The Probiotic Strains Bifidοbacterium lactis, Lactobacillus acidophilus, Lactiplantibacillus plantarum and Saccharomyces boulardii Regulate Wound Healing and Chemokine Responses in Human Intestinal Subepithelial Myofibroblasts
Abstract
1. Introduction
2. Results
2.1. Viability of the Probiotic Strains
2.2. Gram Staining of the Probiotic Strains
2.3. Probiotic Stimulation Affects cSEMF Responses
2.3.1. The Probiotic Strains Regulate the Chemokine mRNA Expression on cSEMFs
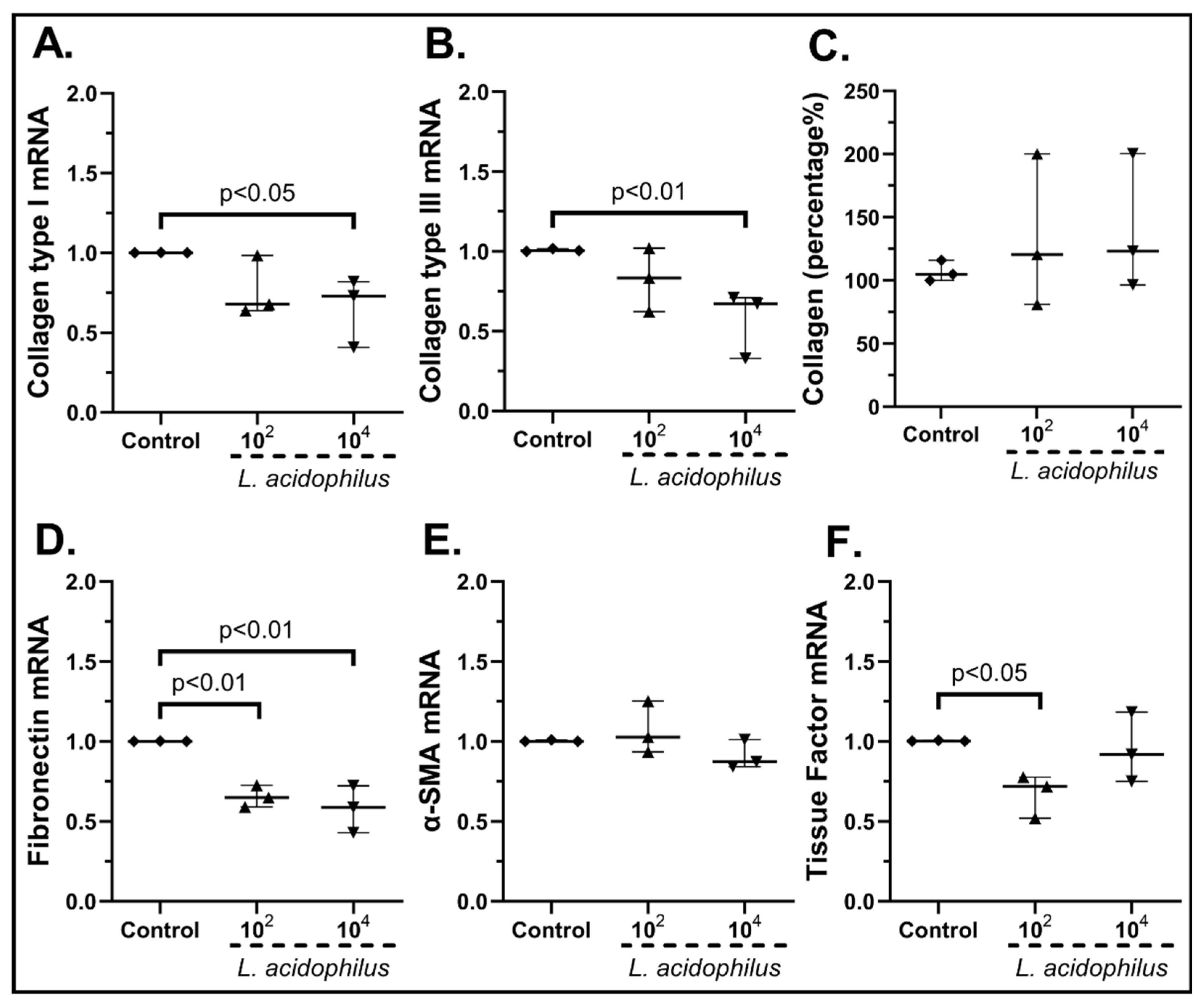
2.3.2. The Probiotic Strains Regulate the Expression of Wound-Healing-Related Factors on cSEMFs
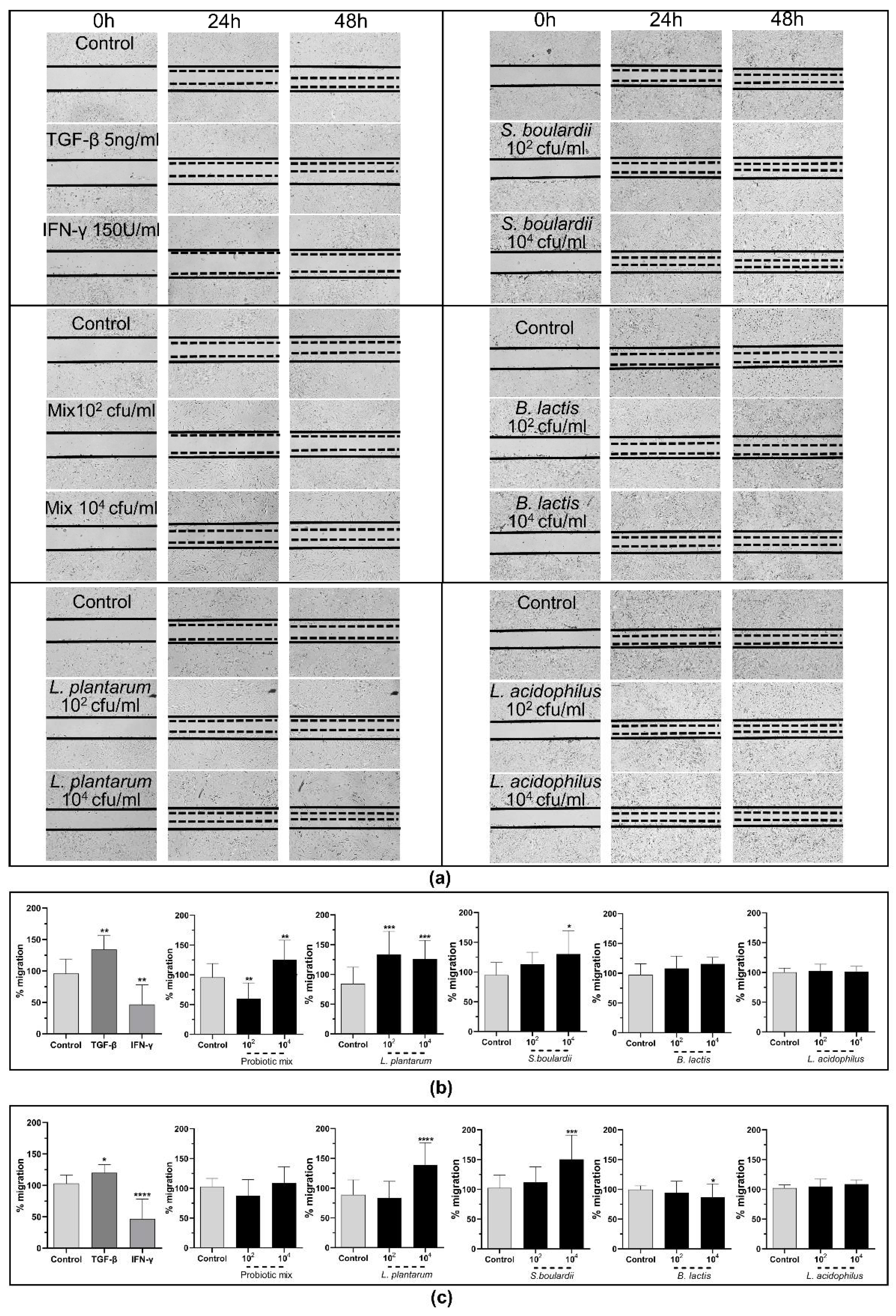
2.3.3. The Probiotic Strains Promote Wound Healing through the Induction of cSEMFs’ Migration
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Colonic Subepithelial Myofibroblast Isolation and Culture
4.3. Probiotics
4.4. Probiotic Viability Assay
4.5. Gram Staining
4.6. cSEMF Probiotics Stimulation
4.7. RNA Extraction, cDNA Synthesis and Real-Time PCR
4.8. Enzyme-Linked Immunosorbent Assay (ELISA)
4.9. Wound Healing Assay after Probiotic Stimulation
4.10. Collagen Production after Probiotic Stimulation
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Chen, T.; Shi, L.; Wang, D.; Tang, D. Regulatory role of short-chain fatty acids in inflammatory bowel disease. Cell Commun. Signal 2022, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Agraib, L.M.; Yamani, M.I.; Rayyan, Y.M.; Abu-Sneineh, A.T.; Tamimi, T.A.; Tayyem, R.F. The probiotic supplementation role in improving the immune system among people with ulcerative colitis: A narrative review. Drug Metab. Pers. Ther. 2021, 37, 7–19. [Google Scholar] [CrossRef]
- Filidou, E.; Kolios, G. Probiotics in Intestinal Mucosal Healing: A New Therapy or an Old Friend? Pharmaceuticals 2021, 14, 1181. [Google Scholar] [CrossRef]
- Damaskos, D.; Kolios, G. Probiotics and prebiotics in inflammatory bowel disease: Microflora ‘on the scope’. Br. J. Clin. Pharmacol. 2008, 65, 453–467. [Google Scholar] [CrossRef]
- Aghamohammad, S.; Sepehr, A.; Miri, S.T.; Najafi, S.; Pourshafie, M.R.; Rohani, M. Anti-inflammatory and immunomodulatory effects of Lactobacillus spp. as a preservative and therapeutic agent for IBD control. Immun. Inflamm. Dis. 2022, 10, e635. [Google Scholar] [CrossRef]
- Le, B.; Yang, S.H. Efficacy of Lactobacillus plantarum in prevention of inflammatory bowel disease. Toxicol Rep. 2018, 5, 314–317. [Google Scholar] [CrossRef]
- Štofilová, J.; Langerholc, T.; Botta, C.; Treven, P.; Gradišnik, L.; Salaj, R.; Šoltésová, A.; Bertková, I.; Hertelyová, Z.; Bomba, A. Cytokine production in vitro and in rat model of colitis in response to Lactobacillus plantarum LS/07. Biomed. Pharmacother. 2017, 94, 1176–1185. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.; Ahn, J.B.; Kim, J.H.; Ma, H.W.; Seo, D.H.; Che, X.; Park, K.C.; Jeon, J.Y.; Kim, S.Y.; et al. Lactobacillus plantarum CBT LP3 ameliorates colitis via modulating T cells in mice. Int. J. Med. Microbiol. 2020, 310, 151391. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, J.; Sun, R.; Wang, M.; Wang, K.; Li, Y.; Shang, H.; Hou, J.; Jiang, Z. Lactobacillus plantarum 23-1 improves intestinal inflammation and barrier function through the TLR4/NF-κB signaling pathway in obese mice. Food Funct. 2022, 13, 5971–5986. [Google Scholar] [CrossRef] [PubMed]
