Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence
Abstract
:1. Introduction
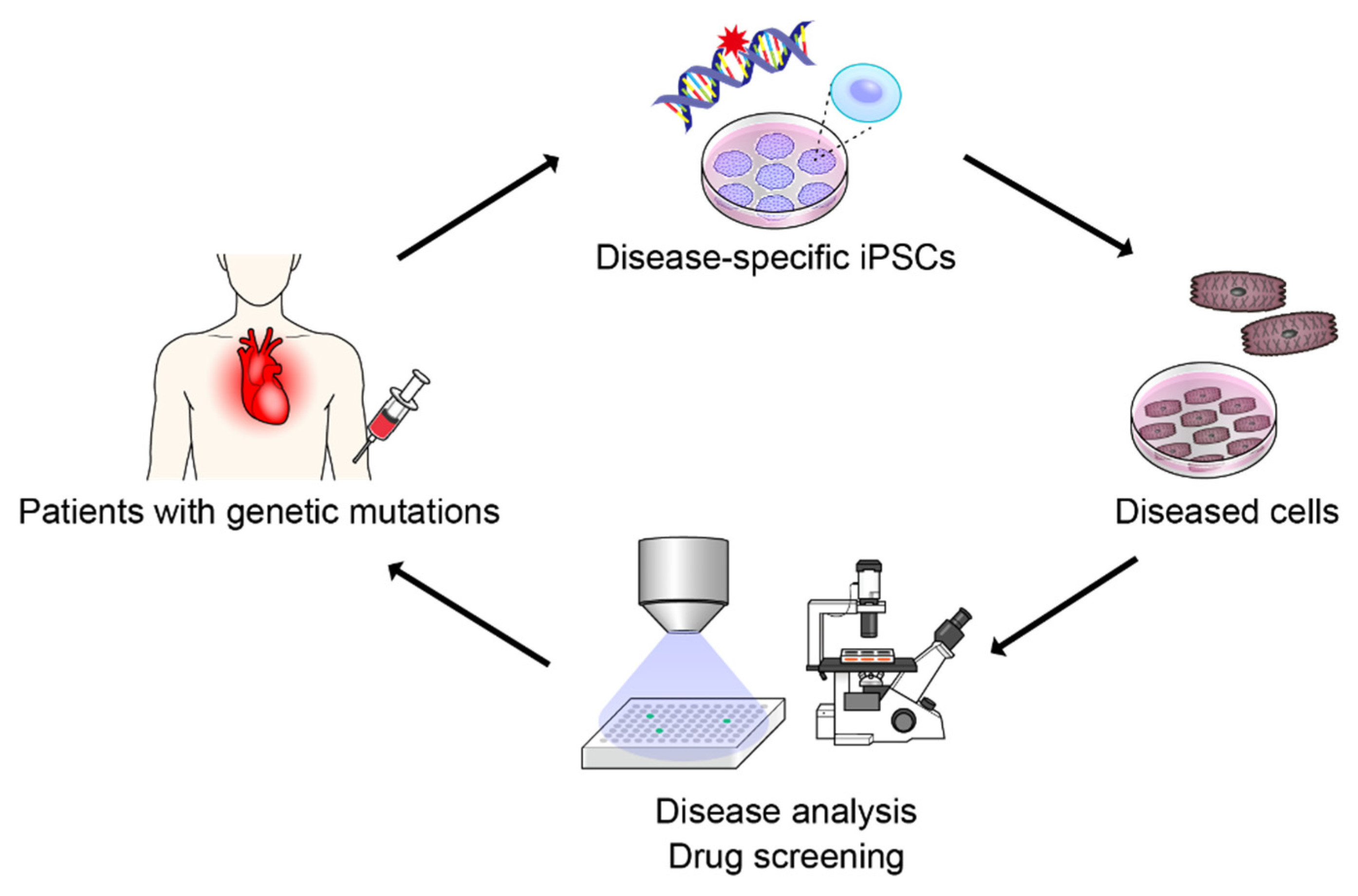
2. Patient-Specific iPSCs
3. Development of AI Technology
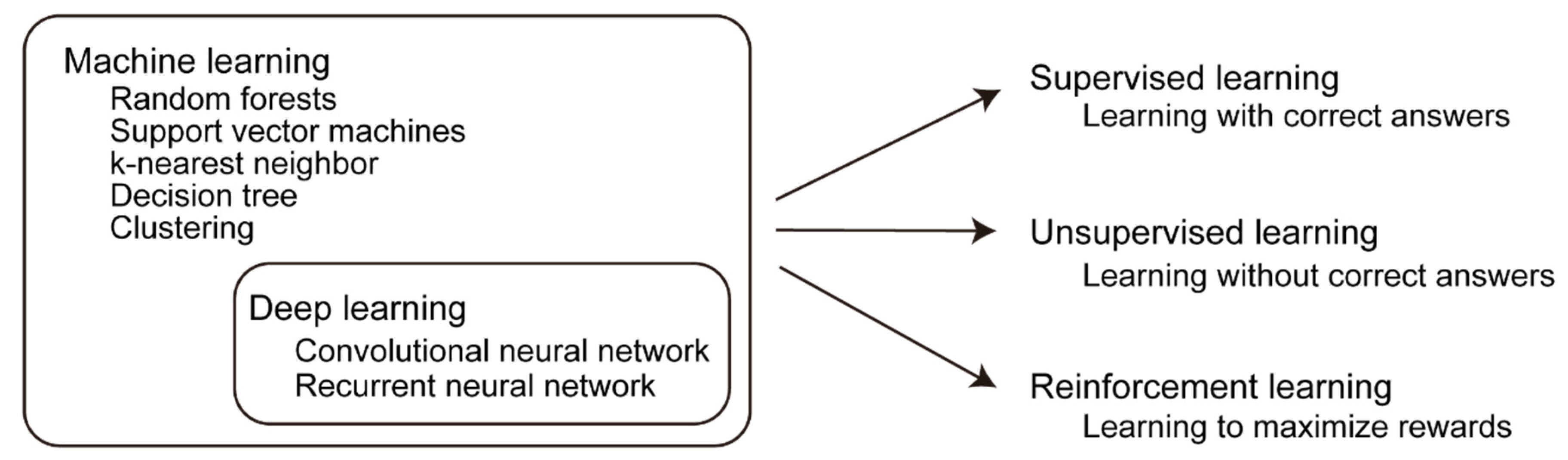
3.1. Development of Machine Learning Technology
3.2. Supervised Learning, Unsupervised Learning, and Reinforcement Learning
3.3. Deep Neural Network
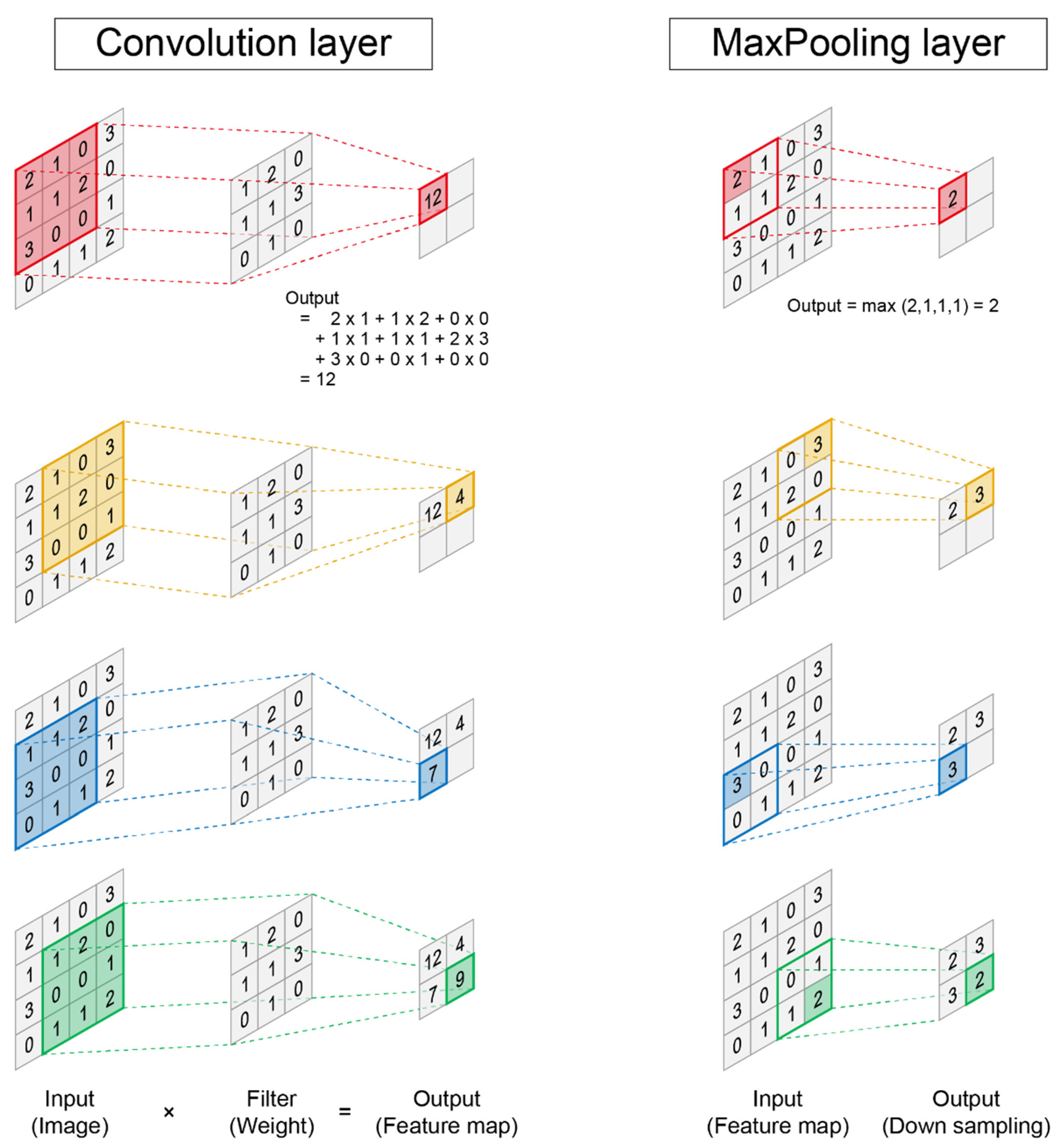
3.4. Convolutional Neural Network
4. AI Technology in Stem Cell Biology
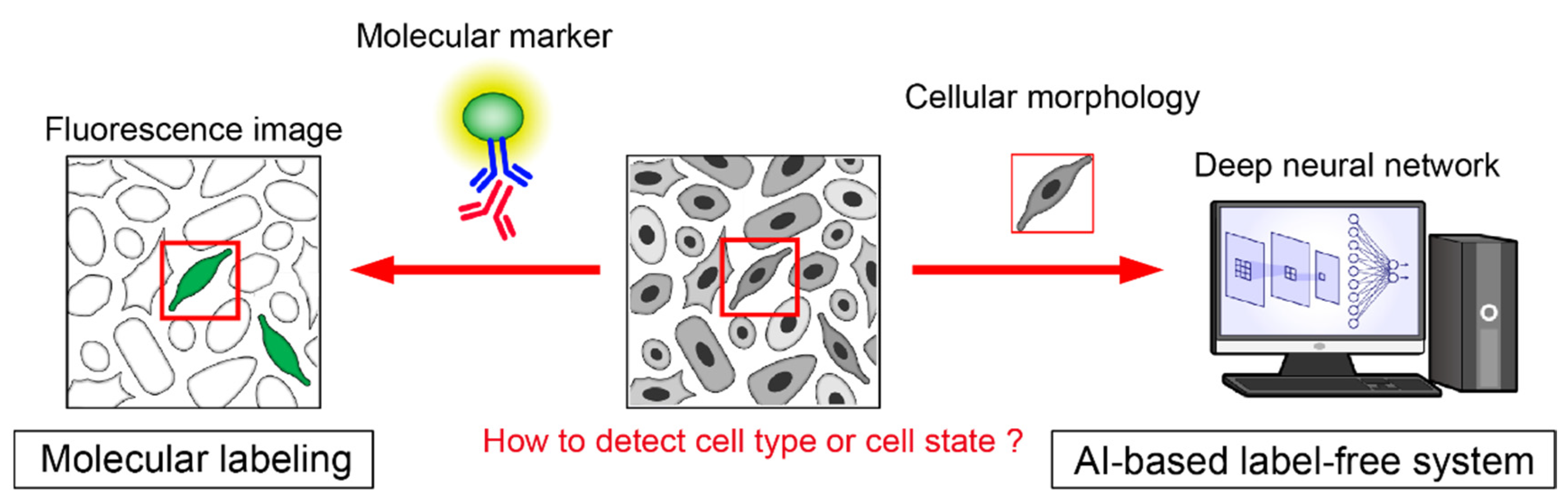
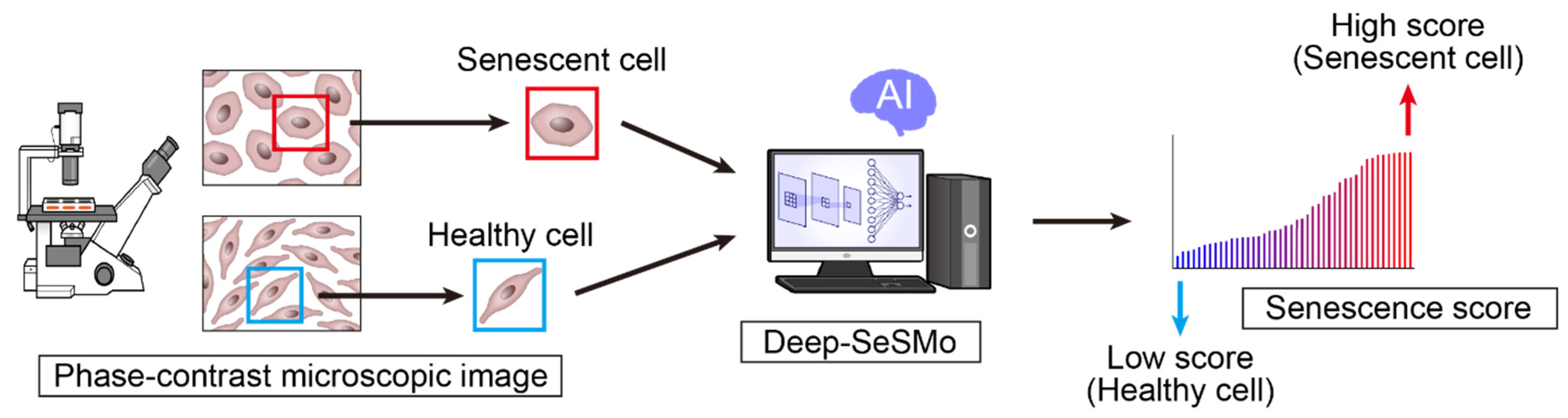
4.1. AI for Cell Recognition Based on Morphology
4.2. AI for Bioinformatics Tool
4.3. AI for iPSC and iPSC-Derived Differentiation Cell
5. AI for Drug Screening
5.1. Disease Evaluation Using AI
5.2. Drug Screening Using AI
5.3. Disease-Specific iPSCs and AI
6. Novel Technology for Disease Modeling with iPSCs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagoshi, N.; Okano, H. Applications of induced pluripotent stem cell technologies in spinal cord injury. J. Neurochem. 2017, 141, 848–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuasa, S.; Fukuda, K. Recent advances in cardiovascular regenerative medicine: The induced pluripotent stem cell era. Expert Rev. Cardiovasc. Ther. 2008, 6, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, S.; Fukuda, K. Cardiac Regenerative Medicine. Circ. J. 2008, 72, A49–A55. [Google Scholar] [CrossRef] [Green Version]
- Sinnecker, D.; Goedel, A.; Dorn, T.; Dirschinger, R.J.; Moretti, A.; Laugwitz, K.L. Modeling long-QT syndromes with iPS cells. J. Cardiovasc. Transl. Res. 2013, 6, 31–36. [Google Scholar] [CrossRef]
- Shimojima, M.; Yuasa, S.; Motoda, C.; Yozu, G.; Nagai, T.; Ito, S.; Lachmann, M.; Kashimura, S.; Takei, M.; Kusumoto, D.; et al. Emerin plays a crucial role in nuclear invagination and in the nuclear calcium transient. Sci. Rep. 2017, 7, 44312. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Yuasa, S.; Mearini, G.; Egashira, T.; Seki, T.; Kodaira, M.; Kusumoto, D.; Kuroda, Y.; Okata, S.; Suzuki, T.; et al. Endothelin-1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell-derived cardiomyocytes. J. Am. Heart Assoc. 2014, 3, e001263. [Google Scholar] [CrossRef] [Green Version]
- Gu, M.; Shao, N.Y.; Sa, S.; Li, D.; Termglinchan, V.; Ameen, M.; Karakikes, I.; Sosa, G.; Grubert, F.; Lee, J.; et al. Patient-Specific iPSC-Derived Endothelial Cells Uncover Pathways that Protect against Pulmonary Hypertension in BMPR2 Mutation Carriers. Cell Stem Cell 2017, 20, 490–504.e495. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Yuasa, S.; Node, K.; Fukuda, K. Cardiovascular Disease Modeling Using Patient-Specific Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2015, 16, 18894–18922. [Google Scholar] [CrossRef] [Green Version]
