Comparative Analysis of Urso- and Tauroursodeoxycholic Acid Neuroprotective Effects on Retinal Degeneration Models
Abstract
:1. Introduction
2. Results
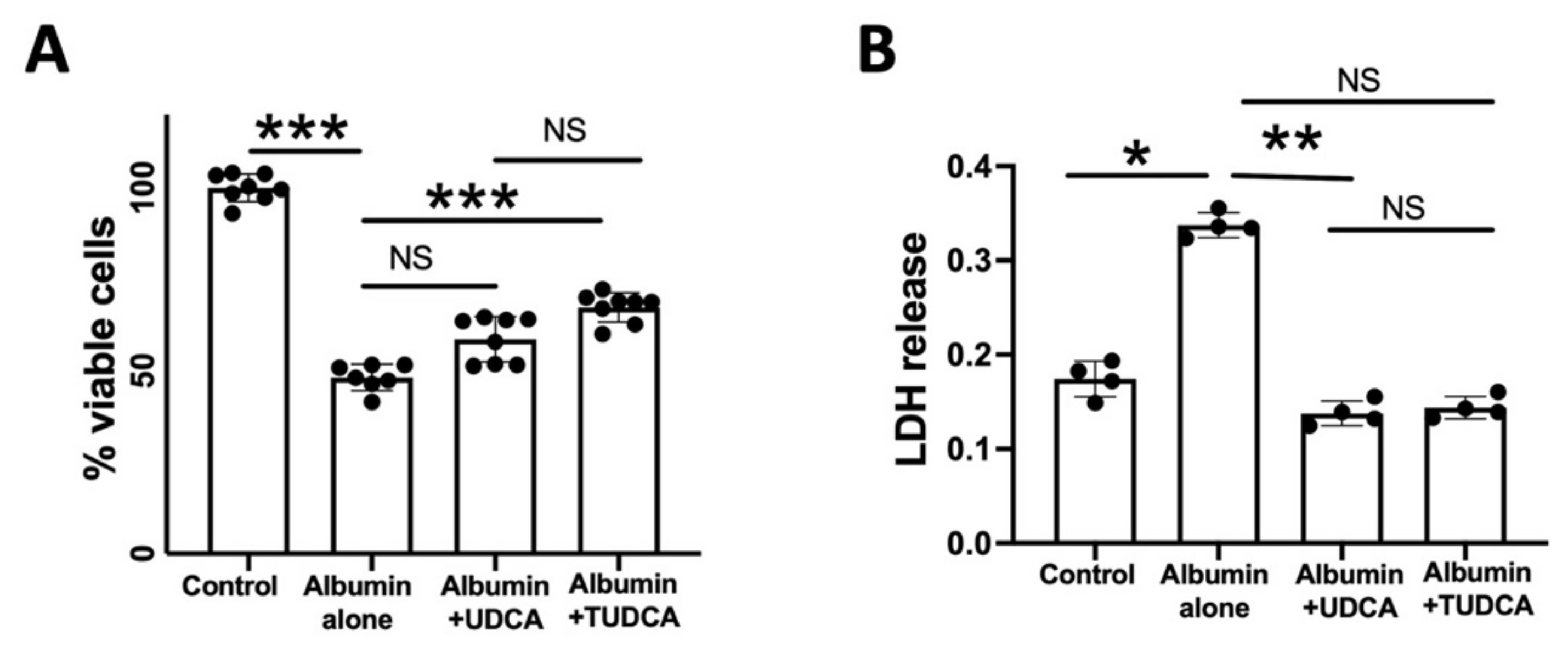
2.1. UDCA and TUDCA Protect against In Vitro Albumin-Induced Cone Photoreceptors Death
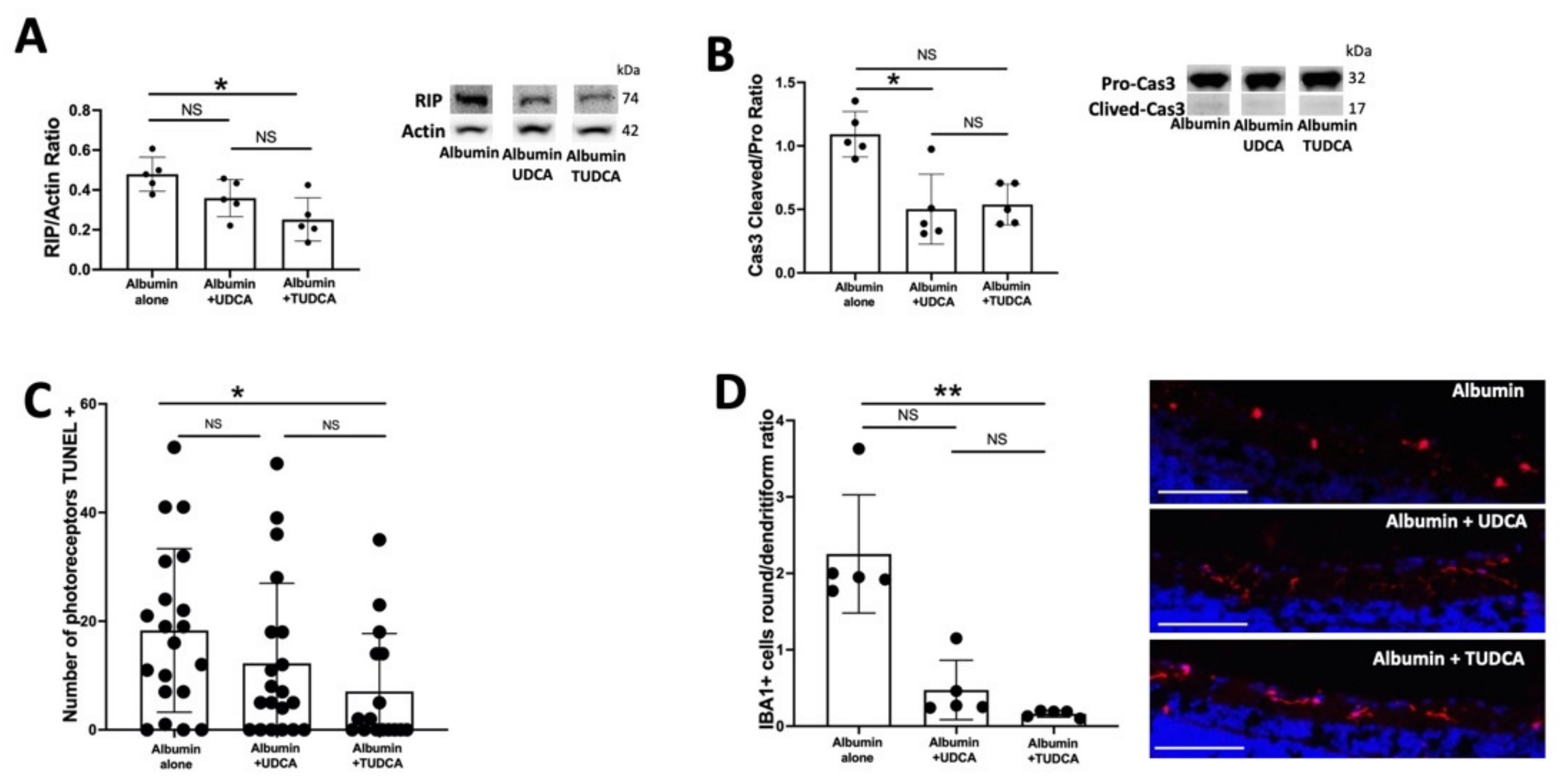
2.2. UDCA and TUDCA Protect against Albumin-Induced Cell Death on Ex Vivo Retina Organoculture
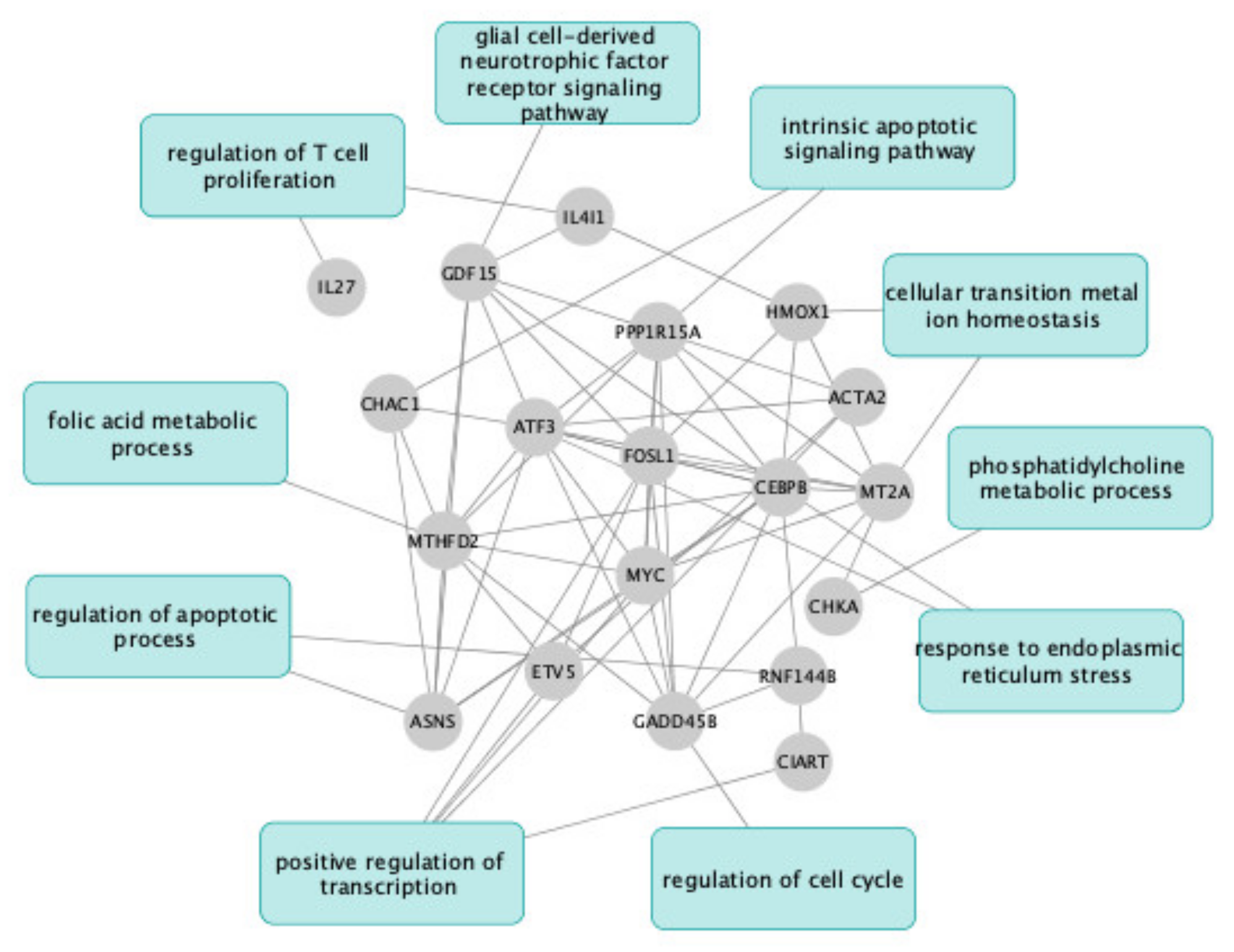
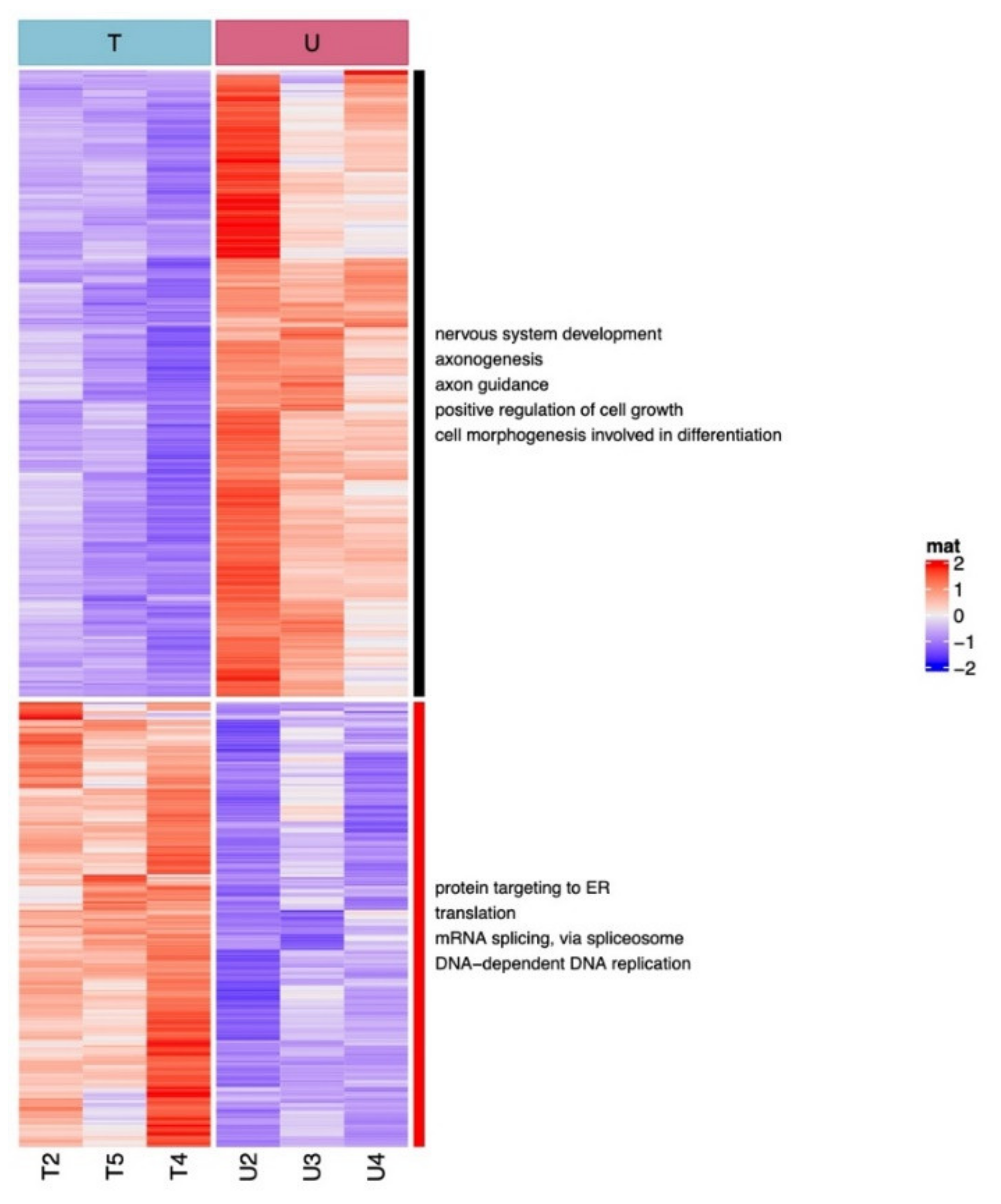
2.3. UDCA and TUDCA Induced Differential Transcriptional Regulation Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Line and In Vitro Model
4.2. Animals and Ex Vivo Model of RD
4.3. Western Immunoblotting Analysis
4.4. Immunohistochemistry and Fluorescence Intensity Evaluation
4.5. Ribonucleic Acid (RNA) Sequencing and RNA-Seq Data Analysis
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, F.; Pariante, C.M.; Borsini, A. From Dried Bear Bile to Molecular Investigation: A Systematic Review of the Effect of Bile Acids on Cell Apoptosis, Oxidative Stress and Inflammation in the Brain, across Pre-Clinical Models of Neurological, Neurodegenerative and Neuropsychiatric Disorders. Brain. Behav. Immun. 2022, 99, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Daruich, A.; Picard, E.; Boatright, J.H.; Behar-Cohen, F. The Bile Acids Urso- and Tauroursodeoxycholic Acid as Neuroprotective Therapies in Retinal Disease. Mol. Vis. 2019, 25, 610. [Google Scholar] [PubMed]