- Uchinaka, A.; Azuma, N.; Mizumoto, H.; Nakano, S.; Minamiya, M.; Yoneda, M.; Aoyama, K.; Komatsu, Y.; Yamada, Y.; Murohara, T.; et al. Anti-inflammatory effects of heat-killed Lactobacillus plantarum L-137 on cardiac and adipose tissue in rats with metabolic syndrome. Sci. Rep. 2018, 8, 8156. [Google Scholar] [CrossRef] [PubMed]
- Paszti-Gere, E.; Szeker, K.; Csibrik-Nemeth, E.; Csizinszky, R.; Marosi, A.; Palocz, O.; Farkas, O.; Galfi, P. Metabolites of Lactobacillus plantarum 2142 prevent oxidative stress-induced overexpression of proinflammatory cytokines in IPEC-J2 cell line. Inflammation 2012, 35, 1487–1499. [Google Scholar] [CrossRef]
- Pellon, A.; Barriales, D.; Peña-Cearra, A.; Castelo-Careaga, J.; Palacios, A.; Lopez, N.; Atondo, E.; Pascual-Itoiz, M.A.; Martín-Ruiz, I.; Sampedro, L.; et al. The commensal bacterium Lactiplantibacillus plantarum imprints innate memory-like responses in mononuclear phagocytes. Gut Microbes 2021, 13, 1939598. [Google Scholar] [CrossRef] [PubMed]
- Satish, L.; Gallo, P.H.; Johnson, S.; Yates, C.C.; Kathju, S. Local Probiotic Therapy with Lactobacillus plantarum Mitigates Scar Formation in Rabbits after Burn Injury and Infection. Surg. Infect. (Larchmt.) 2017, 18, 119–127. [Google Scholar] [CrossRef]
- Dubey, A.K.; Podia, M.; Raut, S.; Singh, S.; Pinnaka, A.K.; Khatri, N. Insight Into the Beneficial Role of Lactiplantibacillus plantarum Supernatant Against Bacterial Infections, Oxidative Stress, and Wound Healing in A549 Cells and BALB/c Mice. Front. Pharmacol. 2021, 12, 728614. [Google Scholar] [CrossRef]
- Argañaraz Aybar, J.N.; Ortiz Mayor, S.; Olea, L.; Garcia, J.J.; Nisoria, S.; Kolling, Y.; Melian, C.; Rachid, M.; Torres Dimani, R.; Werenitzky, C.; et al. Topical Administration of Lactiplantibacillus plantarum Accelerates the Healing of Chronic Diabetic Foot Ulcers through Modifications of Infection, Angiogenesis, Macrophage Phenotype and Neutrophil Response. Microorganisms 2022, 10, 634. [Google Scholar] [CrossRef]
- Park, J.S.; Choi, J.W.; Jhun, J.; Kwon, J.Y.; Lee, B.I.; Yang, C.W.; Park, S.H.; Cho, M.L. Lactobacillus acidophilus Improves Intestinal Inflammation in an Acute Colitis Mouse Model by Regulation of Th17 and Treg Cell Balance and Fibrosis Development. J. Med. Food 2018, 21, 215–224. [Google Scholar] [CrossRef]
- Kang, Y.; Kang, X.; Yang, H.; Liu, H.; Yang, X.; Liu, Q.; Tian, H.; Xue, Y.; Ren, P.; Kuang, X.; et al. Lactobacillus acidophilus ameliorates obesity in mice through modulation of gut microbiota dysbiosis and intestinal permeability. Pharmacol Res. 2022, 175, 106020. [Google Scholar] [CrossRef]
