S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2
Abstract
1. Introduction
2. Results
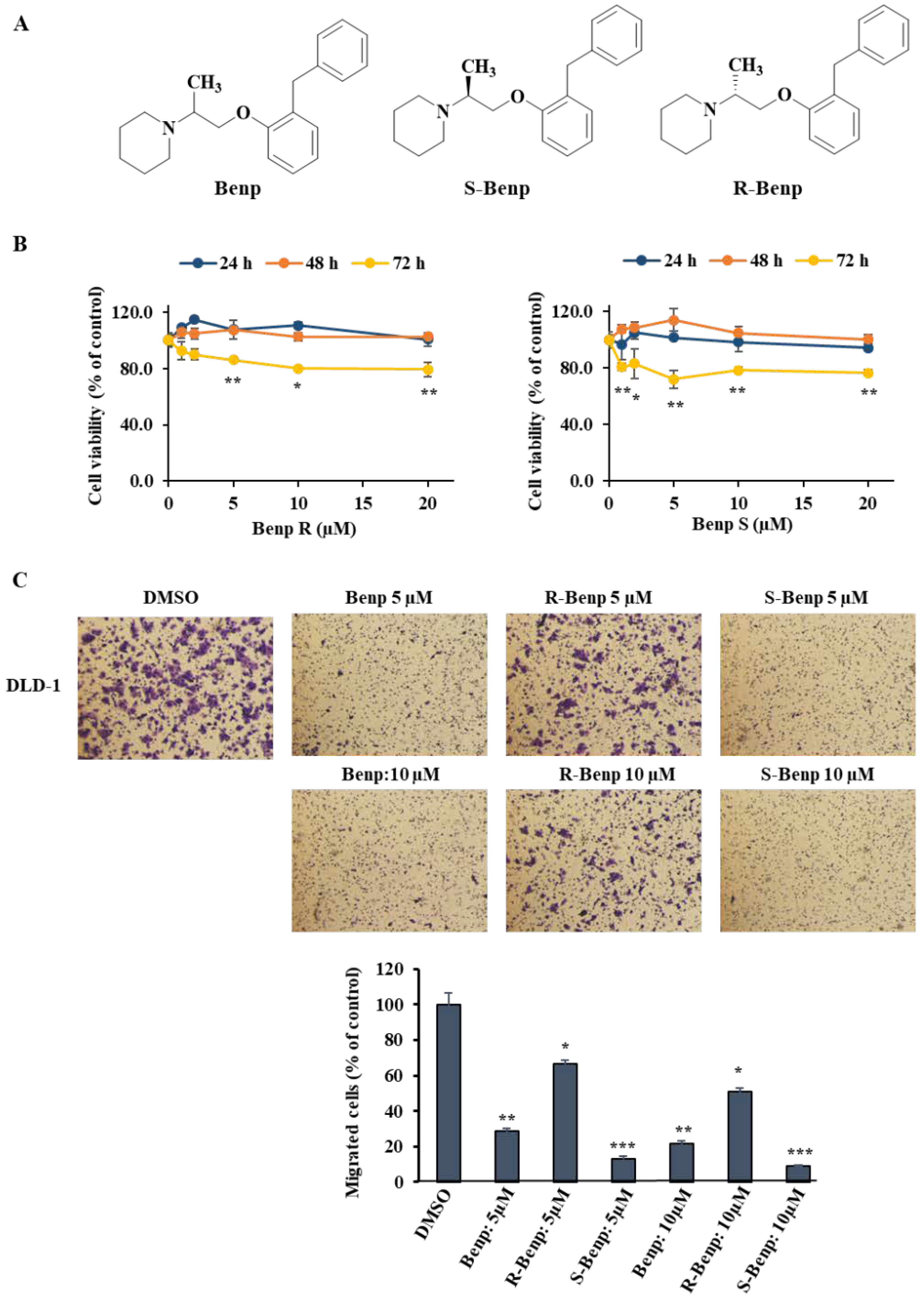
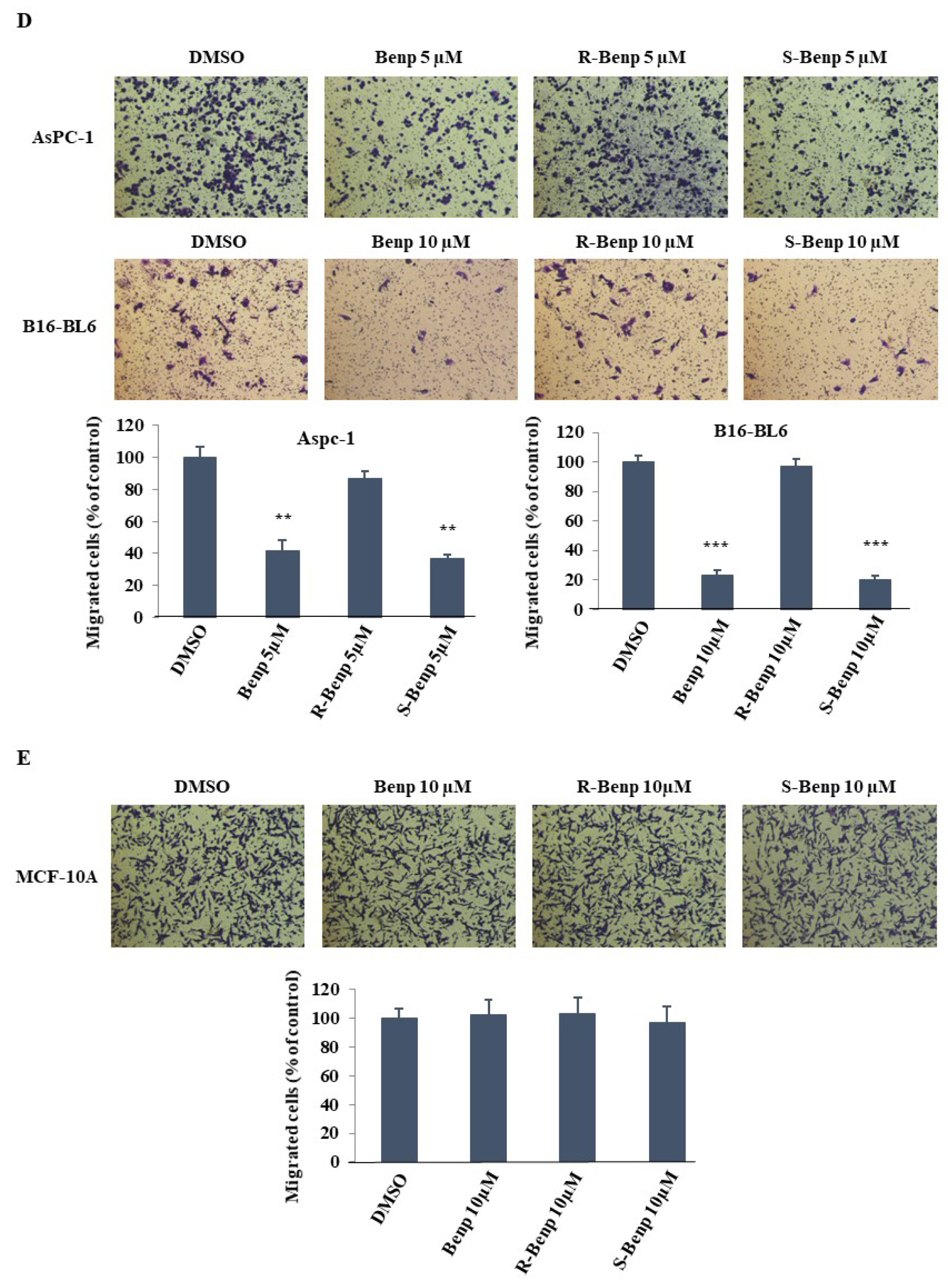
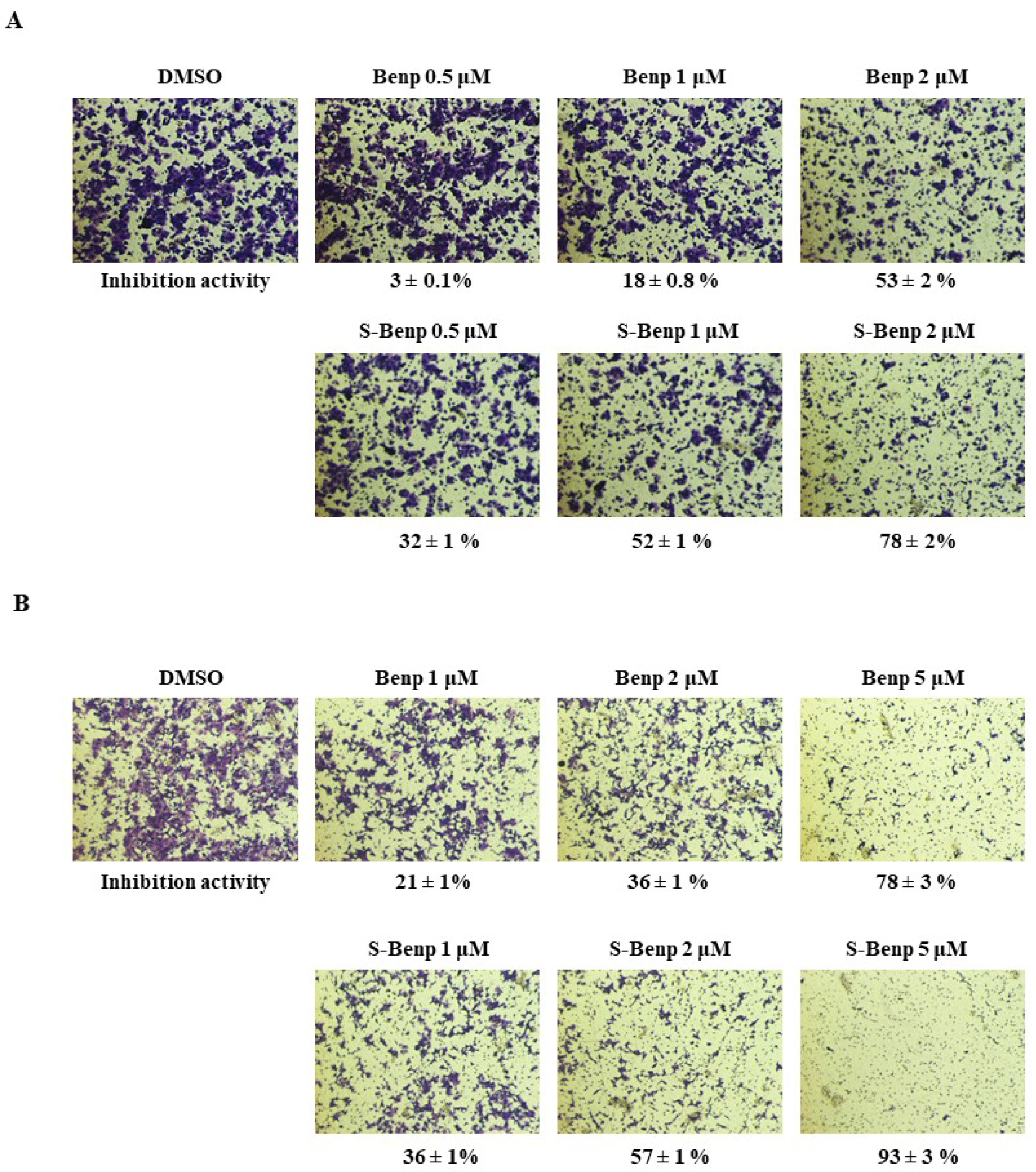
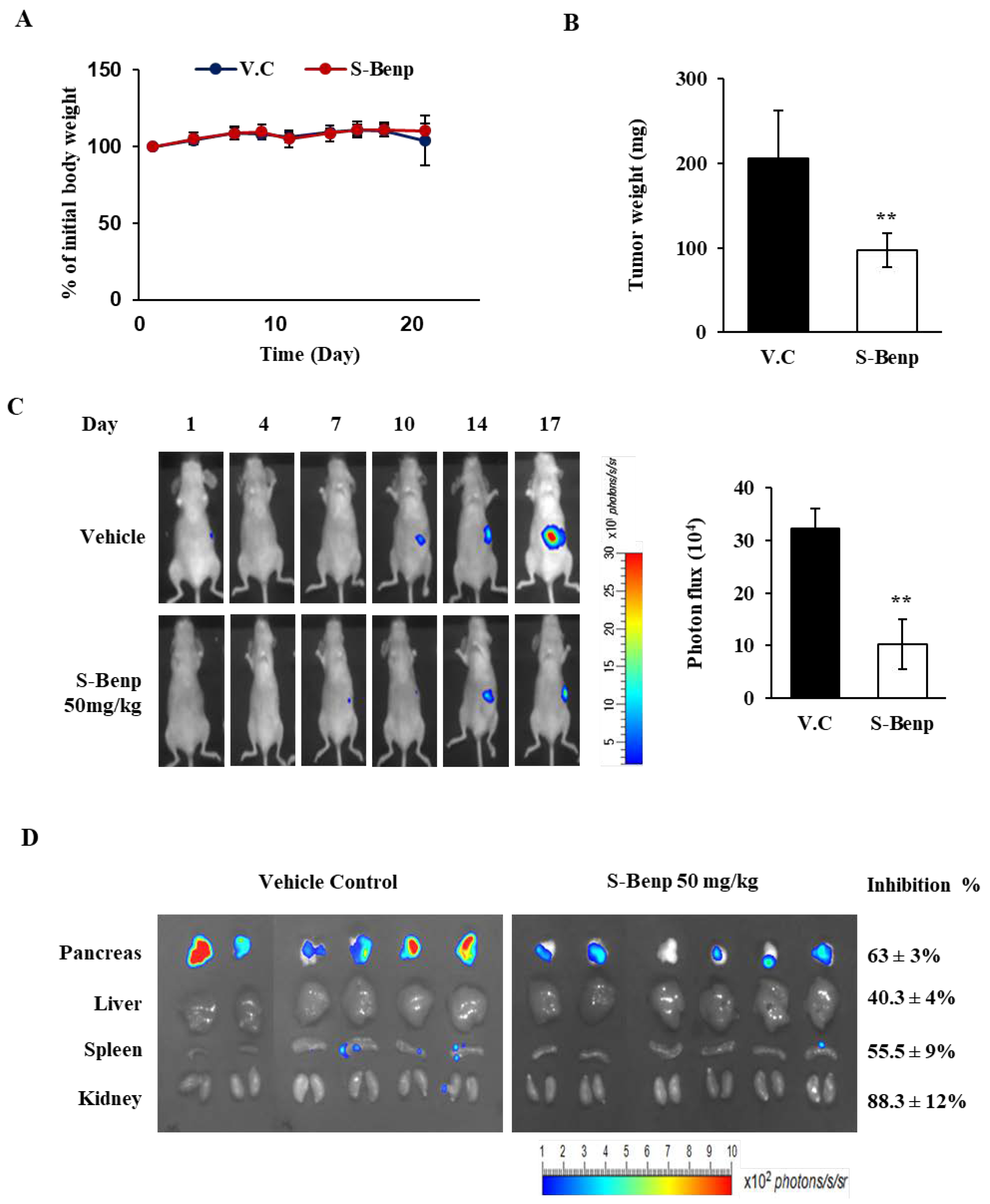
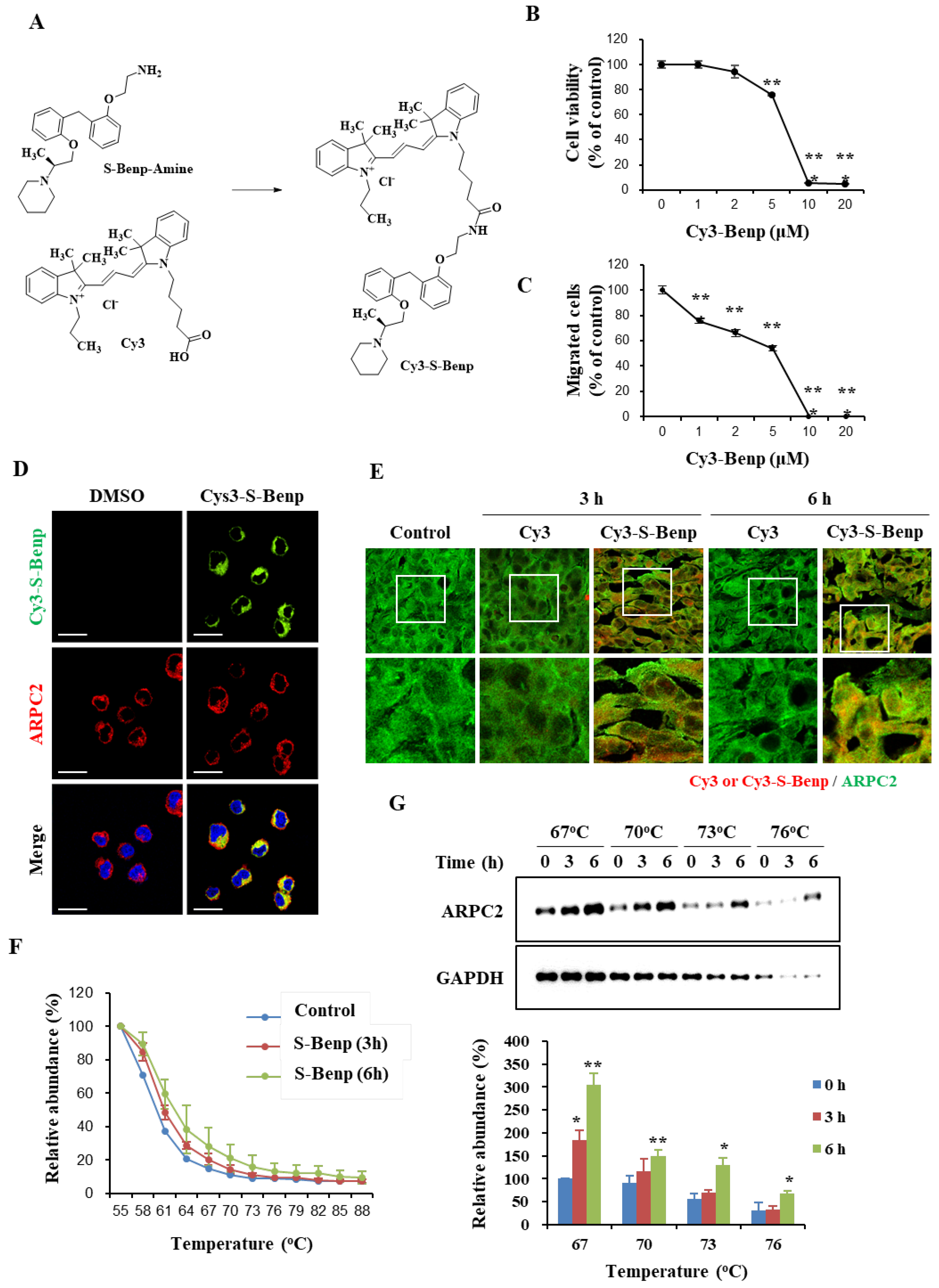
2.1. Antimetastatic Effect of S-Benproperine
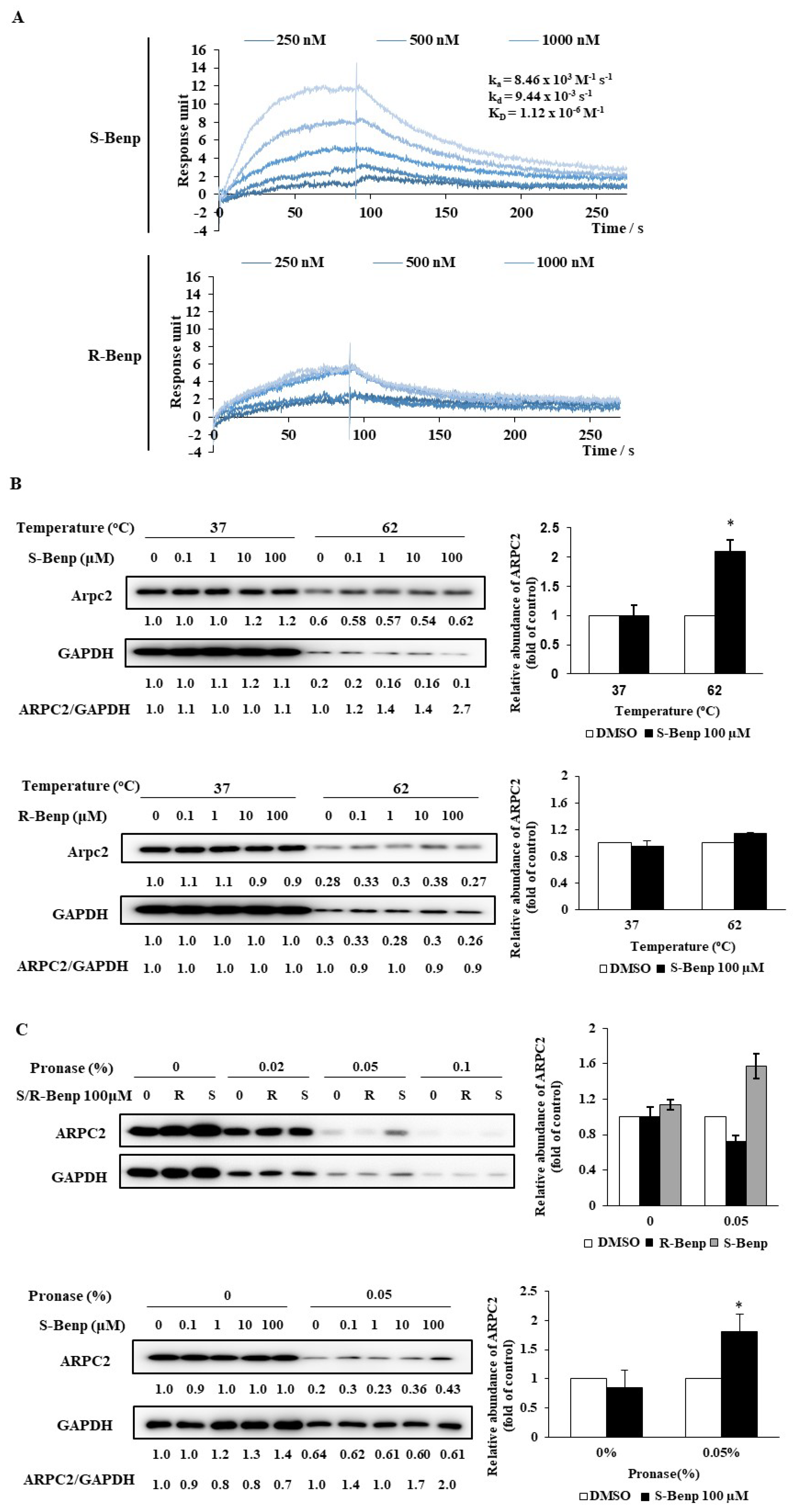
2.2. S-Benproperine Directly Binds to ARPC2
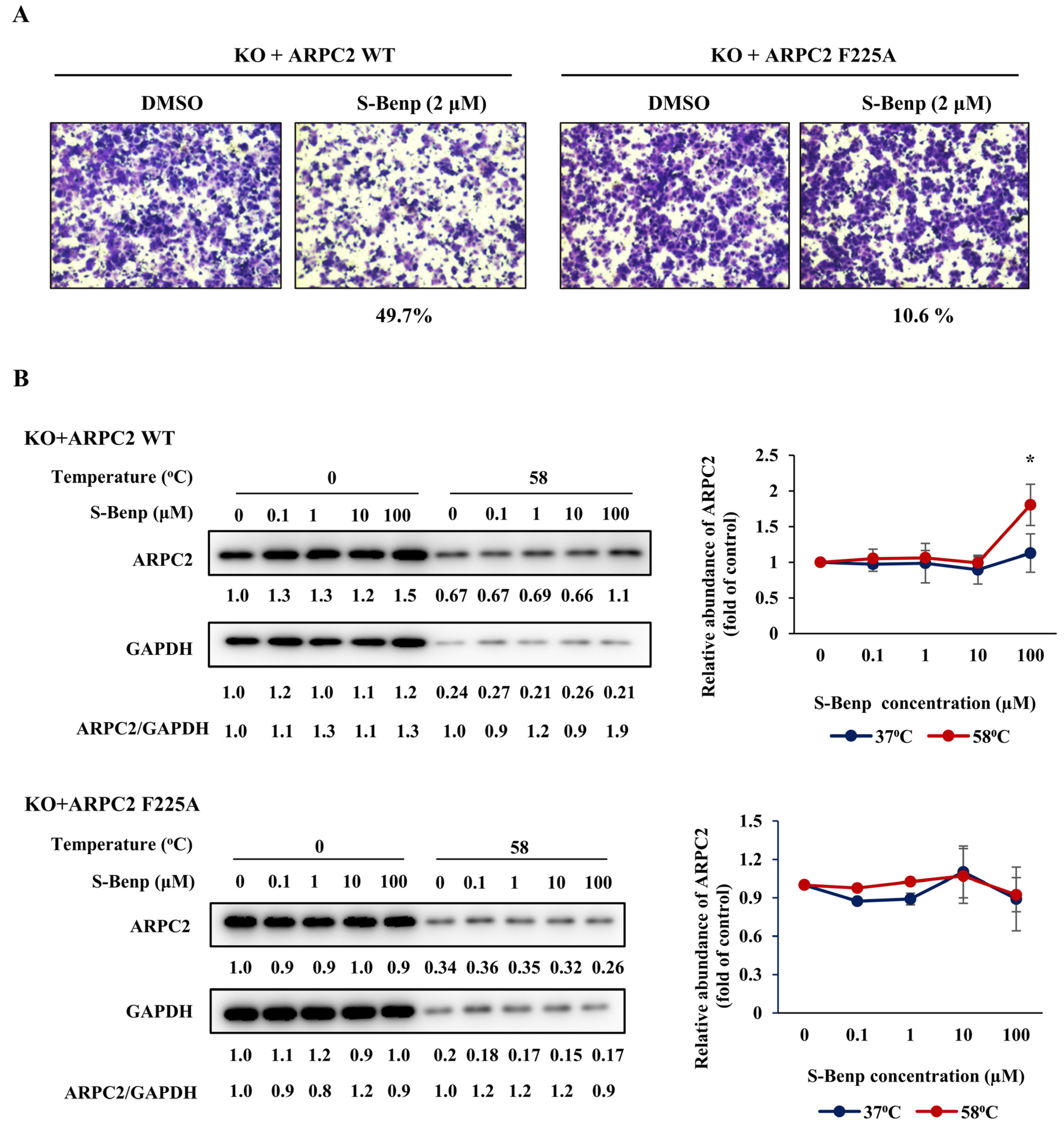
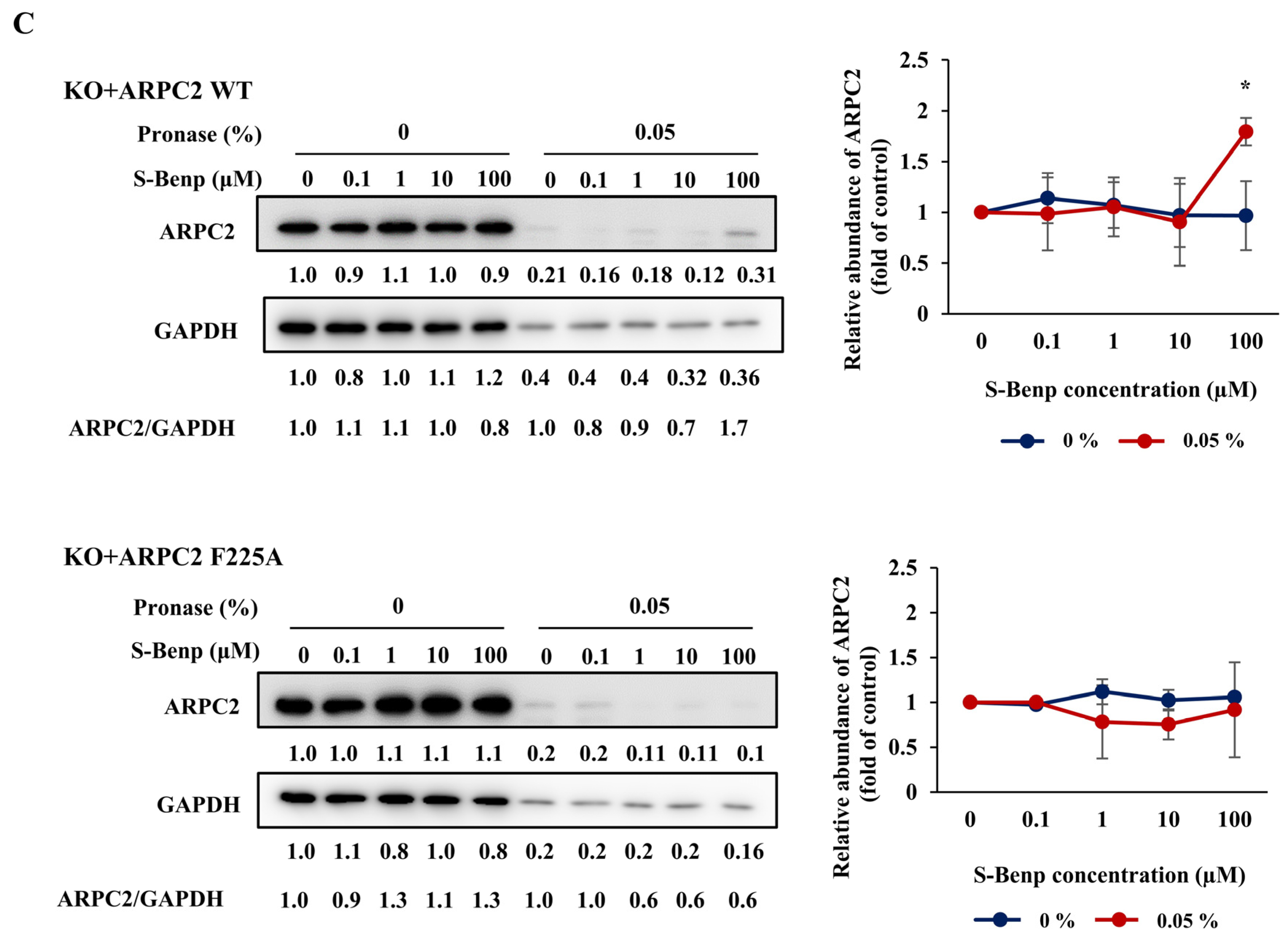
2.3. Validation of Binding Sites of S-Benproperine in ARPC2 Using Mutated Cancer Cells
2.4. S-Benproperine Localization and Binding to ARPC2 In Vitro and In Vivo
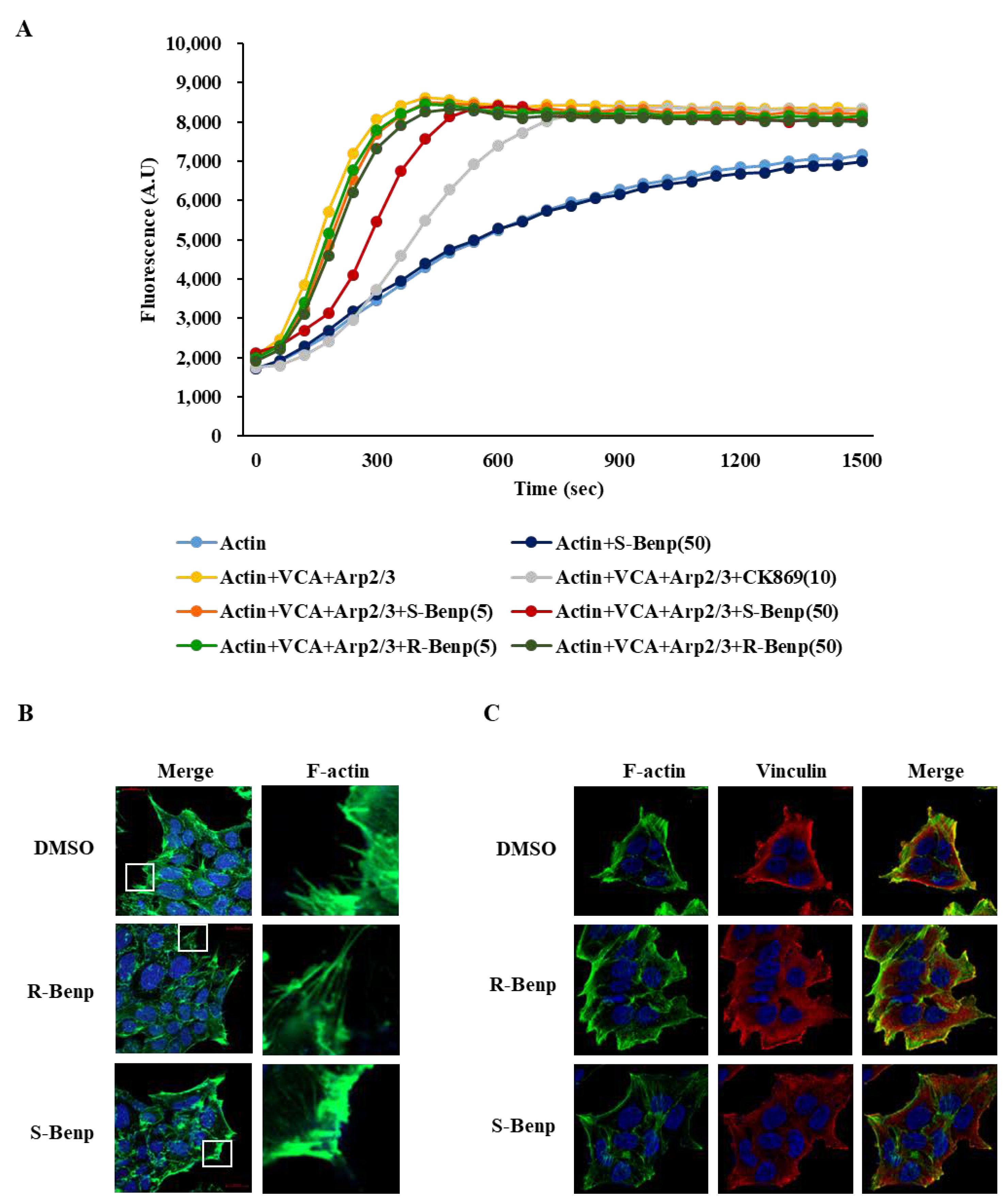
2.5. S-Benproperine Delayed Actin Polymerization
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Proliferation Assay
4.2. Transwell Migration and Invasion Assay
4.3. In Vivo Xenograft and In Vivo Metastasis Assay
4.4. Purification of the Recombinant Protein
4.5. Surface Plasmon Resonance Analysis
4.6. Immunoblotting
4.7. Drug Affinity Responsive Target Stability (DARTS)
4.8. Cellular Thermal Shift Assay (CETSA)
4.9. In Vitro Actin Polymerization Assay
4.10. Immunocytochemistry and Immunohistochemistry
4.11. Synthesis of S-Benp, R-Benp, and S-Benp-Cy3