- Daruich, A.; Jaworski, T.; Henry, H.; Zola, M.; Youale, J.; Parenti, L.; Naud, M.-C.; Delaunay, K.; Bertrand, M.; Berdugo, M.; et al. Oral Ursodeoxycholic Acid Crosses the Blood Retinal Barrier in Patients with Retinal Detachment and Protects Against Retinal Degeneration in an Ex Vivo Model. Neurotherapeutics 2021, 18, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Walker, T.A.; Choi, H.-Y.; Faulkner, A.E.; Kim, M.K.; Sidney, S.S.; Boyd, A.P.; Nickerson, J.M.; Boatright, J.H.; Pardue, M.T. Tauroursodeoxycholic Acid Preservation of Photoreceptor Structure and Function in the Rd10 Mouse through Postnatal Day 30. Invest. Ophthalmol. Vis. Sci. 2008, 49, 2148–2155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Vicente, V.; Lax, P.; Fernández-Sánchez, L.; Rondón, N.; Esquiva, G.; Germain, F.; de la Villa, P.; Cuenca, N. Neuroprotective Effect of Tauroursodeoxycholic Acid on N-Methyl-D-Aspartate-Induced Retinal Ganglion Cell Degeneration. PLoS ONE 2015, 10, e0137826. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Aung, M.H.; Prunty, M.C.; Hanif, A.M.; Hutson, L.M.; Boatright, J.H.; Pardue, M.T. Tauroursodeoxycholic Acid Protects Retinal and Visual Function in a Mouse Model of Type 1 Diabetes. Pharmaceutics 2021, 13, 1154. [Google Scholar] [CrossRef]
- Boatright, J.H.; Moring, A.G.; McElroy, C.; Phillips, M.J.; Do, V.T.; Chang, B.; Hawes, N.L.; Boyd, A.P.; Sidney, S.S.; Stewart, R.E.; et al. Tool from Ancient Pharmacopoeia Prevents Vision Loss. Mol. Vis. 2006, 12, 1706–1714. [Google Scholar]
- Mantopoulos, D.; Murakami, Y.; Comander, J.; Thanos, A.; Roh, M.; Miller, J.W.; Vavvas, D.G. Tauroursodeoxycholic Acid (TUDCA) Protects Photoreceptors from Cell Death after Experimental Retinal Detachment. PLoS ONE 2011, 6, e24245. [Google Scholar] [CrossRef]
- Gaspar, J.M.; Martins, A.; Cruz, R.; Rodrigues, C.M.P.; Ambrósio, A.F.; Santiago, A.R. Tauroursodeoxycholic Acid Protects Retinal Neural Cells from Cell Death Induced by Prolonged Exposure to Elevated Glucose. Neuroscience 2013, 253, 380–388. [Google Scholar] [CrossRef]