- Kye, Y.J.; Lee, S.Y.; Kim, H.R.; Lee, B.H.; Park, J.H.; Park, M.S.; Ji, G.E.; Sung, M.K. Lactobacillus acidophilus PIN7 paraprobiotic supplementation ameliorates DSS-induced colitis through anti-inflammatory and immune regulatory effects. J. Appl. Microbiol. 2022, 132, 3189–3200. [Google Scholar] [CrossRef]
- Hu, T.; Wang, H.; Xiang, C.; Mu, J.; Zhao, X. Preventive Effect of Lactobacillus acidophilus XY27 on DSS-Induced Ulcerative Colitis in Mice. Drug Des. Devel Ther. 2020, 14, 5645–5657. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.K.; Han, D.H.; Jang, Y.J.; Park, S.; Jang, S.J.; Lee, G.; Han, H.S.; Ko, G. Alleviation of DSS-induced colitis via Lactobacillus acidophilus treatment in mice. Food Funct. 2021, 12, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, F.; Palumbo, P.; Mattei, A.; Augello, F.R.; Cifone, M.G.; Giuliani, M.; Cinque, B. Soluble Fraction from Lysates of Selected Probiotic Strains Differently Influences Re-Epithelialization of HaCaT Scratched Monolayer through a Mechanism Involving Nitric Oxide Synthase 2. Biomolecules 2019, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dong, B.; Wang, Y.; Zhang, Q.; Wang, B.; Feng, S.; Zhu, Y. The role of Bacillus acidophilus in osteoporosis and its roles in proliferation and differentiation. J. Clin. Lab. Anal. 2020, 34, e23471. [Google Scholar] [CrossRef] [PubMed]
- Bahr, H.I.; Hamad, R.; Ismail, S.A. The impact of Lactobacillus acidophilus on hepatic and colonic fibrosis induced by ethephon in a rat model. Iran J. Basic. Med. Sci. 2019, 22, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Liu, C.; Dai, M.; Wang, Q.; Li, C.; Hung, W. Bifidobacterium lactis BL-99 modulates intestinal inflammation and functions in zebrafish models. PLoS ONE 2022, 17, e0262942. [Google Scholar] [CrossRef]
- Dong, Y.; Liao, W.; Tang, J.; Fei, T.; Gai, Z.; Han, M. Bifidobacterium BLa80 mitigates colitis by altering gut microbiota and alleviating inflammation. AMB Express 2022, 12, 67. [Google Scholar] [CrossRef]
- Meng, H.; Ba, Z.; Lee, Y.; Peng, J.; Lin, J.; Fleming, J.A.; Furumoto, E.J.; Roberts, R.F.; Kris-Etherton, P.M.; Rogers, C.J. Consumption of Bifidobacterium animalis subsp. lactis BB-12 in yogurt reduced expression of TLR-2 on peripheral blood-derived monocytes and pro-inflammatory cytokine secretion in young adults. Eur. J. Nutr. 2017, 56, 649–661. [Google Scholar] [CrossRef]
- Neyrinck, A.M.; Rodriguez, J.; Taminiau, B.; Amadieu, C.; Herpin, F.; Allaert, F.A.; Cani, P.D.; Daube, G.; Bindels, L.B.; Delzenne, N.M. Improvement of gastrointestinal discomfort and inflammatory status by a synbiotic in middle-aged adults: A double-blind randomized placebo-controlled trial. Sci. Rep. 2021, 11, 2627. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Guo, X.K. Adhesion and immunomodulatory effects of Bifidobacterium lactis HN019 on intestinal epithelial cells INT-407. World J. Gastroenterol. 2010, 16, 2283–2290. [Google Scholar] [CrossRef]