4.12. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Dillekas, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.N.; Mei, Y.; Zhang, J. Cancer metastasis: Issues and challenges. Chin. J. Cancer 2017, 36, 38. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Hodgson, L.; Condeelis, J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012, 24, 277–283. [Google Scholar] [CrossRef]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef]
- Wang, H.R.; Zhang, Y.; Ozdamar, B.; Ogunjimi, A.A.; Alexandrova, E.; Thomsen, G.H.; Wrana, J.L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 2003, 302, 1775–1779. [Google Scholar] [CrossRef]
- Biber, G.; Ben-Shmuel, A.; Sabag, B.; Barda-Saad, M. Actin regulators in cancer progression and metastases: From structure and function to cytoskeletal dynamics. Int. Rev. Cell Mol. Biol. 2020, 356, 131–196. [Google Scholar] [CrossRef]
- Machesky, L.M.; Mullins, R.D.; Higgs, H.N.; Kaiser, D.A.; Blanchoin, L.; May, R.C.; Hall, M.E.; Pollard, T.D. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA 1999, 96, 3739–3744. [Google Scholar] [CrossRef]
- Nurnberg, A.; Kitzing, T.; Grosse, R. Nucleating actin for invasion. Nat. Rev. Cancer 2011, 11, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, D.; Zhuang, L.; Sun, L.; Wu, J. Identification of Arp2/3 Complex Subunits as Prognostic Biomarkers for Hepatocellular Carcinoma. Front. Mol. Biosci. 2021, 8, 690151. [Google Scholar] [CrossRef] [PubMed]
- Gautreau, A.; Krause, M. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Yu, C.J.; Dai, F.; Xiong, J.; Li, H.J.; Wu, Z.S.; Ding, R.; Wang, H. Role of ARPC2 in Human Gastric Cancer. Mediators Inflamm. 2017, 2017, 5432818. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Wei, W.; Wu, Z.; Wang, J.; Ding, X.; Sheng, Y.; Han, Y.; Wu, Q. ARPC2 promotes breast cancer proliferation and metastasis. Oncol. Rep. 2019, 41, 3189–3200. [Google Scholar] [CrossRef]
- Choi, J.; Lee, Y.J.; Yoon, Y.J.; Kim, C.H.; Park, S.J.; Kim, S.Y.; Doo Kim, N.; Cho Han, D.; Kwon, B.M. Pimozide suppresses cancer cell migration and tumor metastasis through binding to ARPC2, a subunit of the Arp2/3 complex. Cancer Sci. 2019, 110, 3788–3801. [Google Scholar] [CrossRef]
- Hetrick, B.; Han, M.S.; Helgeson, L.A.; Nolen, B.J. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 2013, 20, 701–712. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Han, Y.M.; Choi, J.; Lee, Y.J.; Yun, J.; Lee, S.K.; Lee, C.W.; Kang, J.S.; Chi, S.W.; Moon, J.H.; et al. Benproperine, an ARPC2 inhibitor, suppresses cancer cell migration and tumor metastasis. Biochem. Pharmacol. 2019, 163, 46–59. [Google Scholar] [CrossRef]
