrossRef]
- Das, S.; Ng, W.K.; Tan, R.B.H. Are nanostructured lipid carriers (NLCs) better than solid lipid nanoparticles (SLNs): Development, characterizations and comparative evaluations of clotrimazole-loaded SLNs and NLCs? Eur. J. Pharm. Sci. 2012, 47, 139–151. [Google Scholar] [CrossRef]
- Kaur, S.; Nautyal, U.; Singh, R.; Singh, S.; Devi, A. Nanostructure lipid carrier (NLC): The new generation of lipid nanoparticles. Asian Pac. J. Health Sci. 2015, 2, 76–93. [Google Scholar] [CrossRef]
- Zaki, I.; Abou-Elkhair, R.A.I.; Almaaty, A.H.A.; Ali, O.A.A.; Fayad, E.; Gaafar, A.G.A.; Zakaria, M.Y. Design and Synthesis of Newly Synthesized Acrylamide Derivatives as Potential Chemotherapeutic Agents against MCF-7 Breast Cancer Cell Line Lodged on PEGylated Bilosomal Nano-Vesicles for Improving Cytotoxic Activity. Pharmaceuticals 2021, 14, 1021. [Google Scholar] [CrossRef]
- El-Halim, S.M.A.; Abdelbary, G.A.; Amin, M.M.; Zakaria, M.Y.; Shamsel-Din, H.A.; Ibrahim, A.B. Stabilized oral nanostructured lipid carriers of Adefovir Dipivoxil as a potential liver targeting: Estimation of liver function panel and uptake following intravenous injection of radioiodinated indicator. DARU J. Pharm. Sci. 2020, 28, 517–532. [Google Scholar] [CrossRef]
- Aburahma, M.H.; Badr-Eldin, S.M. Compritol 888 ATO: A multifunctional lipid excipient in drug delivery systems and nanopharmaceuticals. Expert Opin. Drug Deliv. 2014, 11, 1865–1883. [Google Scholar] [CrossRef]
- Poonia, N.; Kharb, R.; Lather, V.; Pandita, D. Nanostructured lipid carriers: Versatile oral delivery vehicle. Future Sci. 2016, 2, FSO135. [Google Scholar] [CrossRef] [Green Version]
- Sznitowska, M.; Wolska, E.; Baranska, H.; Cal, K.; Pietkiewicz, J. The effect of a lipid composition and a surfactant on the characteristics of the solid lipid microspheres and nanospheres (SLM and SLN). Eur. J. Pharm. Biopharm. 2017, 110, 24–30. [Google Scholar] [CrossRef]
- El-Zaafarany, G.M.; Soliman, M.E.; Mansour, S.; Awad, G.A.S. Identifying lipidic emulsomes for improved oxcarbazepine brain targeting: In vitro and rat in vivo studies. Int. J. Pharm. 2016, 503, 127–140. [Google Scholar] [CrossRef]
- Zakaria, M.Y.; Fayad, E.; Althobaiti, F.; Zaki, I.; Almaaty, A.H.A. Statistical optimization of bile salt deployed nanovesicles as a potential platform for oral delivery of piperine: Accentuated antiviral and anti-inflammatory activity in MERS-CoV challenged mice. Drug Deliv. 2021, 28, 1150–1165. [Google Scholar] [CrossRef]
- Abdelhaleem, E.F.; Abdelhameid, M.K.; Kassab, A.E.; Kandeel, M.M. Design and synthesis of thienopyrimidine urea derivatives with potential cytotoxic and pro-apoptotic activity against breast cancer cell line MCF-7. Eur. J. Med. Chem. 2018, 143, 1807–1825. [Google Scholar] [CrossRef]




| Compound No. | IC50 Value (μM) | |
|---|---|---|
| MDA-MB-231 | MCF-10A | |
| 2 | 31.59 ± 1.07 | NT |
| 3 | 6.08 ± 0.42 | NT |
| 4a | 23.70 ± 1.13 | NT |
| 4b | 16.10 ± 0.83 | NT |
| 4c | 38.82 ± 0.93 | NT |
| 4d | 5.98 ± 0.28 | NT |
| 4e | 7.10 ± 1.02 | NT |
| 4f | 4.52 ± 0.36 | NT |
| 4g | 3.43 ± 0.22 | NT |
| 4h | 1.27 ± 0.18 | 30.83 ± 2.32 |