- Kessler, T.; Vilne, B.; Schunkert, H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol. Med. 2016, 8, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-Specific Induced Pluripotent Stem Cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, K.; Jaenisch, R. Technical Challenges in Using Human Induced Pluripotent Stem Cells to Model Disease. Cell Stem Cell 2009, 5, 584–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusumoto, D.; Yuasa, S. The application of convolutional neural network to stem cell biology. Inflamm. Regen. 2019, 39, 14. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, D.; Lachmann, M.; Kunihiro, T.; Yuasa, S.; Kishino, Y.; Kimura, M.; Katsuki, T.; Itoh, S.; Seki, T.; Fukuda, K. Automated Deep Learning-Based System to Identify Endothelial Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, D.; Seki, T.; Sawada, H.; Kunitomi, A.; Katsuki, T.; Kimura, M.; Ito, S.; Komuro, J.; Hashimoto, H.; Fukuda, K.; et al. Anti-senescent drug screening by deep learning-based morphology senescence scoring. Nat. Commun. 2021, 12, 257. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flügel, L.; Dorn, T.; Goedel, A.; Höhnke, C.; Hofmann, F.; et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef] [Green Version]
- Takaki, T.; Inagaki, A.; Chonabayashi, K.; Inoue, K.; Miki, K.; Ohno, S.; Makiyama, T.; Horie, M.; Yoshida, Y. Optical Recording of Action Potentials in Human Induced Pluripotent Stem Cell-Derived Cardiac Single Cells and Monolayers Generated from Long QT Syndrome Type 1 Patients. Stem Cells Int. 2019, 2019, 7532657. [Google Scholar] [CrossRef]
- Kuroda, Y.; Yuasa, S.; Watanabe, Y.; Ito, S.; Egashira, T.; Seki, T.; Hattori, T.; Ohno, S.; Kodaira, M.; Suzuki, T.; et al. Flecainide ameliorates arrhythmogenicity through NCX flux in Andersen-Tawil syndrome-iPS cell-derived cardiomyocytes. Biochem. Biophys. Rep. 2017, 9, 245–256. [Google Scholar] [CrossRef]
- Schwartz, P.J.; Gnecchi, M.; Dagradi, F.; Castelletti, S.; Parati, G.; Spazzolini, C.; Sala, L.; Crotti, L. From patient-specific induced pluripotent stem cells to clinical translation in long QT syndrome Type 2. Eur. Heart. J. 2019, 40, 1832–1836. [Google Scholar] [CrossRef]
- Lan, F.; Lee, A.S.; Liang, P.; Sanchez-Freire, V.; Nguyen, P.K.; Wang, L.; Han, L.; Yen, M.; Wang, Y.; Sun, N.; et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013, 12, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toepfer, C.N.; Garfinkel, A.C.; Venturini, G.; Wakimoto, H.; Repetti, G.; Alamo, L.; Sharma, A.; Agarwal, R.; Ewoldt, J.F.; Cloonan, P.; et al. Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy. Circulation 2020, 141, 828–842. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; Van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.J.; Chiou, S.J.; Wong, Y.H.; Wang, Y.H.; Lai, Y.; Chou, C.H.; Wang, C.; Loh, J.K.; Lieu, A.S.; Cheng, J.T.; et al. GSKIP-Mediated Anchoring Increases Phosphorylation of Tau by PKA but Not by GSK3beta via cAMP/PKA/GSKIP/GSK3/Tau Axis Signaling in Cerebrospinal Fluid and iPS Cells in Alzheimer Disease. J. Clin. Med. 2019, 8, 1751. [Google Scholar] [CrossRef] [Green Version]