- Chen, S.; Min, L.; Li, Y.; Li, W.; Zhong, D.; Kong, W. Anti-tussive activity of benproperine enantiomers on citric-acid-induced cough in conscious guinea-pigs. J. Pharm. Pharmacol. 2004, 56, 277–280. [Google Scholar] [CrossRef]
- Sandblad, P.; Arnell, R.; Samuelsson, J.; Fornstedt, T. Approach for reliable evaluation of drug proteins interactions using surface plasmon resonance technology. Anal. Chem. 2009, 81, 3551–3559. [Google Scholar] [CrossRef]
- Frostell, A.; Vinterback, L.; Sjobom, H. Protein-Ligand Interactions Using SPR Systems. Methods Mol. Biol. 2013, 1008, 139–165. [Google Scholar] [CrossRef] [PubMed]
- Martinez Molina, D.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A.; et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef]
- Suraneni, P.; Rubinstein, B.; Unruh, J.R.; Durnin, M.; Hanein, D.; Li, R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012, 197, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, J.; Zhang, Y.; Ma, S.; Yi, W.; Liu, S.; Yu, X.; Wang, J.; Chen, Y. Coronin 2B regulates dendrite outgrowth by modulating actin dynamics. FEBS Lett. 2020, 594, 2975–2987. [Google Scholar] [CrossRef]