| 4i | 3.46 ± 0.51 | NT |
| 4j | 2.84 ± 0.18 | NT |
| 4k | 4.80 ± 0.17 | NT |
| 4l | 7.30 ± 0.64 | NT |
| 5a | 20.15 ± 0.89 | NT |
| 5b | 13.98 ± 1.19 | NT |
| 5c | 20.73 ± 1.84 | NT |
| 5d | 1.98 ± 0.19 | NT |
| 5e | 9.52 ± 1.18 | NT |
| 5f | 8.34 ± 1.22 | NT |
| 5g | 3.39 ± 0.26 | NT |
| CA-4 | 0.54 ± 0.04 | 8.86 ± 0.67 |
| Formula | A (Lipid Type) | B (Lipid Conc. %w/v) | C (Surfactant Conc. %w/v) | Y1 (EE%) | Y2 (PS) | Y3 (PDI) | Y4 (ZP) |
|---|---|---|---|---|---|---|---|
| F1 | Precirol | 2 | 1 | 80.9 ± 3.5 | 142.5 ± 12.2 | 0.34 ± 0.04 | −24.6 ± 4.1 |
| F2 | Geleol | 2 | 1 | 51.2 ± 2.8 | 311.2 ± 19.1 | 0.31 ± 0.05 | −16.4 ± 1.2 |
| F3 | Precirol | 6 | 1 | 95.6 ± 4.1 | 222.4 ± 11.7 | 0.23 ± 0.06 | −39.2 ± 3.4 |
| F4 | Geleol | 6 | 1 | 83.7 ± 4.6 | 354.3 ± 17.3 | 0.27 ± 0.05 | −29.8 ± 2.2 |
| F5 | Precirol | 2 | 2 | 54.3 ± 1.9 | 100.7 ± 8.1 | 0.54 ± 0.08 | −18.3 ± 1.9 |
| F6 | Geleol | 2 | 2 | 37.4 ± 1.8 | 253.4 ± 9.5 | 0.64 ± 0.17 | −12.9 ± 1.8 |
| F7 | Precirol | 6 | 2 | 86.8 ± 3.3 | 189.7 ± 12.6 | 0.36 ± 0.05 | −31.2 ± 3.3 |
| F8 | Geleol | 6 | 2 | 67.2 ± 2.4 | 269.5 ± 23.2 | 0.28 ± 0.02 | 21.7 ± 2.5 |
| Responses | EE(%) | PS (nm) | ZP (Mv) |
|---|---|---|---|
| R2 | 0.961 | 0.954 | 0.981 |
| Adjusted R2 | 0.93 | 0.92 | 0.966 |
| Predicted R2 | 0.845 | 0.818 | 0.92 |
| Adequate precision | 16.9 | 14.5 | 23.6 |
| Significant factors | A, B, C | A, B, C | A, B, C |
| Observed value of the optimal formula (F3) | 95.6 | 222.4 | −39.2 |
| Predicted value of the optimal formula (F3) | 95.7 | 200.6 | −35.5 |
| Absolute deviation % | 0.1 | 9.8 | 9.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bukhari, S.N.A.; Zakaria, M.Y.; Munir, M.U.; Ahmad, N.; Elsherif, M.A.; Badr, R.E.; Hassan, A.K.; Almaaty, A.H.A.; Zaki, I. Design, Synthesis, In Vitro Biological Activity Evaluation and Stabilized Nanostructured Lipid Carrier Formulation of Newly Synthesized Schiff Bases-Based TMP Moieties. Pharmaceuticals 2022, 15, 679. https://doi.org/10.3390/ph15060679
Bukhari SNA, Zakaria MY, Munir MU, Ahmad N, Elsherif MA, Badr RE, Hassan AK, Almaaty AHA, Zaki I. Design, Synthesis, In Vitro Biological Activity Evaluation and Stabilized Nanostructured Lipid Carrier Formulation of Newly Synthesized Schiff Bases-Based TMP Moieties. Pharmaceuticals. 2022; 15(6):679. https://doi.org/10.3390/ph15060679
Chicago/Turabian StyleBukhari, Syed Nasir Abbas, Mohamed Y. Zakaria, Muhammad Usman Munir, Naveed Ahmad, Mervat A Elsherif, Rasha Emad Badr, Ahmad Khalaf Hassan, Ali H. Abu Almaaty, and Islam Zaki. 2022. "Design, Synthesis, In Vitro Biological Activity Evaluation and Stabilized Nanostructured Lipid Carrier Formulation of Newly Synthesized Schiff Bases-Based TMP Moieties" Pharmaceuticals 15, no. 6: 679. https://doi.org/10.3390/ph15060679
APA StyleBukhari, S. N. A., Zakaria, M. Y., Munir, M. U., Ahmad, N., Elsherif, M. A., Badr, R. E., Hassan, A. K., Almaaty, A. H. A., & Zaki, I. (2022). Design, Synthesis, In Vitro Biological Activity Evaluation and Stabilized Nanostructured Lipid Carrier Formulation of Newly Synthesized Schiff Bases-Based TMP Moieties. Pharmaceuticals, 15(6), 679. https://doi.org/10.3390/ph15060679