- Zhang, T.; Baehr, W.; Fu, Y. Chemical Chaperone TUDCA Preserves Cone Photoreceptors in a Mouse Model of Leber Congenital Amaurosis. Invest. Ophthalmol. Vis. Sci. 2012, 53, 3349–3356. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Zhang, T. Pathophysilogical Mechanism and Treatment Strategies for Leber Congenital Amaurosis. Adv. Exp. Med. Biol. 2014, 801, 791–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalho, R.M.; Nunes, A.F.; Dias, R.B.; Amaral, J.D.; Lo, A.C.; D’Hooge, R.; Sebastião, A.M.; Rodrigues, C.M.P. Tauroursodeoxycholic Acid Suppresses Amyloid β-Induced Synaptic Toxicity in Vitro and in APP/PS1 Mice. Neurobiol. Aging 2013, 34, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Luan, J.; Huang, T.; Deng, T.; Li, X.; Xiao, Z.; Zhan, J.; Luo, D.; Hou, Y.; Xu, L.; et al. Tauroursodeoxycholic Acid Alleviates Secondary Injury in Spinal Cord Injury Mice by Reducing Oxidative Stress, Apoptosis, and Inflammatory Response. J. Neuroinflammation 2021, 18, 216. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Mei, X.; Zhang, T.; Lu, B.; Ji, L. Ursodeoxycholic Acid Ameliorates Diabetic Retinopathy via Reducing Retinal Inflammation and Reversing the Breakdown of Blood-Retinal Barrier. Eur. J. Pharmacol. 2018, 840, 20–27. [Google Scholar] [CrossRef]
- Chung, Y.-R.; Choi, J.A.; Koh, J.-Y.; Yoon, Y.H. Ursodeoxycholic Acid Attenuates Endoplasmic Reticulum Stress-Related Retinal Pericyte Loss in Streptozotocin-Induced Diabetic Mice. J. Diabetes Res. 2017, 2017, 1763292. [Google Scholar] [CrossRef]
