Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes
Abstract
:1. Introduction
2. Results
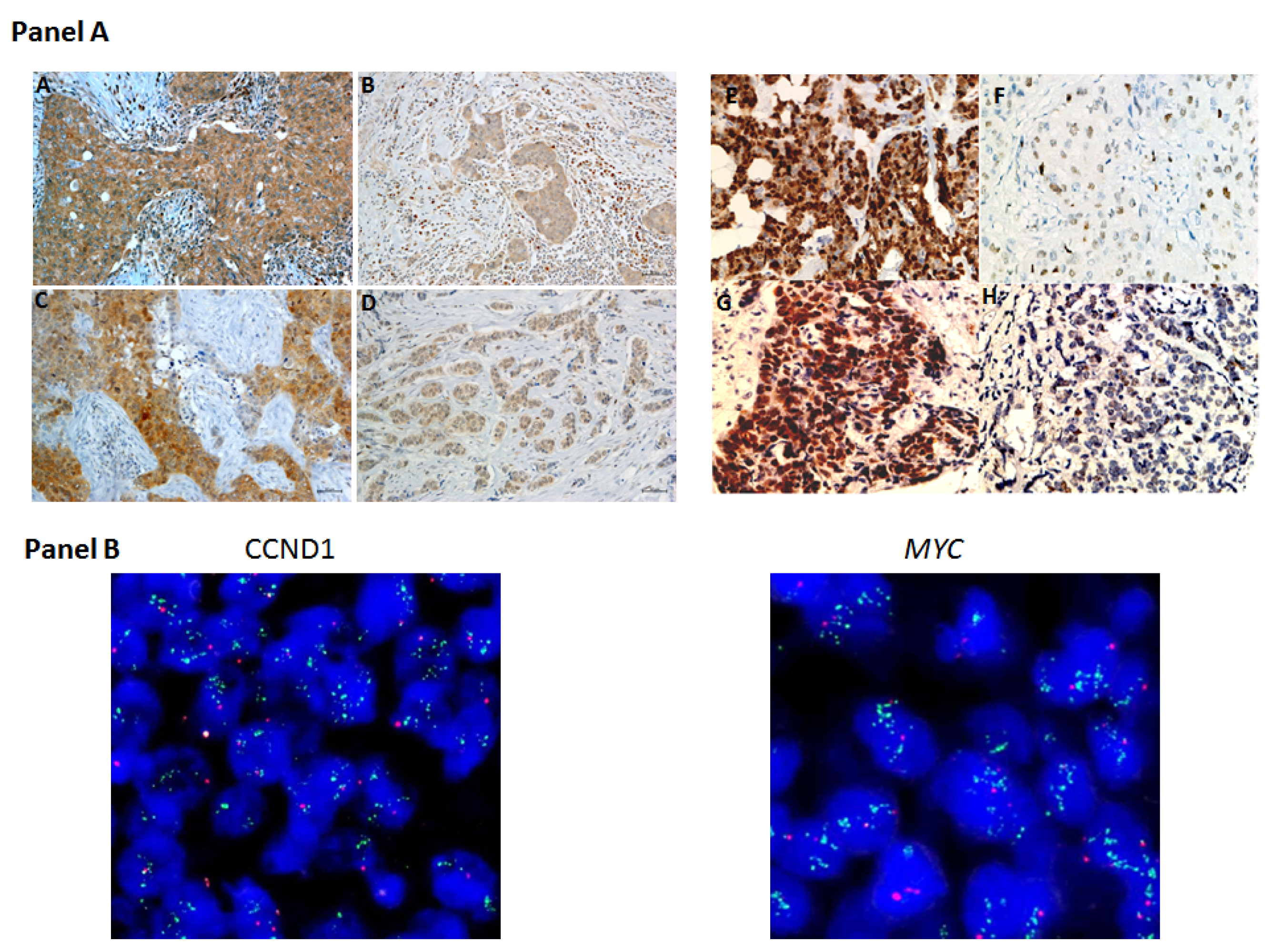
2.1. Protein and Gene Status Profiling
2.2. Relationship between Tumor Marker Alterations and Clinicopathological Characteristics
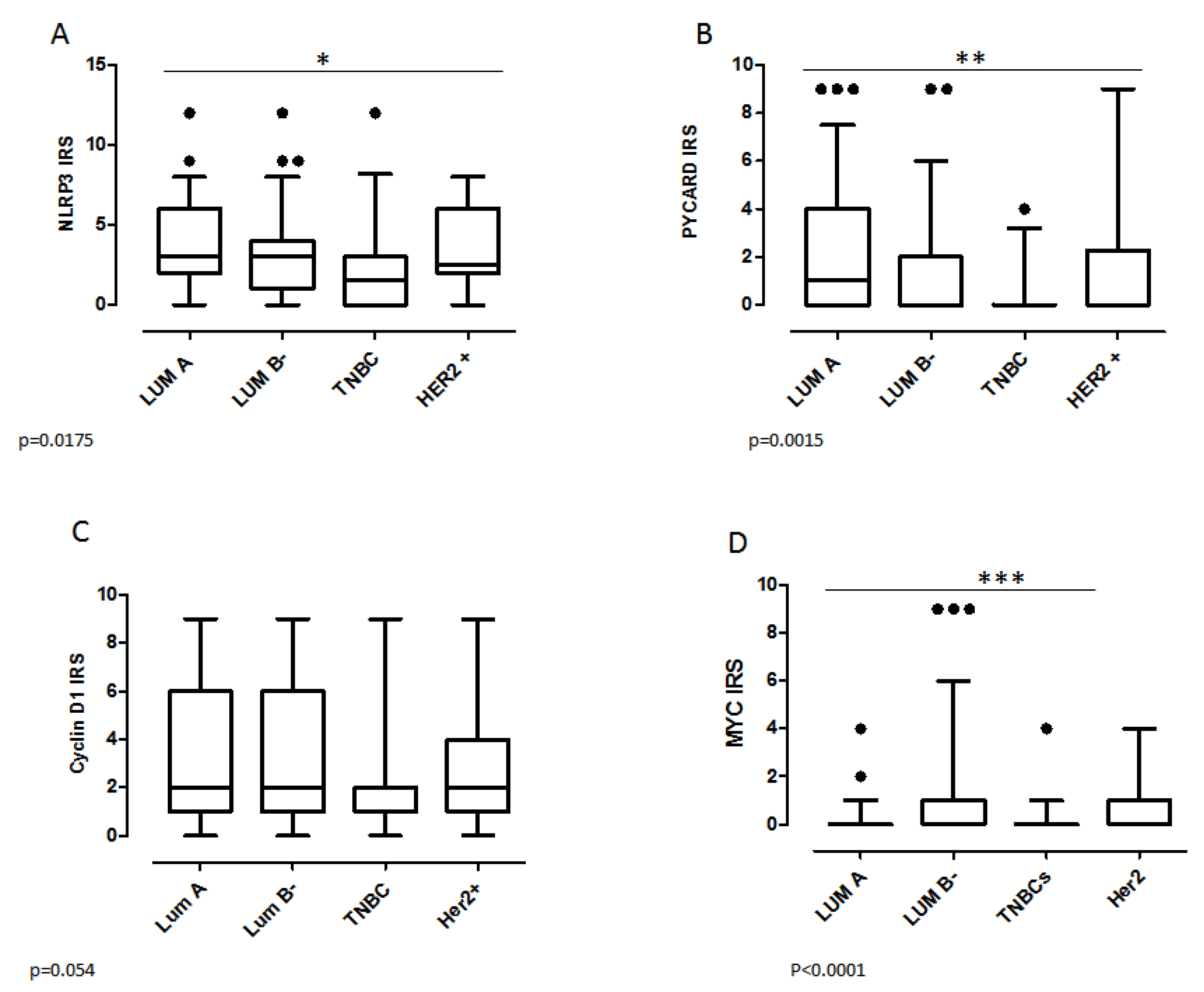
2.3. Biomarker Assessment in Relation to Molecular Phenotype
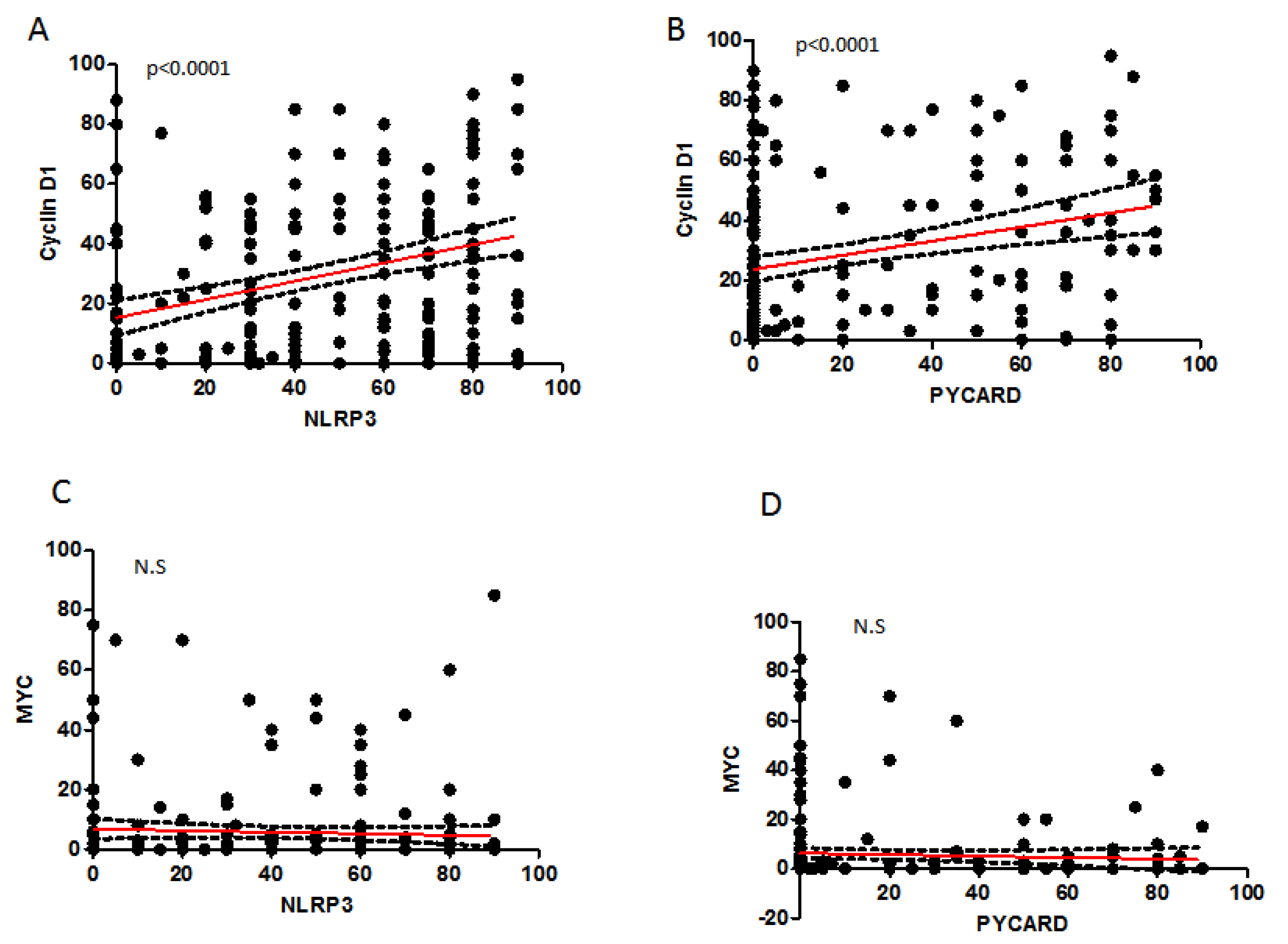
2.4. Correlation between Analyzed Protein Expressions
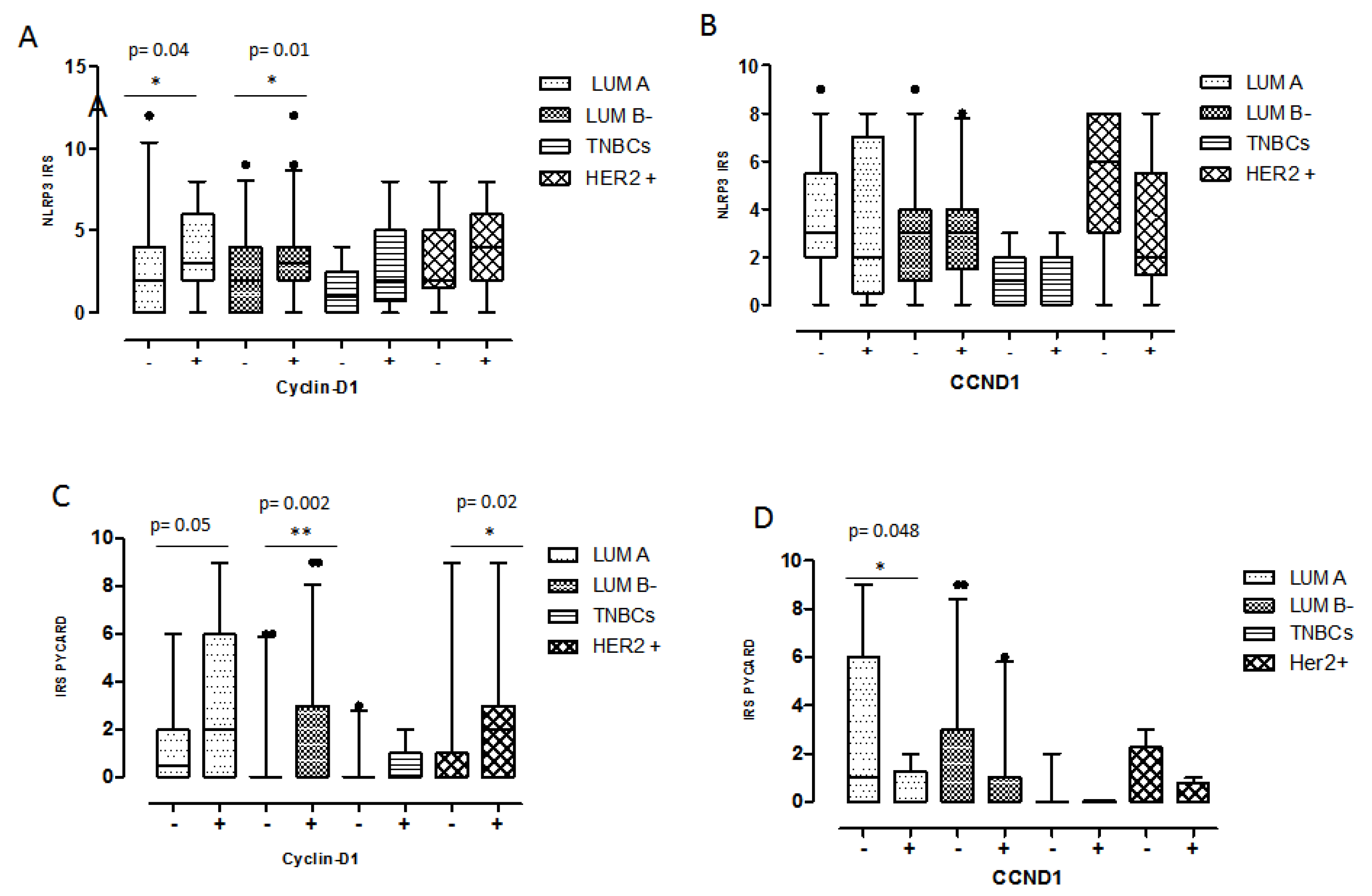
2.5. Relationship between Coupled Biomarkers and Clinicopathological Characteristics
3. Discussion
4. Materials and Methods
4.1. Patients and Clinicopathological Characteristics
4.2. Tissue Microarrays and Immunohistochemistry
4.3. Immunohistochemical Assessment
4.4. Detection of CCND1 and MYC Gene Alterations by FISH
4.5. Follow-Up and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.aiom.it/wp-content (accessed on 1 January 2021).
- Lu, H.; Ouyang, W.; Huang, C. Inflammation, a key event in cancer development. Mol. Cancer Res. 2006, 4, 221–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broz, P.; Dixit, V. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Li, T. Regulation of NLRP3 Inflammasome by Phosphorylation. Front. Immunol. 2018, 9, 2305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, F. Hydroxytyrosol alleviates dextran sodium sulfate-induced colitis by inhibiting NLRP3 inflammasome activation and modulating gut microbiota in vivo. Nutrition 2021, 97, 11579. [Google Scholar] [CrossRef]
- Mao, J.; Ma, X.; Zhu, J.; Zhang, H. Ginsenoside Rg1 ameliorates psoriasis-like skin lesions by suppressing proliferation and NLRP3 inflammasomes in keratinocytes. J. Food Biochem. 2022, 46, e14053. [Google Scholar] [CrossRef]
- Lu, F.; Zhao, Y.; Pang, Y.; Ji, M.; Sun, Y.; Wang, H.; Zou, J.; Wang, Y.; Li, G.; Sun, T.; et al. NLRP3 inflammasome upregulates PD-L1 expression and contributes to immune suppression in lymphoma. Cancer Lett. 2021, 497, 178–189. [Google Scholar] [CrossRef]
- Tang, D.; Liu, H.; Zhao, Y.; Qian, D.; Luo, S.; Patz, E.F., Jr.; Su, L.; Shen, S.; Christiani, D.C.; Gao, W.; et al. Genetic variants of BIRC3 and NRG1 in the NLRP3 inflammasome pathway are associated with non-small cell lung cancer survival. Am. J. Cancer Res. 2020, 10, 2582–2595. [Google Scholar]
- Wang, B.; Li, H.; Wang, X.; Zhu, X. The association of aberrant expression of NLRP3 and p-S6K1 in colorectal cancer. Pathol. Res. Pract. 2020, 216, 152737. [Google Scholar] [CrossRef]
- Kantono, M.; Guo, B. Inflammasomes and Cancer: The Dynamic Role of the Inflammasome in Tumor Development. Front. Immunol. 2017, 8, 1132. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, H.; Xu, Y.; Peng, T.; Meng, X.; Zou, F. NLRP3 induces the autocrine secretion of IL-1β to promote epithelial–mesenchymal transition and metastasis in breast cancer. Biochem. Biophys. Res. Commun. 2021, 560, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.T.-H.; Pham, D.-V.; Choi, D.-Y.; Park, P.-H. Mitophagy induction and aryl hydrocarbon receptor-mediated redox signaling contribute to the suppression of breast cancer cell growth by raloxifene via regulation of inflammasomes activation. Antioxid. Redox Signal. 2022, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Scarpi, E.; Sonnessa, M.; Cioffi, A.; Buccino, F.; Giotta, F.; Pastena, M.I.; Zito, F.A.; Mangia, A. Prognostic Value of NLRP3 Inflammasome and TLR4 Expression in Breast Cancer Patients. Front. Oncol. 2021, 11, 705331. [Google Scholar] [CrossRef] [PubMed]