- Devine, M.J.; Ryten, M.; Vodicka, P.; Thomson, A.J.; Burdon, T.; Houlden, H.; Cavaleri, F.; Nagano, M.; Drummond, N.J.; Taanman, J.W.; et al. Parkinson’s disease induced pluripotent stem cells with triplication of the α-synuclein locus. Nat. Commun. 2011, 2, 440. [Google Scholar] [CrossRef]
- Barbuti, P.; Antony, P.; Santos, B.; Massart, F.; Cruciani, G.; Dording, C.; Arias, J.; Schwamborn, J.; Krüger, R. Using High-Content Screening to Generate Single-Cell Gene-Corrected Patient-Derived iPS Clones Reveals Excess Alpha-Synuclein with Familial Parkinson’s Disease Point Mutation A30P. Cells 2020, 9, 2065. [Google Scholar] [CrossRef]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lim, R.G.; Kaye, J.A.; Dardov, V.; Coyne, A.N.; Wu, J.; Milani, P.; Cheng, A.; Thompson, T.G.; Ornelas, L.; et al. An integrated multi-omic analysis of iPSC-derived motor neurons from C9ORF72 ALS patients. iScience 2021, 24, 103221. [Google Scholar] [CrossRef]
- Brennand, K.J.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D.; et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225. [Google Scholar] [CrossRef]
- Topol, A.; English, J.A.; Flaherty, E.; Rajarajan, P.; Hartley, B.J.; Gupta, S.; Desland, F.; Zhu, S.; Goff, T.; Friedman, L.; et al. Increased abundance of translation machinery in stem cell-derived neural progenitor cells from four schizophrenia patients. Transl. Psychiatry 2015, 5, e662. [Google Scholar] [CrossRef] [Green Version]
- Hamauchi, S.; Shichinohe, H.; Uchino, H.; Yamaguchi, S.; Nakayama, N.; Kazumata, K.; Osanai, T.; Abumiya, T.; Houkin, K.; Era, T. Cellular Functions and Gene and Protein Expression Profiles in Endothelial Cells Derived from Moyamoya Disease-Specific iPS Cells. PLoS ONE 2016, 11, e0163561. [Google Scholar] [CrossRef] [PubMed]
- Ameku, T.; Taura, D.; Sone, M.; Numata, T.; Nakamura, M.; Shiota, F.; Toyoda, T.; Matsui, S.; Araoka, T.; Yasuno, T.; et al. Identification of MMP1 as a novel risk factor for intracranial aneurysms in ADPKD using iPSC models. Sci. Rep. 2016, 6, 30013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soga, M.; Ishitsuka, Y.; Hamasaki, M.; Yoneda, K.; Furuya, H.; Matsuo, M.; Ihn, H.; Fusaki, N.; Nakamura, K.; Nakagata, N.; et al. HPGCD outperforms HPBCD as a potential treatment for Niemann-Pick disease type C during disease modeling with iPS cells. Stem Cells 2015, 33, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Korogi, Y.; Gotoh, S.; Ikeo, S.; Yamamoto, Y.; Sone, N.; Tamai, K.; Konishi, S.; Nagasaki, T.; Matsumoto, H.; Ito, I.; et al. In Vitro Disease Modeling of Hermansky-Pudlak Syndrome Type 2 Using Human Induced Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Rep. 2019, 12, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.Y.; Matsa, E.; Wu, J.C. Induced pluripotent stem cells: At the heart of cardiovascular precision medicine. Nat. Rev. Cardiol. 2016, 13, 333–349. [Google Scholar] [CrossRef] [Green Version]
- Vera, E.; Studer, L. When rejuvenation is a problem: Challenges of modeling late-onset neurodegenerative disease. Development 2015, 142, 3085–3089. [Google Scholar] [CrossRef] [Green Version]