- Woo, S.J.; Kim, J.H.; Yu, H.G. Ursodeoxycholic Acid and Tauroursodeoxycholic Acid Suppress Choroidal Neovascularization in a Laser-Treated Rat Model. J. Ocul. Pharmacol. Ther. Off. J. Assoc. Ocul. Pharmacol. Ther. 2010, 26, 223–229. [Google Scholar] [CrossRef]
- Duncan, T.; Fariss, R.N.; Wiggert, B. Confocal Immunolocalization of Bovine Serum Albumin, Serum Retinol-Binding Protein, and Interphotoreceptor Retinoid-Binding Protein in Bovine Retina. Mol. Vis. 2006, 12, 1632–1639. [Google Scholar]
- Ling, X.-B.; Wei, H.-W.; Wang, J.; Kong, Y.-Q.; Wu, Y.-Y.; Guo, J.-L.; Li, T.-F.; Li, J.-K. Mammalian Metallothionein-2A and Oxidative Stress. Int. J. Mol. Sci. 2016, 17, 1483. [Google Scholar] [CrossRef]
- Lu, H.; Hunt, D.M.; Ganti, R.; Davis, A.; Dutt, K.; Alam, J.; Hunt, R.C. Metallothionein Protects Retinal Pigment Epithelial Cells against Apoptosis and Oxidative Stress. Exp. Eye Res. 2002, 74, 83–92. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Wang, S.; Liu, S.; Zhao, Y. Neuronal Uptake of Serum Albumin Is Associated with Neuron Damage during the Development of Epilepsy. Exp. Ther. Med. 2016, 12, 695–701. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Barrios, A.; Álvarez, L.; García, M.; Artime, E.; Pereiro, R.; González-Iglesias, H. Antioxidant Defenses in the Human Eye: A Focus on Metallothioneins. Antioxidants 2021, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Facchinetti, M.M. Heme-Oxygenase-1. Antioxid. Redox Signal. 2020, 32, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Chen, X.; Li, J.; Comish, P.; Kang, R.; Tang, D. Transcription Factors in Ferroptotic Cell Death. Cancer Gene Ther. 2020, 27, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Kole, C.; Brommer, B.; Nakaya, N.; Sengupta, M.; Bonet-Ponce, L.; Zhao, T.; Wang, C.; Li, W.; He, Z.; Tomarev, S. Activating