- Houschyar, K.S.; Chelliah, M.P.; Rein, S.; Maan, Z.N.; Weissenberg, K.; Duscher, D.; Branski, L.K.; Siemers, F. Role of Wnt signaling during inflammation and sepsis: A review of the literature. Int. J. Artif. Organs 2018, 41, 247–253. [Google Scholar] [CrossRef]
- Goretsky, T.; Bradford, E.M.; Ryu, H.; Tahir, M.; Moyer, M.P.; Gao, T.; Li, L.; Barrett, T.A. A Cytosolic Multiprotein Complex Containing p85α Is Required for β-Catenin Activation in Colitis and Colitis-associated Cancer. J. Biol. Chem. 2016, 291, 4166–4177. [Google Scholar] [CrossRef] [Green Version]
- Betten, R.; Scharner, B.; Probst, S.; Edemir, B.; Wolff, N.A.; Langelueddecke, C.; Lee, W.K.; Thévenod, F. Tonicity inversely modulates lipocalin-2 (Lcn2/24p3/NGAL) receptor (SLC22A17) and Lcn2 expression via Wnt/β-catenin signaling in renal inner medullary collecting duct cells: Implications for cell fate and bacterial infection. Cell Commun. Signal. 2018, 16, 74. [Google Scholar] [CrossRef] [Green Version]
- Monga, P.S. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef] [Green Version]
- Haybar, H.; Khodadi, E.; Shahrabi, S. Wnt/β-catenin in ischemic myocardium: Interactions and signaling pathways as a therapeutic target. Heart Fail. Rev. 2019, 24, 411–419. [Google Scholar] [CrossRef]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef]
- Segditsas, S.; Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25, 7531–7537. [Google Scholar] [CrossRef] [Green Version]
- Palacios, J.; Gamallo, C. Mutations in the β-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res. 1998, 58, 1344–1347. [Google Scholar] [PubMed]
- Lin, S.Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.C. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.-X.; Pan, H.-R.; Song, X.-Y.; Chang, Q.-Q.; Zhang, D.-D. Hedyotis diffusa plus Scutellaria barbata Suppress the Growth of Non-Small-Cell Lung Cancer via NLRP3/NF- κ B/MAPK Signaling Pathways. Evid. Based Complementary Altern. Med. 2021, 2021, 6666499. [Google Scholar]
- Li, S.; Liang, X.; Ma, L.; Shen, L.; Li, T.; Zheng, L.; Sun, A.; Shang, W.; Chen, C.; Zhao, W.; et al. MiR-22 sustains NLRP3 expression and attenuates H. pylori-induced gastric carcinogenesis. Oncogene 2017, 37, 884–896. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Y.; Sheng, Y.; Wang, J.; Li, W.; Zhou, X.; Ruan, S.; Han, C. Galloflavin Relieves the Malignant Behavior of Colorectal Cancer Cells in the Inflammatory Tumor Microenvironment. Front. Pharmacol. 2021, 12, 752118. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, C.; Hua, M.; Wang, R.; Zhong, C.; Yu, J.; Han, F.; He, N.; Zhao, Y.; Liu, G.; et al. NLRP3 inflammasome activation plays a carcinogenic role through effector cytokine IL-18 in lymphoma. Oncotarget 2017, 8, 108571–108583. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Yépez, E.A.; Ayala-Sumuano, J.T.; Lezama, R.; Meza, I. A novel β-catenin signaling pathway activated by IL-1β leads to the onset of epithelial–mesenchymal transition in breast cancer cells. Cancer Lett. 2014, 354, 164–171. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.P.; Harris, C.C. Inflammation and cancer: An ancient link with novel potentials. Int. J. Cancer 2007, 121, 2373–2380. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Ghanawat, M.; Arjmand, B.; Rahim, F. The Pro-tumor and Anti-tumor Effects of NLRP3 Inflammasome as a New Therapeutic Option for Colon Cancer: A Meta-analysis of Pre-clinical Studies. J. Gastrointest. Cancer 2022. [Google Scholar] [CrossRef] [PubMed]
- Guey, B.; Bodnar-Wachtel, M.; Drouillard, A.; Eberhardt, A.; Pratviel, M.; Goutagny, N.; Bendriss-Vermare, N.; Puisieux, I.; Caux, C.; Walzer, T.; et al. Inflammasome Deletion Promotes Anti-Tumor NK Cell Function in an IL-1/IL-18 Independent Way in Murine Invasive Breast Cancer. Front. Oncol. 2020, 10, 1683. [Google Scholar] [CrossRef] [PubMed]
- Muthusami, S.; Ramachandran, I.K.; Babu, K.N.; Krishnamoorthy, S.; Guruswamy, A.; Queimado, L.; Chaudhuri, G.; Ramachandran, I. Role of Inflammation in the Development of Colorectal Cancer. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Ershaid, N.; Sharon, Y.; Doron, H.; Raz, Y.; Shani, O.; Cohen, N.; Monteran, L.; Leider-Trejo, L.; Ben-Shmuel, A.; Yassin, M.; et al. NLRP3 Inflammasome in Fibroblasts Links Tissue Damage With Inflammation in Breast Cancer Progression and Metastasis. Nat. Commun. 2019, 10, 4375. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.G.; Wu, X.X.; Hua, T.; Xin, X.Y.; Feng, D.L.; Chi, S.Q.; Wang, X.X.; Wang, H.B. NLRP3 inflammasome activation by estrogen promotes the progression of human endometrial cancer. Onco Targets Ther. 2019, 12, 6927–6936. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Fan, W.; Gao, X.; Huang, K.; Ding, C.; Ma, G.; Yan, L.; Song, S. Estrogen receptor alpha regulates the Wnt/β-catenin signaling pathway in colon cancer by targeting the NOD-like receptors. Cell. Signal. 2019, 61, 86–92. [Google Scholar] [CrossRef]
- Fan, W.; Gao, X.; Ding, C.; Lv, Y.; Shen, T.; Ma, G.; Yan, L.; Song, S. Estrogen receptors participate in carcinogenesis signaling pathways by directly regulating NOD-like receptors. Biochem. Biophys. Res. Commun. 2019, 511, 468–475. [Google Scholar] [CrossRef]
- Raut, P.-K.; Kim, S.-H.; Choi, D.-Y.; Jeong, G.-S.; Park, P.-H. Growth of breast cancer cells by leptin is mediated via activation of the inflammasome: Critical roles of estrogen receptor signaling and reactive oxygen species production. Biochem. Pharmacol. 2019, 161, 73–88. [Google Scholar] [CrossRef]
- Yang, H.; Liu, J.; Chen, X.; Li, G. Angptl2 gene knockdown is critical for abolishing angiotensin II-induced vascular smooth muscle cell proliferation and migration. Biochem. Cell Biol. 2022, 100, 59–67. [Google Scholar] [CrossRef]
- Koizumi, M.; Watanabe, T.; Masumoto, J.; Sunago, K.; Imamura, Y.; Kanemitsu, K.; Kumagi, T.; Hiasa, Y. Apoptosis-associated speck-like protein containing a CARD regulates the growth of pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 22351. [Google Scholar] [CrossRef]