- Goertzel, B. Human-level artificial general intelligence and the possibility of a technological singularity: A reaction to Ray Kurzweil’s The Singularity Is Near, and McDermott’s critique of Kurzweil. Artif. Intell. 2007, 171, 1161–1173. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Chen, M.C.; Ball, R.L.; Yang, L.; Moradzadeh, N.; Chapman, B.E.; Larson, D.B.; Langlotz, C.P.; Amrhein, T.J.; Lungren, M.P. Deep Learning to Classify Radiology Free-Text Reports. Radiology 2018, 286, 845–852. [Google Scholar] [CrossRef]
- Mor-Yosef, S.; Samueloff, A.; Modan, B.; Navot, D.; Schenker, J.G. Ranking the risk factors for cesarean: Logistic regression analysis of a nationwide study. Obstet. Gynecol. 1990, 75, 944–947. [Google Scholar]
- Gorodeski, E.Z.; Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Hsich, E.; Zhang, Z.M.; Vitolins, M.Z.; Manson, J.E.; Curb, J.D.; Martin, L.W.; et al. Use of hundreds of electrocardiographic biomarkers for prediction of mortality in postmenopausal women: The Women’s Health Initiative. Circ. Cardiovasc. Qual. Outcomes 2011, 4, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Heylman, C.; Datta, R.; Sobrino, A.; George, S.; Gratton, E. Supervised Machine Learning for Classification of the Electrophysiological Effects of Chronotropic Drugs on Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. PLoS ONE 2015, 10, e0144572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsich, E.; Gorodeski, E.Z.; Blackstone, E.H.; Ishwaran, H.; Lauer, M.S. Identifying important risk factors for survival in patient with systolic heart failure using random survival forests. Circ. Cardiovasc. Qual. Outcomes 2011, 4, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Cunningham, P.; Cord, M.; Delany, S.J. Supervised Learning. In Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval; Cord, M., Cunningham, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 21–49. [Google Scholar]
- Barlow, H.B. Unsupervised Learning. Neural Comput. 1989, 1, 295–311. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Rusu, A.A.; Veness, J.; Bellemare, M.G.; Graves, A.; Riedmiller, M.; Fidjeland, A.K.; Ostrovski, G.; et al. Human-level control through deep reinforcement learning. Nature 2015, 518, 529. Available online: https://www.nature.com/articles/nature14236#supplementary-information (accessed on 25 February 2015). [CrossRef]
- Silver, D.; Schrittwieser, J.; Simonyan, K.; Antonoglou, I.; Huang, A.; Guez, A.; Hubert, T.; Baker, L.; Lai, M.; Bolton, A.; et al. Mastering the game of Go without human knowledge. Nature 2017, 550, 354–359. [Google Scholar] [CrossRef]
- Mahmud, M.; Kaiser, M.S.; Hussain, A.; Vassanelli, S. Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 2063–2079. [Google Scholar] [CrossRef] [Green Version]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533. [Google Scholar] [CrossRef]