Transcription Factor 3 (ATF3) Protects Retinal Ganglion Cells and Promotes Functional Preservation After Optic Nerve Crush. Invest. Ophthalmol. Vis. Sci. 2020, 61, 31. [Google Scholar] [CrossRef] [Green Version]
- Samardzija, M.; Wariwoda, H.; Imsand, C.; Huber, P.; Heynen, S.R.; Gubler, A.; Grimm, C. Activation of Survival Pathways in the Degenerating Retina of Rd10 Mice. Exp. Eye Res. 2012, 99, 17–26. [Google Scholar] [CrossRef]
- Zeitler, L.; Fiore, A.; Meyer, C.; Russier, M.; Zanella, G.; Suppmann, S.; Gargaro, M.; Sidhu, S.S.; Seshagiri, S.; Ohnmacht, C.; et al. Anti-Ferroptotic Mechanism of IL4i1-Mediated Amino Acid Metabolism. eLife 2021, 10, e64806. [Google Scholar] [CrossRef]
- Yue, Y.; Huang, W.; Liang, J.; Guo, J.; Ji, J.; Yao, Y.; Zheng, M.; Cai, Z.; Lu, L.; Wang, J. IL4I1 Is a Novel Regulator of M2 Macrophage Polarization That Can Inhibit T Cell Activation via L-Tryptophan and Arginine Depletion and IL-10 Production. PLoS ONE 2015, 10, e0142979. [Google Scholar] [CrossRef]
- Peng, J.-J.; Song, W.-T.; Yao, F.; Zhang, X.; Peng, J.; Luo, X.-J.; Xia, X.-B. Involvement of Regulated Necrosis in Blinding Diseases: Focus on Necroptosis and Ferroptosis. Exp. Eye Res. 2020, 191, 107922. [Google Scholar] [CrossRef]
- Torp, R.; Su, J.H.; Deng, G.; Cotman, C.W. GADD45 Is Induced in Alzheimer’s Disease, and Protects against Apoptosis in Vitro. Neurobiol. Dis. 1998, 5, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Sinha, T.; Ikelle, L.; Naash, M.I.; Al-Ubaidi, M.R. The Intersection of Serine Metabolism and Cellular Dysfunction in Retinal Degeneration. Cells 2020, 9, 674. [Google Scholar] [CrossRef] [Green Version]