- Lan, H.; Liu, W.H.; Zheng, H.; Feng, H.; Zhao, W.; Hung, W.L.; Li, H. Bifidobacterium lactis BL-99 protects mice with osteoporosis caused by colitis via gut inflammation and gut microbiota regulation. Food Funct. 2022, 13, 1482–1494. [Google Scholar] [CrossRef]
- Turner, R.B.; Woodfolk, J.A.; Borish, L.; Steinke, J.W.; Patrie, J.T.; Muehling, L.M.; Lahtinen, S.; Lehtinen, M.J. Effect of probiotic on innate inflammatory response and viral shedding in experimental rhinovirus infection - a randomised controlled trial. Benef. Microbes 2017, 8, 207–215. [Google Scholar] [CrossRef]
- Huang, F.C.; Huang, S.C. The different effects of probiotics treatment on Salmonella-induced interleukin-8 response in intestinal epithelia cells via PI3K/Akt and NOD2 expression. Benef. Microbes 2016, 7, 739–748. [Google Scholar] [CrossRef]
- Martín, R.; Laval, L.; Chain, F.; Miquel, S.; Natividad, J.; Cherbuy, C.; Sokol, H.; Verdu, E.F.; van Hylckama Vlieg, J.; Bermudez-Humaran, L.G.; et al. Bifidobacterium animalis ssp. lactis CNCM-I2494 Restores Gut Barrier Permeability in Chronically Low-Grade Inflamed Mice. Front. Microbiol. 2016, 7, 608. [Google Scholar] [CrossRef]
- Wang, N.; Wang, S.; Xu, B.; Liu, F.; Huo, G.; Li, B. Alleviation Effects of Bifidobacterium animalis subsp. lactis XLTG11 on Dextran Sulfate Sodium-Induced Colitis in Mice. Microorganisms 2021, 9, 2093. [Google Scholar] [CrossRef]
- Li, M.; Zhu, L.; Xie, A.; Yuan, J. Oral administration of Saccharomyces boulardii ameliorates carbon tetrachloride-induced liver fibrosis in rats via reducing intestinal permeability and modulating gut microbial composition. Inflammation 2015, 38, 170–179. [Google Scholar] [CrossRef]
- Dalmasso, G.; Cottrez, F.; Imbert, V.; Lagadec, P.; Peyron, J.-F.; Rampal, P.; Czerucka, D.; Groux, H. Saccharomyces boulardii Inhibits Inflammatory Bowel Disease by Trapping T Cells in Mesenteric Lymph Nodes. Gastroenterology 2006, 131, 1812–1825. [Google Scholar] [CrossRef]
- Thomas, S.; Metzke, D.; Schmitz, J.; Dörffel, Y.; Baumgart, D.C. Anti-inflammatory effects of Saccharomyces boulardii mediated by myeloid dendritic cells from patients with Crohn’s disease and ulcerative colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G1083–G1092. [Google Scholar] [CrossRef]
- Thomas, S.; Przesdzing, I.; Metzke, D.; Schmitz, J.; Radbruch, A.; Baumgart, D.C. Saccharomyces boulardii inhibits lipopolysaccharide-induced activation of human dendritic cells and T cell proliferation. Clin. Exp. Immunol. 2009, 156, 78–87. [Google Scholar] [CrossRef]
- Singh, A.; Mandal, U.K.; Narang, R.K. Development and In Vivo Evaluation of Pectin Based Enteric Coated Microparticles Loaded with Mesalamine and Saccharomyces boulardii for Management of Ulcerative Colitis. Assay Drug Dev. Technol. 2022, 20, 22–34. [Google Scholar] [CrossRef]
- Gao, H.; Li, Y.; Sun, J.; Xu, H.; Wang, M.; Zuo, X.; Fu, Q.; Guo, Y.; Chen, Z.; Zhang, P.; et al. Saccharomyces boulardii Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice by Regulating NF-κB and Nrf2 Signaling Pathways. Oxid. Med. Cell Longev. 2021, 2021, 1622375. [Google Scholar] [CrossRef]