- Bengio, Y.; Lamblin, P.; Popovici, D.; Larochelle, H. Greedy layer-wise training of deep networks. In Proceedings of the 19th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 4–7 December 2006; MIT Press: Cambridge, MA, USA, 2006; pp. 153–160. [Google Scholar]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Ranzato, M.A.; Poultney, C.; Chopra, S.; LeCun, Y. Efficient learning of sparse representations with an energy-based model. In Proceedings of the 19th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 4–7 December 2006; MIT Press: Cambridge, MA, USA, 2006; pp. 1137–1144. [Google Scholar]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25; Curran Associates, Inc.: Red Hook, NY, USA, 2012. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. arXiv 2014, arXiv:1409.4842. [Google Scholar]
- Zeng, X.; Ouyang, W.; Yan, J.; Li, H.; Xiao, T.; Wang, K.; Liu, Y.; Zhou, Y.; Yang, B.; Wang, Z.; et al. Crafting GBD-Net for Object Detection. arXiv 2016, arXiv:1610.02579. [Google Scholar] [CrossRef] [Green Version]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Christiansen, E.M.; Yang, S.J.; Ando, D.M.; Javaherian, A.; Skibinski, G.; Lipnick, S.; Mount, E.; O’Neil, A.; Shah, K.; Lee, A.K.; et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell 2018, 173, 792–803.e719. [Google Scholar] [CrossRef] [Green Version]
- Edlund, C.; Jackson, T.R.; Khalid, N.; Bevan, N.; Dale, T.; Dengel, A.; Ahmed, S.; Trygg, J.; Sjögren, R. LIVECell-A large-scale dataset for label-free live cell segmentation. Nat. Methods 2021, 18, 1038–1045. [Google Scholar] [CrossRef]
- Guo, Y.; Shen, D.; Zhou, Y.; Yang, Y.; Liang, J.; Zhou, Y.; Li, N.; Liu, Y.; Yang, G.; Li, W. Deep Learning-Based Morphological Classification of Endoplasmic Reticulum Under Stress. Front. Cell Dev. Biol. 2021, 9, 767866. [Google Scholar] [CrossRef] [PubMed]
- Sarti, M.; Parlani, M.; Diaz-Gomez, L.; Mikos, A.G.; Cerveri, P.; Casarin, S.; Dondossola, E. Deep Learning for Automated Analysis of Cellular and Extracellular Components of the Foreign Body Response in Multiphoton Microscopy Images. Front. Bioeng. Biotechnol. 2021, 9, 797555. [Google Scholar] [CrossRef] [PubMed]
- Niioka, H.; Asatani, S.; Yoshimura, A.; Ohigashi, H.; Tagawa, S.; Miyake, J. Classification of C2C12 cells at differentiation by convolutional neural network of deep learning using phase contrast images. Hum. Cell 2018, 31, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buggenthin, F.; Buettner, F.; Hoppe, P.S.; Endele, M.; Kroiss, M.; Strasser, M.; Schwarzfischer, M.; Loeffler, D.; Kokkaliaris, K.D.; Hilsenbeck, O.; et al. Prospective identification of hematopoietic lineage choice by deep learning. Nat. Methods 2017, 14, 403–406. [Google Scholar] [CrossRef]