- Stone, A.-R.; Bobo, W.; Brat, D.-J.; Devi, N.-S.; Van Meir, E.-G.; Vertino, P.-M. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am. J. Pathol. 2004, 165, 1151–1161. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Saiki, Y.; Arai, K.; Ishizawa, K.; Fukushige, S.; Aoki, K.; Abe, J.; Takahashi, S.; Sato, I.; Sakurada, A.; et al. S100A10 upregulation associates with poor prognosis in lung squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2018, 505, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.-E.; McConnell, B.-B.; Bowring, C.-E.; Donald, C.-D.; Warren, S.-T.; Vertino, P.-M. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000, 60, 6236–6242. [Google Scholar] [PubMed]
- Guan, X.; Sagara, J.; Yokoyama, T.; Koganehira, Y.; Oguchi, M.; Saida, T.; Taniguchi, S.I. ASC/TMS1, a caspase-1 activating adaptor, is downregulated by aberrant methylation in human melanoma. Int. J. Cancer 2003, 107, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, S.; Wang, X. The Metabolic Mechanisms of Breast Cancer Metastasis. Front. Oncol. 2021, 10, 602416. [Google Scholar] [CrossRef]
- Qu, J.; Zhao, X.; Wang, J.; Liu, X.; Yan, Y.; Liu, L.; Cai, H.; Qu, H.; Lu, N.; Sun, Y.; et al. MYC overexpression with its prognostic and clinicopathological significance in breast cancer. Oncotarget 2017, 8, 93998–94008. [Google Scholar] [CrossRef] [PubMed]
- Gillett, C.; Smith, P.; Gregory, W.; Richards, M.; Millis, R.; Peters, G.; Barnes, D. Cyclin D1 and prognosis in human breast cancer. Int. J. Cancer 1996, 69, 92–99. [Google Scholar] [CrossRef]
- Kenny, F.S.; Hui, R.; Musgrove, E.A.; Gee, J.M.; Blamey, R.W.; Nicholson, R.I.; Sutherland, R.L.; Robertson, J.F. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin. Cancer Res. 1999, 5, 2069–2076. [Google Scholar]
- McIntosh, G.G.; Anderson, J.J.; Milton, I.; Steward, M.; Parr, A.H.; Thomas, M.D.; Henry, J.A.; Angus, B.; Lennard, T.W.; Horne, C.H. Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene 1995, 11, 885–891. [Google Scholar]
- Saponaro, C.; Vagheggini, A.; Scarpi, E.; Centonze, M.; Catacchio, I.; Popescu, O.; Pastena, M.I.; Giotta, F.; Silvestris, N.; Mangia, A. NHERF1 and Tumor Microenvironment: A New Scene in Invasive Breast Carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 96. [Google Scholar] [CrossRef] [Green Version]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| N. (%) | |
|---|---|
| Age (years): median value (range 29–80) | 53 |
| ≤53 | 121 (50) |
| >53 | 119 (50) |
| Histotype | |
| IDC | 205 (86.0) |
| ILC | 17 (7.0) |
| Other | 17 (7.0) |
| unknown | 1 |
| Tumor size (cm) | |
| ≤2.0 | 137 (58) |
| >2.0 | 99 (42) |
| Unknown | 4 |
| Node | |
| Negative | 149 (63) |
| Positive | 86 (37) |
| unknown | 5 |
| Grade | |
| 1 | 18 (8) |
| 2 | 107 (45) |
| 3 | 113 (47) |
| unknown | 2 |
| ER (%) | |
| <1 | 85 (36) |
| ≥1 | 152 (64) |
| unknown | 3 |
| PgR (%) | |
| <1 | 100 (42) |
| ≥1 | 137 (58) |
| unknown | 3 |
| Ki67 (%) | |
| <14 | 83 (35) |
| ≥14 | 154 (65) |
| unknown | 3 |
| HER2 | |
| Negative | 204 (86) |
| Positive | 33 (14) |
| unknown | 3 |
| Molecular Subtype | |
| Luminal A | 75 (32) |