- Li, B.; Zhang, H.; Shi, L.; Li, R.; Luo, Y.; Deng, Y.; Li, S.; Li, R.; Liu, Z. Saccharomyces boulardii alleviates DSS-induced intestinal barrier dysfunction and inflammation in humanized mice. Food Funct. 2022, 13, 102–112. [Google Scholar] [CrossRef]
- Canonici, A.; Pellegrino, E.; Siret, C.; Terciolo, C.; Czerucka, D.; Bastonero, S.; Marvaldi, J.; Lombardo, D.; Rigot, V.; André, F. Saccharomyces boulardii improves intestinal epithelial cell restitution by inhibiting αvβ5 integrin activation state. PLoS ONE 2012, 7, e45047. [Google Scholar] [CrossRef]
- Canonici, A.; Siret, C.; Pellegrino, E.; Pontier-Bres, R.; Pouyet, L.; Montero, M.P.; Colin, C.; Czerucka, D.; Rigot, V.; André, F. Saccharomyces boulardii improves intestinal cell restitution through activation of the α2β1 integrin collagen receptor. PLoS ONE 2011, 6, e18427. [Google Scholar] [CrossRef]
- Terciolo, C.; Dobric, A.; Ouaissi, M.; Siret, C.; Breuzard, G.; Silvy, F.; Marchiori, B.; Germain, S.; Bonier, R.; Hama, A.; et al. Saccharomyces boulardii CNCM I-745 Restores intestinal Barrier Integrity by Regulation of E-cadherin Recycling. J. Crohns. Colitis. 2017, 11, 999–1010. [Google Scholar] [CrossRef]
- Roulis, M.; Flavell, R.A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 2016, 92, 116–131. [Google Scholar] [CrossRef]
- Valatas, V.; Filidou, E.; Drygiannakis, I.; Kolios, G. Stromal and immune cells in gut fibrosis: The myofibroblast and the scarface. Ann. Gastroenterol. 2017, 30, 393–404. [Google Scholar] [CrossRef]
- Kandilogiannakis, L.; Filidou, E.; Drygiannakis, I.; Tarapatzi, G.; Didaskalou, S.; Koffa, M.; Arvanitidis, K.; Bamias, G.; Valatas, V.; Paspaliaris, V.; et al. Development of a Human Intestinal Organoid Model for In Vitro Studies on Gut Inflammation and Fibrosis. Stem. Cells Int. 2021, 2021, 9929461. [Google Scholar] [CrossRef]
- Lei, N.Y.; Jabaji, Z.; Wang, J.; Joshi, V.S.; Brinkley, G.J.; Khalil, H.; Wang, F.; Jaroszewicz, A.; Pellegrini, M.; Li, L.; et al. Intestinal subepithelial myofibroblasts support the growth of intestinal epithelial stem cells. PLoS ONE 2014, 9, e84651. [Google Scholar] [CrossRef]
- Filidou, E.; Valatas, V.; Drygiannakis, I.; Arvanitidis, K.; Vradelis, S.; Kouklakis, G.; Kolios, G.; Bamias, G. Cytokine Receptor Profiling in Human Colonic Subepithelial Myofibroblasts: A Differential Effect of Th Polarization-Associated Cytokines in Intestinal Fibrosis. Inflamm. Bowel. Dis. 2018, 24, 2224–2241. [Google Scholar] [CrossRef]
- Drygiannakis, I.; Valatas, V.; Sfakianaki, O.; Bourikas, L.; Manousou, P.; Kambas, K.; Ritis, K.; Kolios, G.; Kouroumalis, E. Proinflammatory cytokines induce crosstalk between colonic epithelial cells and subepithelial myofibroblasts: Implication in intestinal fibrosis. J. Crohns. Colitis. 2013, 7, 286–300. [Google Scholar] [CrossRef]