- Wahlig, S.; Lovatt, M.; Mehta, J.S. Functional Role of Peroxiredoxin 6 in the Eye. Free Radic. Biol. Med. 2018, 126, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Saïd, W.; Fradot, V.; Ivkovic, I.; Sahel, J.-A.; Picaud, S.; Froger, N. Taurine Promotes Retinal Ganglion Cell Survival Through GABAB Receptor Activation. Adv. Exp. Med. Biol. 2017, 975 Pt 2, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Castelli, V.; Paladini, A.; d’Angelo, M.; Allegretti, M.; Mantelli, F.; Brandolini, L.; Cocchiaro, P.; Cimini, A.; Varrassi, G. Taurine and Oxidative Stress in Retinal Health and Disease. CNS Neurosci. Ther. 2021, 27, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Lobysheva, E.; Taylor, C.M.; Marshall, G.R.; Kisselev, O.G. Tauroursodeoxycholic Acid Binds to the G-Protein Site on Light Activated Rhodopsin. Exp. Eye Res. 2018, 170, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ripps, H.; Shen, W. Review: Taurine: A “Very Essential” Amino Acid. Mol. Vis. 2012, 18, 2673–2686. [Google Scholar]
- Claudel, T.; Staels, B.; Kuipers, F. The Farnesoid X Receptor: A Molecular Link between Bile Acid and Lipid and Glucose Metabolism. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 2020–2030. [Google Scholar] [CrossRef]
- Murase, H.; Tsuruma, K.; Shimazawa, M.; Hara, H. TUDCA Promotes Phagocytosis by Retinal Pigment Epithelium via MerTK Activation. Invest. Ophthalmol. Vis. Sci. 2015, 56, 2511–2518. [Google Scholar] [CrossRef] [Green Version]
- Couturier, A.; Bousquet, E.; Zhao, M.; Naud, M.-C.; Klein, C.; Jonet, L.; Tadayoni, R.; de Kozak, Y.; Behar-Cohen, F. Anti-Vascular Endothelial Growth Factor Acts on Retinal Microglia/Macrophage Activation in a Rat Model of Ocular Inflammation. Mol. Vis. 2014, 20, 908–920. [Google Scholar]