| Luminal B− | 89 (38) |
| Her2+ | 32 (13) |
| TNBC | 41 (17) |
| unknown | 3 |
| NLRP3 | |
| Negative | 112 (50) |
| Positive | 113 (50) |
| unknown | 15 |
| PYCARD | |
| Negative | 154 (69) |
| Positive | 68 (31) |
| unknown | 18 |
| CyclinD1 | |
| Negative | 120 (52) |
| Positive | 109 (48) |
| unknown | 11 |
| MYC | |
| Negative | 158 (68) |
| Positive | 74 (32) |
| unknown | 8 |
| CCND1 | |
| Negative | 106 (70) |
| Positive | 46 (30) |
| unknown | 88 |
| MYC | |
| Negative | 89 (60) |
| Positive | 59 (40) |
| unknown | 92 |
| Protein Expression | Gene Alteration | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NLRP3 | PYCARD | Cyclin D1 | MYC | CCND1 | Myc | |||||||||||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |||||||
| N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | |
| Age | ||||||||||||||||||
| ≤53 | 61 (54.5) | 52 (46) | 0.2052 | 79 (71) | 75 (68) | 0.56 | 67 (56) | 46 (42) | 71 (45) | 44 (59) | 0.039 | 46 (43) | 27 (59) | 46 (52) | 31 (52) | 0.918 | ||
| >53 | 51 (45.5) | 61 (54) | 32 (29) | 36 (32) | 53 (44) | 63 (58) | 0.039 | 87 (55) | 30 (41) | 60 (57) | 19 (41) | 0.08 | 43 (48) | 28 (48) | ||||
| Histotype | ||||||||||||||||||
| IDC | 91 (85) | 99 (88) | 0.012 | 130 (85) | 60 (88) | 0.20 | 100 (84) | 95 (87) | 0.55 | 133 (85) | 65 (88) | 0.035 | 87 (82) | 42 (91) | 75 (84) | 53 (90) | 0.359 | |
| ILC | 4 (4) | 11 (10) | 9 (6) | 6 (9) | 8 (7) | 8 (7) | 15 (9) | 1 (1) | 7 (7) | 1 (2) | 6 (7) | 1 (2) | ||||||
| Other | 12 (11) | 3 (2) | 14 (9) | 2 (3) | 11 (9) | 6 (6) | 9 (6) | 8 (11) | 12 (11) | 3 (7) | 0.32 | 8 (9) | 5 (8) | |||||
| T. size (cm) | ||||||||||||||||||
| ≤2.0 | 63 (57) | 65 (59) | 0.78 | 93 (61) | 34 (51.5) | 0.20 | 71 (61) | 59 (54) | 0.282 | 93 (60) | 41 (56) | 0.583 | 55 (53) | 29 (63) | 54 (61) | 28 (48) | 0.118 | |
| >2.0 | 48 (43) | 46 (41) | 60 (39) | 32 (48.5) | 45 (39) | 50 (46) | 62 (40) | 32 (44) | 49 (47) | 17 (37) | 0.247 | 34 (39) | 30 (52) | |||||
| Node | ||||||||||||||||||
| Negative | 70 (64) | 69 (62) | 0.68 | 96 (64) | 41 (60) | 0.56 | 81 (69) | 62 (58) | 0.114 | 95 (62) | 52 (71) | 0.159 | 66 (63) | 27 (59) | 58 (66) | 35 (60) | 0.493 | |
| Positive | 39 (36) | 43 (38) | 53 (36) | 27 (40) | 37 (31) | 44 (42) | 59 (38) | 21 (29) | 38 (37) | 19 (41) | 0.57 | 30 (34) | 23 (40) | |||||
| Grade | ||||||||||||||||||
| 1-2 | 55 (49.6) | 64 (57) | 0.25 | 77 (50.3) | 39 (58) | 0.28 | 57 (48) | 63 (58) | 0.115 | 90 (57) | 33 (45) | 0.086 | 60 (58) | 25 (54) | 61 (74) | 21 (36) | <0.0001 | |
| 3 | 56 (50.4) | 48 (43) | 76 (49.7) | 28 (42) | 62 (52) | 45 (42) | 67 (43) | 40 (55) | 44 (42) | 21 (46) | 0.703 | 21 (26) | 37 (64) | |||||
| ER (%) | ||||||||||||||||||
| <1 | 58(52) | 21 (19) | 66 (44) | 9 (13) | 59 (50) | 20 (18) | <0.0001 | 48 (31) | 33 (45) | 0.033 | 26 (25) | 17 (37) | 16 (18) | 27 (46) | 0.0002 | |||
| ≥1 | 53 (48) | 90 (81) | <0.0001 | 85 (56) | 59 (87) | <0.0001 | 59 (50) | 88 (82) | 108 (69) | 40 (55) | 79 (75) | 29 (63) | 0.126 | 72 (92) | 31 (54) | |||
| PgR (%) | ||||||||||||||||||
| <1 | 62 (56) | 32 (29) | 72 (48) | 18 (26) | 65 (55) | 29 (27) | <0.0001 | 61 (39) | 35 (48) | 0.206 | 37 (35) | 18 (39) | 24 (27) | 30 (52) | 0.0027 | |||