- Ota, S.; Horisaki, R.; Kawamura, Y.; Ugawa, M.; Sato, I.; Hashimoto, K.; Kamesawa, R.; Setoyama, K.; Yamaguchi, S.; Fujiu, K.; et al. Ghost cytometry. Science 2018, 360, 1246–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugawa, M.; Kawamura, Y.; Toda, K.; Teranishi, K.; Morita, H.; Adachi, H.; Tamoto, R.; Nomaru, H.; Nakagawa, K.; Sugimoto, K.; et al. In silico-labeled ghost cytometry. eLife 2021, 10, e67660. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Zhang, S.; Zhang, Y.; Lu, J.; Holcombe, M.; Zhang, X. A Machine Learning Assisted, Label-free, Non-invasive Approach for Somatic Reprogramming in Induced Pluripotent Stem Cell Colony Formation Detection and Prediction. Sci. Rep. 2017, 7, 13496. [Google Scholar] [CrossRef] [Green Version]
- Sommer, C.; Gerlich, D.W. Machine learning in cell biology—Teaching computers to recognize phenotypes. J. Cell Sci. 2013, 126, 5529–5539. [Google Scholar] [CrossRef] [Green Version]
- Juhola, M.; Joutsijoki, H.; Varpa, K.; Saarikoski, J.; Rasku, J.; Iltanen, K.; Laurikkala, J.; Hyyro, H.; Avalos-Salguero, J.; Siirtola, H.; et al. On computation of calcium cycling anomalies in cardiomyocytes data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2014, 2014, 1444–1447. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, aah7111. [Google Scholar] [CrossRef] [Green Version]
- Danter, W.R. DeepNEU: Cellular reprogramming comes of age—A machine learning platform with application to rare diseases research. Orphanet J. Rare Dis. 2019, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Joutsijoki, H.; Haponen, M.; Rasku, J.; Aalto-Setälä, K.; Juhola, M. Machine Learning Approach to Automated Quality Identification of Human Induced Pluripotent Stem Cell Colony Images. Comput. Math. Methods Med. 2016, 2016, 3091039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungvari, Z.; Tarantini, S.; Donato, A.J.; Galvan, V.; Csiszar, A. Mechanisms of Vascular Aging. Circ. Res. 2018, 123, 849–867. [Google Scholar] [CrossRef] [PubMed]
- Childs, B.G.; Durik, M.; Baker, D.J.; van Deursen, J.M. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 2015, 21, 1424–1435. [Google Scholar] [CrossRef] [Green Version]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Schiff, L.; Migliori, B.; Chen, Y.; Carter, D.; Bonilla, C.; Hall, J.; Fan, M.; Tam, E.; Ahadi, S.; Fischbacher, B.; et al. Integrating deep learning and unbiased automated high-content screening to identify complex disease signatures in human fibroblasts. Nat. Commun. 2022, 13, 1590. [Google Scholar] [CrossRef]
- Le Pelletier, L.; Mantecon, M.; Gorwood, J.; Auclair, M.; Foresti, R.; Motterlini, R.; Laforge, M.; Atlan, M.; Fève, B.; Capeau, J.; et al. Metformin alleviates stress-induced cellular senescence of aging human adipose stromal cells and the ensuing adipocyte dysfunction. eLife 2021, 10, e62635. [Google Scholar] [CrossRef]
- Khaidizar, F.D.; Bessho, Y.; Nakahata, Y. Nicotinamide Phosphoribosyltransferase as a Key Molecule of the Aging/Senescence Process. Int. J. Mol. Sci. 2021, 22, 3709. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e613. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, J.; Zhang, C.; Wang, S. SSGraphCPI: A Novel Model for Predicting Compound-Protein Interactions Based on Deep Learning. Int. J. Mol. Sci. 2022, 23, 3780. [Google Scholar] [CrossRef]