- Vetuschi, A.; Battista, N.; Pompili, S.; Cappariello, A.; Prete, R.; Taticchi, A.; Selvaggini, R.; Latella, G.G.; Corsetti, A.; Sferra, R. The antiinflammatory and antifibrotic effect of olive phenols and Lactiplantibacillus plantarum IMC513 in dextran sodium sulfate-induced chronic colitis. Nutrition 2022, 94, 111511. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, F.; Augello, F.R.; Palumbo, P.; Mollsi, E.; Giuliani, M.; Cimini, A.M.; Cifone, M.G.; Cinque, B. Soluble Fraction from Lysate of a High Concentration Multi-Strain Probiotic Formulation Inhibits TGF-β1-Induced Intestinal Fibrosis on CCD-18Co Cells. Nutrients 2021, 13, 882. [Google Scholar] [CrossRef] [PubMed]
- Uribe, G.; Villéger, R.; Bressollier, P.; Dillard, R.N.; Worthley, D.L.; Wang, T.C.; Powell, D.W.; Urdaci, M.C.; Pinchuk, I.V. Lactobacillus rhamnosus GG increases cyclooxygenase-2 expression and prostaglandin E2 secretion in colonic myofibroblasts via a MyD88-dependent mechanism during homeostasis. Cell Microbiol. 2018, 20, e12871. [Google Scholar] [CrossRef]
- Pierzchalska, M.; Panek, M.; Czyrnek, M.; Gielicz, A.; Mickowska, B.; Grabacka, M. Probiotic Lactobacillus acidophilus bacteria or synthetic TLR2 agonist boost the growth of chicken embryo intestinal organoids in cultures comprising epithelial cells and myofibroblasts. Comp. Immunol. Microbiol. Infect. Dis. 2017, 53, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Dong, M.; Zhang, H.; Xu, H.; Wang, Q.; Yan, C.; Ye, R.; Jiang, X.; Zhou, H.; Chen, L.; et al. Oral delivery of a Lactococcus lactis expressing extracellular TGFβR2 alleviates hepatic fibrosis. Appl. Microbiol. Biotechnol. 2021, 105, 6007–6018. [Google Scholar] [CrossRef] [PubMed]
- Lamousé-Smith, E.; Kelly, D.; De Cremoux, I. Designing bugs as drugs: Exploiting the gut microbiome. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 320, G295–G303. [Google Scholar] [CrossRef] [PubMed]
- Sun, J. Intestinal organoid as an in vitro model in studying host-microbial interactions. Front. Biol. 2017, 12, 94–102. [Google Scholar] [CrossRef]
- Saxami, G.; Karapetsas, A.; Chondrou, P.; Vasiliadis, S.; Lamprianidou, E.; Kotsianidis, I.; Ypsilantis, P.; Botaitis, S.; Simopoulos, C.; Galanis, A. Potentially probiotic Lactobacillus strains with anti-proliferative activity induce cytokine/chemokine production and neutrophil recruitment in mice. Benef. Microbes. 2017, 8, 615–623. [Google Scholar] [CrossRef]
- Wang, B.; Li, J.; Chen, J.; Huang, Q.; Li, N.; Li, J. Effect of live Lactobacillus plantarum L2 on TNF-alpha-induced MCP-1 production in Caco-2 cells. Int. J. Food Microbiol. 2010, 142, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Gabryszewski, S.J.; Bachar, O.; Dyer, K.D.; Percopo, C.M.; Killoran, K.E.; Domachowske, J.B.; Rosenberg, H.F. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J. Immunol. 2011, 186, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Tomasik, J.; Yolken, R.H.; Bahn, S.; Dickerson, F.B. Immunomodulatory Effects of Probiotic Supplementation in Schizophrenia Patients: A Randomized, Placebo-Controlled Trial. Biomark. Insights. 