| ≥1 | 49 (44) | 79 (71) | <0.0001 | 79 (52) | 50 (74) | 0.003 | 53 (45) | 79 (73) | 95 (61) | 38 (52) | 68 (65) | 28 (61) | 0.647 | 64 (73) | 28 (48) | |||
| Ki67 (%) | ||||||||||||||||||
| <14 | 32 (29) | 42 (38) | 43 (28) | 32 (47) | 38 (32) | 40 (37) | 0.445 | 64 (41) | 16 (22) | 0.0047 | 44 (42) | 12 (26) | 44 (50) | 12 (21) | 0.0004 | |||
| ≥14 | 79 (71) | 69 (62) | 0.15 | 108 (72) | 36 (53) | 0.007 | 80 (68) | 68 | 92 (59) | 57 (78) | 61 (58) | 34 (74) | 0.064 | 44 (50) | 46 (79) | |||
| HER2 | ||||||||||||||||||
| Negative | 96 (86) | 95 (85) | 0.84 | 130 (86) | 58 (85) | 0.87 | 104 (88) | 92 (85) | 0.5 | 134 (86) | 64 (86) | 0.84 | 93 (86) | 38 (83) | 0.31 | 79 (90) | 48 (83) | 0.21 |
| Positive | 15 (14) | 16 (15) | 21 (14) | 10 (15) | 14 (12) | 16 (15) | 22 (14) | 10 (14) | 12 (14) | 8 (17) | 9 (10) | 10 (17) | ||||||
| Protein Expression | Gene Alteration | |||||||||||||||||
| NLRP3 | PYCARD | Cyclin D1 | MYC | CCND1 | Myc | |||||||||||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |||||||
| N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | |
| Luminal A | ||||||||||||||||||
| no | 81 (73) | 73 (72) | 0.24 | 113 (75) | 39 (57) | 0.009 | 83 (70) | 72 (66) | 0.55 | 99 (63) | 59 (80) | 0.01 | 63 (60) | 36 (78) | 0.02 | 48 (55) | 47 (81) | 0.001 |
| yes | 30 (27) | 38 (28) | 38 (25) | 29 (43) | 35 (30) | 36 (34) | 57 (37) | 15 (20) | 42 (40) | 10 (22) | 40 (45) | 11 (19) | ||||||
| Luminal B- | ||||||||||||||||||
| no | 71(64) | 65(59) | 0.40 | 87(58) | 45(66) | 0.23 | 77(65) | 62(57) | 0.22 | 112 (72) | 31(42) | <0.0001 | 62 (59) | 25 (54) | 0.59 | 58 (66) | 32 (55) | 0.19 |
| yes | 40(36) | 46(41) | 64(42) | 23(34) | 41(35) | 46(43) | 44(28) | 43(58) | 43 (41) | 21 (46) | 30 (34) | 26 (45) | ||||||
| HER2+ | ||||||||||||||||||
| no | 96 (86) | 96 (86) | 1.00 | 131 (87) | 58 (85) | 0.77 | 104 (88) | 92 (85) | 0.51 | 135 (86) | 64 (86) | 0.99 | 93 (88) | 38 (82) | 0.31 | 79 (90) | 48 (83) | 0.21 |
| yes | 15 (14) | 15 (14) | 20 (13) | 10 (15) | 14 (12) | 16 (15) | 21 (14) | 10 (14) | 12 (12) | 8 (18) | 9 (10) | 10 (17) | ||||||
| TNBC | ||||||||||||||||||
| no | 85 (76) | 99 (89) | 0.01 | 122 (81) | 62 (91) | 0.052 | 90 (76) | 98 (91) | 0.003 | 122 (78) | 68 (92) | 0.01 | 97 (92) | 39 (85) | 0.15 | 79 (90) | 47 (81) | 0.13 |
| yes | 26 (24) | 12 (11) | 29 (19) | 6 (9) | 28 (24) | 10 (9) | 34 (22) | 6 (8) | 8 (8) | 7 (15) | 9 (10) | 11 (19) | ||||||
| NLRP3 | PYCARD | CyclinD1 | MYC | |||||
|---|---|---|---|---|---|---|---|---|
| r | p-Value | r | p-Value | r | p-Value | r | p-Value | |
| NLRP3 | 0.322 | <0.0001 | 0.366 | <0.0001 | −0.127 | 0.059 | ||
| PYCARD | 0.285 | <0.0001 | 0.003 | 0.957 | ||||
| Cyclin-D1 | −0.037 | 0.577 | ||||||
| (A) Protein expression | (B) Gene expression | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NLRP3/Cyclin D1 | PYCARD/Cyclin D1 | CCND1 | MYC | |||||||||
| Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | |||||
| N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | N (%) | N (%) | p | |
| Age | ||||||||||||
| ≤53 | 42 (64) | 29 (45) | 51 (57) | 20 (44) | 24 (53) | 13 (68) | 29 (47,5) | 4 (50) | ||||