- Lee, E.K.; Kurokawa, Y.K.; Tu, R.; George, S.C.; Khine, M. Machine learning plus optical flow: A simple and sensitive method to detect cardioactive drugs. Sci. Rep. 2015, 5, 11817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, K.; Yada, Y.; Izumi, Y.; Morita, M.; Kawata, A.; Arisato, T.; Nagahashi, A.; Enami, T.; Tsukita, K.; Kawakami, H.; et al. Prediction Model of Amyotrophic Lateral Sclerosis by Deep Learning with Patient Induced Pluripotent Stem Cells. Ann. Neurol. 2021, 89, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, T.; Imamura, K.; Hioki, T.; Takagi, T.; Giga, Y.; Giga, M.H.; Nishimura, Y.; Kawahara, Y.; Hayashi, S.; Niki, T.; et al. Prediction of Compound Bioactivities Using Heat-Diffusion Equation. Patterns 2020, 1, 100140. [Google Scholar] [CrossRef] [PubMed]
- Teles, D.; Kim, Y.; Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Machine Learning Techniques to Classify Healthy and Diseased Cardiomyocytes by Contractility Profile. ACS Biomater. Sci. Eng. 2021, 7, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Juhola, M.; Joutsijoki, H.; Penttinen, K.; Shah, D.; Aalto-Setälä, K. On computational classification of genetic cardiac diseases applying iPSC cardiomyocytes. Comput. Methods Programs Biomed. 2021, 210, 106367. [Google Scholar] [CrossRef] [PubMed]
- Monzel, A.S.; Smits, L.M.; Hemmer, K.; Hachi, S.; Moreno, E.L.; van Wuellen, T.; Jarazo, J.; Walter, J.; Brüggemann, I.; Boussaad, I.; et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017, 8, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [Green Version]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.R.; Ueno, Y.; Zheng, Y.W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Huang, W.K.; Wong, S.Z.H.; Pather, S.R.; Nguyen, P.T.T.; Zhang, F.; Zhang, D.Y.; Zhang, Z.; Lu, L.; Fang, W.; Chen, L.; et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell 2021, 28, 1657–1670.e1610. [Google Scholar] [CrossRef]
- Tang, X.Y.; Xu, L.; Wang, J.; Hong, Y.; Wang, Y.; Zhu, Q.; Wang, D.; Zhang, X.Y.; Liu, C.Y.; Fang, K.H.; et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Investig. 2021, 131, e135763. [Google Scholar] [CrossRef]
- Arber, C.; Lovejoy, C.; Harris, L.; Willumsen, N.; Alatza, A.; Casey, J.M.; Lines, G.; Kerins, C.; Mueller, A.K.; Zetterberg, H.; et al. Familial Alzheimer’s Disease Mutations in PSEN1 Lead to Premature Human Stem Cell Neurogenesis. Cell Rep. 2021, 34, 108615. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, H.J.; Choi, H.; Chang, Y.; Park, H.; Shin, J.; Kim, J.; Lengner, C.J.; Lee, Y.K.; Kim, J. Modeling G2019S-LRRK2 Sporadic Parkinson’s Disease in 3D Midbrain Organoids. Stem Cell Rep. 2019, 12, 518–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, A.J.; Dye, B.R.; Ferrer-Torres, D.; Hill, D.R.; Overeem, A.W.; Shea, L.D.; Spence, J.R. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 2019, 14, 518–540. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.L.; Elhendawi, M.; Morlock, M.; Liu, W.; Liu, S.; Palakkan, A.; Seidl, L.F.; Hohenstein, P.; Sjögren, A.K.; Davies, J.A. Human iPSC-derived renal organoids engineered to report oxidative stress can predict drug-induced toxicity. iScience 2022, 25, 103884. [Google Scholar] [CrossRef]