- Mahaffy, R.E.; Pollard, T.D. Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry 2008, 47, 6460–6467. [Google Scholar] [CrossRef]
- Omachi, T.; Ichikawa, T.; Kimura, Y.; Ueda, K.; Kioka, N. Vinculin association with actin cytoskeleton is necessary for stiffness-dependent regulation of vinculin behavior. PLoS ONE 2017, 12, e0175324. [Google Scholar] [CrossRef]
- Bays, J.L.; DeMali, K.A. Vinculin in cell-cell and cell-matrix adhesions. Cell Mol. Life Sci. 2017, 74, 2999–3009. [Google Scholar] [CrossRef]
- Boujemaa-Paterski, R.; Martins, B.; Eibauer, M.; Beales, C.T.; Geiger, B.; Medalia, O. Talin-activated vinculin interacts with branched actin networks to initiate bundles. eLife 2020, 9, e53990. [Google Scholar] [CrossRef]
- Iiizumi, M.; Liu, W.; Pai, S.K.; Furuta, E.; Watabe, K. Drug development against metastasis-related genes and their pathways: A rationale for cancer therapy. Biochim. Biophys. Acta 2008, 1786, 87–104. [Google Scholar] [CrossRef]
- Lin, G.-Q.; You, Q.-D.; Cheng, J.-F.; ProQuest. Chiral Drugs: Chemistry and Biological Action; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Petrie, R.J.; Doyle, A.D.; Yamada, K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Ito, T.; Liu, S.; Ando, H.; Sakamoto, S.; Yamaguchi, Y.; Tokunaga, E.; Shibata, N.; Handa, H.; Hakoshima, T. Structural