| >53 | 24 (36) | 36 (55) | 0.0289 | 39 (43) | 25 (56) | 0.180 | 21 (47) | 6 (32) | 0.264 | 32 (52,5) | 4(50) | 0.895 |
| Histotype | ||||||||||||
| IDC | 55 (85) | 58 (89) | 76 (85) | 41 (91) | 35 (78) | 19 (100) | 49 (80) | 8 (100) | ||||
| ILC | 2 (3) | 5 (8) | 3 (4) | 2 (4,5) | 2 (4) | 0 | 2 (3) | 0 | ||||
| Other | 8 (12) | 2 (3) | 0.083 | 10 (11) | 2 (4,5) | 0.418 | 8 (18) | 0 | 0.08 | 10 (17) | 0 | 0.385 |
| T. size (cm) | ||||||||||||
| ≤2.0 | 38 (58,5) | 34 (52) | 56 (63) | 22 (49) | 20 (45) | 9 (47) | 34 (57) | 3 (37,5) | ||||
| >2.0 | 27 (41,5) | 31 (48) | 0.48 | 33 (37) | 23 (51) | 0.119 | 24 (55) | 10 (53) | 0.888 | 26 (43) | 5 (62,5) | 0.306 |
| Node | ||||||||||||
| Negative | 46 (71) | 39 (61) | 59 (67) | 25 (56) | 28 (65) | 8 (42) | 38 (64) | 2 (25) | ||||
| Positive | 19 (29) | 25 (39) | 0.238 | 29 (33) | 20 (44) | 0.193 | 15 (35) | 11 (58) | 0.09 | 21 (36) | 6 (75) | 0.033 |
| Grade | ||||||||||||
| 1-2 | 28 (43) | 35 (55) | 38 (43) | 23 (52) | 25 (57) | 9 (47) | 32 (53) | 2 (25) | ||||
| 3 | 37 (57) | 29 (45) | 0.187 | 51 (57) | 21 (48) | 0.297 | 19 (43) | 10 (53) | 0.489 | 28 (47) | 6 (75) | 0.132 |
| ER (%) | ||||||||||||
| <1 | 22 (34) | 7 (11) | 27 (31) | 3 (7) | 6 (14) | 2 (10,5) | 10 (17) | 0 | ||||
| ≥1 | 43 (66) | 58 (89) | 0.0016 | 61 (69) | 42 (93) | 0.0017 | 38 (86) | 17 (89,5) | 0.733 | 50 (83) | 8 (100) | 0.211 |
| PgR (%) | ||||||||||||
| <1 | 26 (40) | 14 (22) | 31 (35) | 10 (22) | 9 (20,5) | 3 (16) | 13 (22) | 0 | ||||
| ≥1 | 39 (60) | 50 (78) | 0.026 | 57 (65) | 35 (78) | 0.124 | 35 (79,5) | 16 (84) | 0.665 | 47 (78) | 8 (100) | 0.143 |
| Ki67 (%) | ||||||||||||
| <14 | 16 (25) | 22 (34) | 23 (26) | 20 (44) | 19 (43) | 4 (21) | 20 (33) | 3 (37,5) | ||||
| ≥14 | 49 (75) | 42 (66) | 0.224 | 65 (74) | 25 (56) | 0.032 | 25 (57) | 15 (79) | 0.094 | 40 (67) | 5 (62,5) | 0.815 |
| HER2 | ||||||||||||
| Negative | 58 (89) | 55 (86) | 76 (86) | 37 (82) | 42 (95) | 16 (84) | 53 (88) | 6 (75) | ||||
| Positive | 7 (11) | 9 (14) | 0.570 | 12 (14) | 8 (18) | 0.527 | 2 (5) | 3 (16) | 0.129 | 7 (12) | 2 (25) | 0.295 |
| PFS | OS | |||||||||
| N. pts | Events | 5-years DFS | p-value | HR (95% CI) | N pts | Events | 5-years OS | p-value | HR (95% CI) | |
| Overall | ||||||||||
| NLRP3 | 0.050 | 0.441 | ||||||||
| 0 | 107 | 23 | 85.9 | 1.00 | 107 | 8 | 93.5 | 1.000 | ||
| 1 | 111 | 12 | 93.7 | 1.997 (0.987–4.040) | 111 | 5 | 98.2 | 1.554 (0.501–4.821) | ||
| PYCARD | 0.017 | 0.059 | ||||||||
| 0 | 149 | 32 | 85.8 | 1.00 | 149 | 13 | 93.3 | 1.000 | ||
| 1 | 66 | 5 | 93.9 | 2.974 (1.158–7.637) | 66 | 1 | 98.5 | 5.650 (0.737–43.318) | ||
| Ciclina D1 | 0.909 | 0.560 | ||||||||
| 0 | 117 | 21 | 88.9 | 1.00 | 117 | 7 | 94.9 | 1.000 | ||
| 1 | 106 | 17 | 87.6 | 1.038 (0.546–1.975) | 106 | 9 | 94.3 | 0.739 (0.266–2.054) | ||
| MYC | 0.779 | 0.041 | ||||||||
| 0 | 155 | 25 | 87.1 | 1.00 | 155 | 7 | 97.4 | 1.000 | ||
| 1 | 72 | 13 | 91.6 | 0.909 (0.465–1.776) | 72 | 8 | 90.3 | 0.347 (0.120–1.005) | ||
| CCND1 | 0.227 | 0.013 | ||||||||
| 0 | 102 | 12 | 94.1 | 1.00 | 102 | 2 | 98.0 | 1.000 | ||
| 1 | 43 | 8 | 88.1 | 0.580 (0.237–1.419) | 43 | 5 | 93.0 | 0.162 (0.031–0.836) | ||
| Myc | 0.017 | 0.378 | ||||||||
| 0 | 89 | 7 | 96.6 | 1.00 | 89 | 3 | 97.8 | 1.000 | ||