2015, 10, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Roselli, M.; Finamore, A.; Nuccitelli, S.; Carnevali, P.; Brigidi, P.; Vitali, B.; Nobili, F.; Rami, R.; Garaguso, I.; Mengheri, E. Prevention of TNBS-induced colitis by different Lactobacillus and Bifidobacterium strains is associated with an expansion of gammadeltaT and regulatory T cells of intestinal intraepithelial lymphocytes. Inflamm. Bowel. Dis. 2009, 15, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Justino, P.F.; Melo, L.F.; Nogueira, A.F.; Morais, C.M.; Mendes, W.O.; Franco, A.X.; Souza, E.P.; Ribeiro, R.A.; Souza, M.H.; Soares, P.M. Regulatory role of Lactobacillus acidophilus on inflammation and gastric dysmotility in intestinal mucositis induced by 5-fluorouracil in mice. Cancer Chemother. Pharmacol. 2015, 75, 559–567. [Google Scholar] [CrossRef]
- Fukuyama, K.; Islam, M.A.; Takagi, M.; Ikeda-Ohtsubo, W.; Kurata, S.; Aso, H.; Vignolo, G.; Villena, J.; Kitazawa, H. Evaluation of the Immunomodulatory Ability of Lactic Acid Bacteria Isolated from Feedlot Cattle Against Mastitis Using a Bovine Mammary Epithelial Cells In Vitro Assay. Pathogens 2020, 9, 410. [Google Scholar] [CrossRef]
- Kolios, G.; Lira, A.S. Chemokines and their Receptors in Gut Homeostasis and Disease. Curr. Immunol. Rev. (Discontin.) 2011, 7, 271–279. [Google Scholar] [CrossRef]
- Li, S.C.; Hsu, W.F.; Chang, J.S.; Shih, C.K. Combination of Lactobacillus acidophilus and Bifidobacterium animalis subsp. lactis Shows a Stronger Anti-Inflammatory Effect than Individual Strains in HT-29 Cells. Nutrients 2019, 11, 969. [Google Scholar] [CrossRef] [PubMed]
- Borshchev, Y.Y.; Protsak, E.S.; Burovenko, I.Y.; Semenova, N.Y.; Zubkov, I.G.; Galagudza, M.M. Effect of Probiotic Therapy on Hemodynamic Response Associated with Systemic Inflammatory Reaction and Antibiotic-Induced Dysbiosis in Chronic Experiments in Rats. Bull. Exp. Biol. Med. 2022, 172, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.K.; Socca, E.A.R.; de Souza, B.R.; Genaro, S.C.; Durán, N.; Fávaro, W.J. Effects of combined OncoTherad immunotherapy and probiotic supplementation on modulating the chronic inflammatory process in colorectal carcinogenesis. Tissue Cell 2022, 75, 101747. [Google Scholar] [CrossRef]
- Sheikhi, A.; Shakerian, M.; Giti, H.; Baghaeifar, M.; Jafarzadeh, A.; Ghaed, V.; Heibor, M.R.; Baharifar, N.; Dadafarin, Z.; Bashirpour, G. Probiotic Yogurt Culture Bifidobacterium Animalis Subsp. Lactis BB-12 and Lactobacillus Acidophilus LA-5 Modulate the Cytokine Secretion by Peripheral Blood Mononuclear Cells from Patients with Ulcerative Colitis. Drug Res. (Stuttg.) 2016, 66, 300–305. [Google Scholar] [CrossRef][Green Version]