basis of thalidomide enantiomer binding to cereblon. Sci. Rep. 2018, 8, 1294. [Google Scholar] [CrossRef] [PubMed]
- Rauhala, H.E.; Teppo, S.; Niemela, S.; Kallioniemi, A. Silencing of the ARP2/3 complex disturbs pancreatic cancer cell migration. Anticancer Res. 2013, 33, 45–52. [Google Scholar] [PubMed]
- Dayel, M.J.; Holleran, E.A.; Mullins, R.D. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 14871–14876. [Google Scholar] [CrossRef]
- Laboy Cintron, D.; Muir, A.M.; Scott, A.; McDonald, M.; Monaghan, K.G.; Santiago-Sim, T.; Wentzensen, I.M.; De Luca, C.; Italian Undiagnosed Diseases, N.; Brancati, F.; et al. A recurrent, de novo pathogenic variant in ARPC4 disrupts actin filament formation and causes microcephaly and speech delay. HGG Adv. 2022, 3, 100072. [Google Scholar] [CrossRef]
- Zhao, Z.; Ukidve, A.; Kim, J.; Mitragotri, S. Targeting Strategies for Tissue-Specific Drug Delivery. Cell 2020, 181, 151–167. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Polarity proteins in migration and invasion. Oncogene 2008, 27, 6970–6980. [Google Scholar] [CrossRef]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: Integrating signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef]