| 1 | 55 | 12 | 85.5 | 0.339 (0.133–0.864) | 55 | 4 | 96.4 | 0.513 (0.113–2.327) | ||
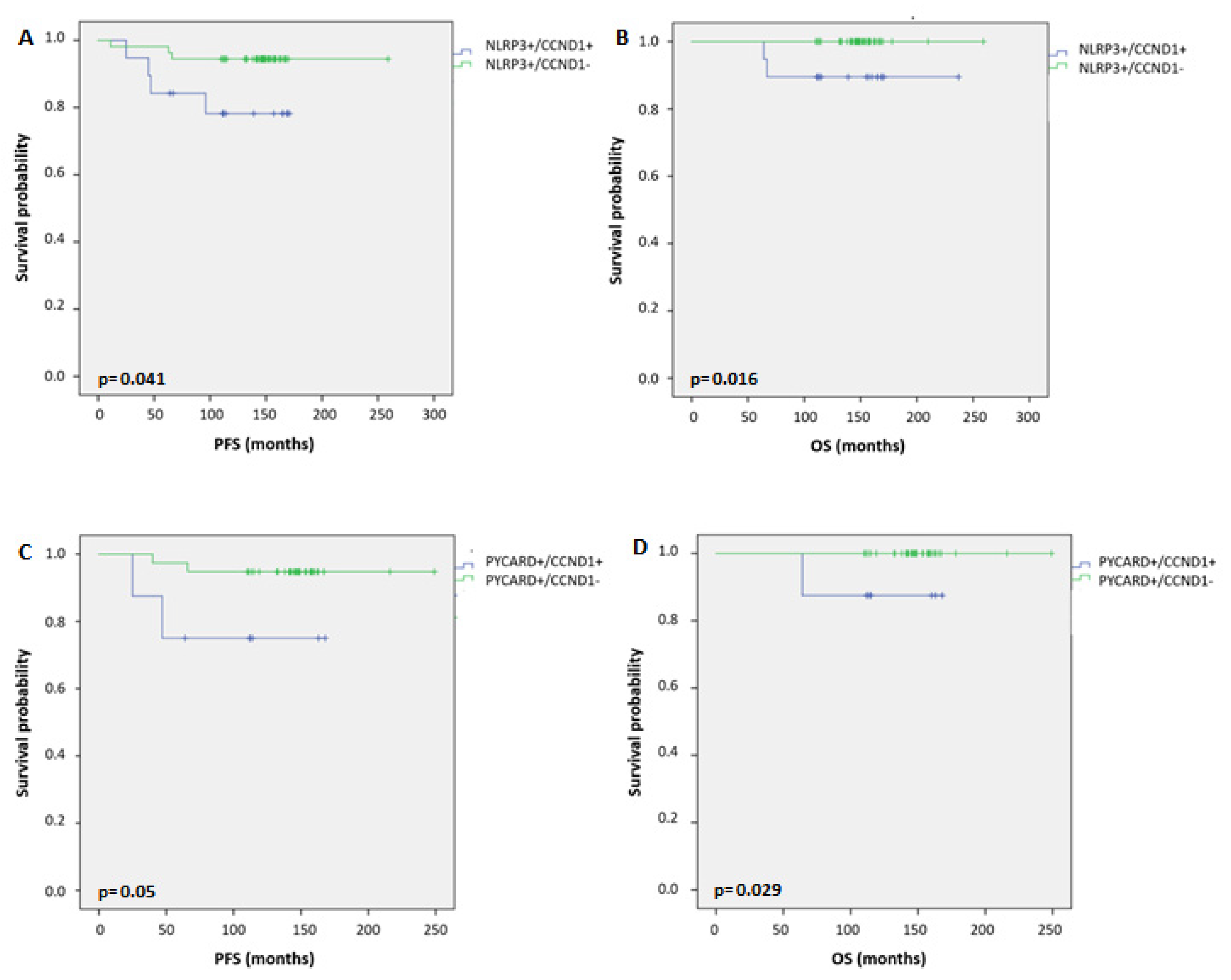
| NLRP3/CCND1 | 0.155 | 0.064 | ||||||||
| 0-0/0 | 43 | 8 | 90.7 | 1.00 | 43 | 2 | 95.3 | 1.000 | ||
| 1-1/1 | 19 | 4 | 84.2 | 0.642 (0.263–1.567) | 19 | 2 | 100 | 2.227 (0.314–15.810) | ||
| 2-1/0 | 54 | 3 | 98.1 | 0.482 (0.156–1.486) | 54 | 0 | 100 | - | ||
| 3-0/1 | 23 | 4 | 91.3 | 1.168 (0.483–2.825) | 23 | 3 | 87.0 | 2.983 (0.498–17.856) | ||
| NLRP3/Ciclina D1 | 0.370 | 0.116 | ||||||||
| 0-0/0 | 63 | 13 | 88.9 | 1.00 | 63 | 2 | 96.8 | 1.000 | ||
| 1-1/1 | 65 | 8 | 92.3 | 1.221 (0.367–4.062) | 65 | 3 | 98.5 | 1.557 (0.252–9.644) | ||
| 2-1/0 | 42 | 4 | 95.2 | 0.280 (0.074–1.055) | 42 | 2 | 97.6 | 1.700 (0.235–12.269) | ||
| 3-0/1 | 39 | 8 | 81.7 | 0.995 (0.299–3.304) | 39 | 6 | 87.2 | 4.886 (0.957–24.938) | ||
| PYCARD/CCND1 | 0.242 | 0.093 | ||||||||
| 0-0/0 | 58 | 9 | 93.1 | 1.00 | 58 | 2 | 96.6 | 1.000 | ||
| 1-1/1 | 8 | 2 | 75.0 | 0.471 (0.158–1.403) | 8 | 1 | 100 | 3.629 (0.329–40.028) | ||
| 2-1/0 | 38 | 2 | 97.4 | 0.259 (0.034–1.948) | 38 | 0 | 100 | - | ||
| 3-0/1 | 34 | 6 | 91.0 | 1.290 (0.625–2.660) | 34 | 4 | 91.2 | 3.549 (0.650–19.377) | ||
| PYCARD/Ciclina D1 | 0.141 | 0.133 | ||||||||
| 0-0/0 | 87 | 17 | 88.5 | 1.00 | 87 | 6 | 94.3 | 1.000 | ||
| 1-1/1 | 44 | 4 | 93.2 | 1.971 (0.424–9.174) | 44 | 1 | 97.7 | 0.355 (0.042–2.978) | ||
| 2-1/0 | 19 | 1 | 94.7 | 0.324 (0.070–1.502) | 19 | 0 | 100 | - | ||
| 3-0/1 | 57 | 13 | 82.1 | 1.206 (0.429–3.390) | 57 | 7 | 89.5 | 1.966 (0.655–5.895 | ||
| 95.0% Cl Per Exp(B) | ||||||
| B | SE | p-Value | Exp(B) | Lower | Upper | |
| PYCARD | 0.847 | 0.842 | 0.314 | 2.333 | 0.448 | 12.147 |
| NLRP3 | 0.679 | 0.728 | 0.351 | 1.973 | 0.474 | 8.213 |
| CyclinD1 | −0.932 | 0.785 | 0.235 | 0.394 | 0.085 | 1.832 |
| MYC | −0.911 | 0.705 | 0.196 | 0.402 | 0.101 | 1.600 |
| CCND1 | −1.130 | 0.683 | 0.098 | 0.323 | 0.085 | 1.231 |
| MYC gene | −1.677 | 0.800 | 0.036 | 0.187 | 0.039 | 0.897 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saponaro, C.; Fanizzi, A.; Sonnessa, M.; Mondelli, P.; Vergara, D.; Loisi, D.; Massafra, R.; Latorre, A.; Zito, F.A.; Schirosi, L. Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes. Pharmaceuticals 2022, 15, 651. https://doi.org/10.3390/ph15060651
Saponaro C, Fanizzi A, Sonnessa M, Mondelli P, Vergara D, Loisi D, Massafra R, Latorre A, Zito FA, Schirosi L. Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes. Pharmaceuticals. 2022; 15(6):651. https://doi.org/10.3390/ph15060651
Chicago/Turabian StyleSaponaro, Concetta, Annarita Fanizzi, Margherita Sonnessa, Paolo Mondelli, Daniele Vergara, Donato Loisi, Raffaella Massafra, Agnese Latorre, Francesco A. Zito, and Laura Schirosi. 2022. "Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes" Pharmaceuticals 15, no. 6: 651. https://doi.org/10.3390/ph15060651
APA StyleSaponaro, C., Fanizzi, A., Sonnessa, M., Mondelli, P., Vergara, D., Loisi, D., Massafra, R., Latorre, A., Zito, F. A., & Schirosi, L. (2022). Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes. Pharmaceuticals, 15(6), 651. https://doi.org/10.3390/ph15060651