- Chaiwiang, N.; Poyomtip, T. Microbial Dysbiosis and Microbiota-Gut-Retina Axis: The Lesson from Brain Neurodegenerative Diseases to Primary Open-Angle Glaucoma Pathogenesis of Autoimmunity. Acta Microbiol. Immunol. Hung. 2019, 66, 541–558. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daruich, A.; Picard, E.; Guégan, J.; Jaworski, T.; Parenti, L.; Delaunay, K.; Naud, M.-C.; Berdugo, M.; Boatright, J.H.; Behar-Cohen, F. Comparative Analysis of Urso- and Tauroursodeoxycholic Acid Neuroprotective Effects on Retinal Degeneration Models. Pharmaceuticals 2022, 15, 334. https://doi.org/10.3390/ph15030334
Daruich A, Picard E, Guégan J, Jaworski T, Parenti L, Delaunay K, Naud M-C, Berdugo M, Boatright JH, Behar-Cohen F. Comparative Analysis of Urso- and Tauroursodeoxycholic Acid Neuroprotective Effects on Retinal Degeneration Models. Pharmaceuticals. 2022; 15(3):334. https://doi.org/10.3390/ph15030334
Chicago/Turabian StyleDaruich, Alejandra, Emilie Picard, Justine Guégan, Thara Jaworski, Léa Parenti, Kimberley Delaunay, Marie-Christine Naud, Marianne Berdugo, Jeffrey H. Boatright, and Francine Behar-Cohen. 2022. "Comparative Analysis of Urso- and Tauroursodeoxycholic Acid Neuroprotective Effects on Retinal Degeneration Models" Pharmaceuticals 15, no. 3: 334. https://doi.org/10.3390/ph15030334
APA StyleDaruich, A., Picard, E., Guégan, J., Jaworski, T., Parenti, L., Delaunay, K., Naud, M.-C., Berdugo, M., Boatright, J. H., & Behar-Cohen, F. (2022). Comparative Analysis of Urso- and Tauroursodeoxycholic Acid Neuroprotective Effects on Retinal Degeneration Models. Pharmaceuticals, 15(3), 334. https://doi.org/10.3390/ph15030334