- Uehara, K.; Koyanagi-Aoi, M.; Koide, T.; Itoh, T.; Aoi, T. Epithelial-derived factors induce muscularis mucosa of human induced pluripotent stem cell-derived gastric organoids. Stem Cell Rep. 2022, 17, 820–834. [Google Scholar] [CrossRef]
- Crespo, M.; Vilar, E.; Tsai, S.Y.; Chang, K.; Amin, S.; Srinivasan, T.; Zhang, T.; Pipalia, N.H.; Chen, H.J.; Witherspoon, M.; et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2017, 23, 878–884. [Google Scholar] [CrossRef]
- Park, J.C.; Jang, S.Y.; Lee, D.; Lee, J.; Kang, U.; Chang, H.; Kim, H.J.; Han, S.H.; Seo, J.; Choi, M.; et al. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commun. 2021, 12, 280. [Google Scholar] [CrossRef]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef]
- Musah, S.; Dimitrakakis, N.; Camacho, D.M.; Church, G.M.; Ingber, D.E. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nat. Protoc. 2018, 13, 1662–1685. [Google Scholar] [CrossRef]
- Tristan, C.A.; Ormanoglu, P.; Slamecka, J.; Malley, C.; Chu, P.H.; Jovanovic, V.M.; Gedik, Y.; Jethmalani, Y.; Bonney, C.; Barnaeva, E.; et al. Robotic high-throughput biomanufacturing and functional differentiation of human pluripotent stem cells. Stem Cell Rep. 2021, 16, 3076–3092. [Google Scholar] [CrossRef] [PubMed]
| Authors | Reference | Cell Line | Disease or Phenotype | Classifier | Input |
|---|---|---|---|---|---|
| Kusumoto D, et al. | [16] | Human Umbilical Vein Endothelial Cells | Cellular senescence | CNN | Phase-contrast Images |
| Schiff L, et al. | [82] | Fibroblasts | Parkinson‘s disease | CNN | Cell painting dyes |
| Lee EK, et al. | [87] | iPSC-derived cardiomyocytes | Cardiotoxisity | SVM | Brightfield (myocardial contraction) |
| Imamura, et al. | [88] | iPSC-derived neurons | Amyotrophic lateral sclerosis (ALS) | CNN | Immunostaining for b3-tubulin |
| Juhola M, et al. | [89] | iPSC-derived cardiomyocytes | Six genetic cardiac disaease | k-NN, Random forest, SVM, etc. | Calcium transient |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusumoto, D.; Yuasa, S.; Fukuda, K. Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence. Pharmaceuticals 2022, 15, 562. https://doi.org/10.3390/ph15050562
Kusumoto D, Yuasa S, Fukuda K. Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence. Pharmaceuticals. 2022; 15(5):562. https://doi.org/10.3390/ph15050562
Chicago/Turabian StyleKusumoto, Dai, Shinsuke Yuasa, and Keiichi Fukuda. 2022. "Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence" Pharmaceuticals 15, no. 5: 562. https://doi.org/10.3390/ph15050562
APA StyleKusumoto, D., Yuasa, S., & Fukuda, K. (2022). Induced Pluripotent Stem Cell-Based Drug Screening by Use of Artificial Intelligence. Pharmaceuticals, 15(5), 562. https://doi.org/10.3390/ph15050562