- Yamazaki, D.; Kurisu, S.; Takenawa, T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005, 96, 379–386. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 2007, 1773, 642–652. [Google Scholar] [CrossRef]
- Jannie, K.M.; Ellerbroek, S.M.; Zhou, D.W.; Chen, S.; Crompton, D.J.; Garcia, A.J.; DeMali, K.A. Vinculin-dependent actin bundling regulates cell migration and traction forces. Biochem. J. 2015, 465, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Rotty, J.D.; Brighton, H.E.; Craig, S.L.; Asokan, S.B.; Cheng, N.; Ting, J.P.; Bear, J.E. Arp2/3 Complex Is Required for Macrophage Integrin Functions but Is Dispensable for FcR Phagocytosis and In Vivo Motility. Dev. Cell 2017, 42, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.E.; King, S.J.; Asokan, S.B.; Rotty, J.D.; Bear, J.E.; Haugh, J.M. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J. Cell Biol. 2015, 208, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.M.; Zhong, D.F.; Kang, Y.; Chen, X.Y. Enantioselective pharmacokinetics of benproperine in healthy volunteers. Yao Xue Xue Bao 2000, 35, 909–912. [Google Scholar] [PubMed]
- Kwon, B.M.; Han, D.C.; Yoon, Y.J.; Lee, Y.J.; Choi, J.Y.; Lee, S. Inhibitors of Cancer Metastasis through Inhibiting Migration and Invasion of Cancer Cell. U.S. Patent WO 2019/212261, 7 November 2019. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, H.-J.; Yoon, Y.J.; Choi, J.; Lee, Y.-J.; Lee, S.; Cho, W.; Byun, W.G.; Park, S.B.; Han, D.C.; Kwon, B.-M. S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2. Pharmaceuticals 2022, 15, 1462. https://doi.org/10.3390/ph15121462
Jang H-J, Yoon YJ, Choi J, Lee Y-J, Lee S, Cho W, Byun WG, Park SB, Han DC, Kwon B-M. S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2. Pharmaceuticals. 2022; 15(12):1462. https://doi.org/10.3390/ph15121462
Chicago/Turabian StyleJang, Hyun-Jin, Yae Jin Yoon, Jiyeon Choi, Yu-Jin Lee, Sangku Lee, Wansang Cho, Wan Gi Byun, Seung Bum Park, Dong Cho Han, and Byoung-Mog Kwon. 2022. "S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2" Pharmaceuticals 15, no. 12: 1462. https://doi.org/10.3390/ph15121462
APA StyleJang, H.-J., Yoon, Y. J., Choi, J., Lee, Y.-J., Lee, S., Cho, W., Byun, W. G., Park, S. B., Han, D. C., & Kwon, B.-M. (2022). S-Benproperine, an Active Stereoisomer of Benproperine, Suppresses Cancer Migration and Tumor Metastasis by Targeting ARPC2. Pharmaceuticals, 15(12), 1462. https://doi.org/10.3390/ph15121462