- Leventogiannis, K.; Gkolfakis, P.; Spithakis, G.; Tsatali, A.; Pistiki, A.; Sioulas, A.; Giamarellos-Bourboulis, E.J.; Triantafyllou, K. Effect of a Preparation of Four Probiotics on Symptoms of Patients with Irritable Bowel Syndrome: Association with Intestinal Bacterial Overgrowth. Probiotics Antimicrob. Proteins 2019, 11, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Stavrou, G.; Giamarellos-Bourboulis, E.J.; Kotzampassi, K. The role of probiotics in the prevention of severe infections following abdominal surgery. Int. J. Antimicrob. Agents. 2015, 46 (Suppl. S1), S2–S4. [Google Scholar] [CrossRef]
- Tzikos, G.; Tsalkatidou, D.; Stavrou, G.; Thoma, G.; Chorti, A.; Tsilika, M.; Michalopoulos, A.; Papavramidis, T.; Giamarellos-Bourboulis, E.J.; Kotzampassi, K. A Four-Probiotic Regime to Reduce Surgical Site Infections in Multi-Trauma Patients. Nutrients 2022, 14, 2620. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Q.; Zhang, Q.; Xu, M.; Qu, Y.; Cai, X.; Lu, L. CXCL6 promotes human hepatocyte proliferation through the CXCR1-NFκB pathway and inhibits collagen I secretion by hepatic stellate cells. Biochem. Cell Biol. 2016, 94, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed]
- Castilho, T.J.C.; Almeida, G.; Mello, E.; Campos, A.C.L. Effect of supplementation with probiotics on colonic anastomoses in rats: Morphological and tensiometric study. Arq. Bras. Cir. Dig. 2021, 33, e1550. [Google Scholar] [CrossRef] [PubMed]
- Tagliari, E.; Campos, L.F.; Campos, A.C.; Costa-Casagrande, T.A.; Noronha, L. EFFECT OF PROBIOTIC ORAL ADMINISTRATION ON SKIN WOUND HEALING IN RATS. Arq. Bras. Cir. Dig. 2019, 32, e1457. [Google Scholar] [CrossRef]
- Moysidis, M.; Stavrou, G.; Cheva, A.; Abba Deka, I.; Tsetis, J.K.; Birba, V.; Kapoukranidou, D.; Ioannidis, A.; Tsaousi, G.; Kotzampassi, K. The 3-D configuration of excisional skin wound healing after topical probiotic application. Injury 2022, 53, 1385–1393. [Google Scholar] [CrossRef]
- Alkushi, A.G.; Elazab, S.T.; Abdelfattah-Hassan, A.; Mahfouz, H.; Salem, G.A.; Sheraiba, N.I.; Mohamed, E.A.A.; Attia, M.S.; El-Shetry, E.S.; Saleh, A.A.; et al. Multi-Strain-Probiotic-Loaded Nanoparticles Reduced Colon Inflammation and Orchestrated the Expressions of Tight Junction, NLRP3 Inflammasome and Caspase-1 Genes in DSS-Induced Colitis Model. Pharmaceutics 2022, 14, 1183. [Google Scholar] [CrossRef]
- Tsai, W.H.; Chou, C.H.; Huang, T.Y.; Wang, H.L.; Chien, P.J.; Chang, W.W.; Lee, H.T. Heat-Killed Lactobacilli Preparations Promote Healing in the Experimental Cutaneous Wounds. Cells 2021, 10, 3264. [Google Scholar] [CrossRef]
- Kazemi, A.; Ataellahi Eshkoor, P.; Saeedi, P.; Halabian, R. Evaluation of antioxidant and antibacterial effects of lactobacilli metabolites- preconditioned bone marrow mesenchymal stem cells in skin lesions amelioration. Bioorg. Chem. 2022, 124, 105797. [Google Scholar] [CrossRef] [PubMed]
- Ghyselinck, J.; Verstrepen, L.; Moens, F.; Van den Abbeele, P.; Said, J.; Smith, B.; Bjarnason, I.; Basit, A.W.; Gaisford, S. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. Int. J. Pharm. 2020, 587, 119648. [Google Scholar] [CrossRef] [PubMed]
- Toumi, R.; Abdelouhab, K.; Rafa, H.; Soufli, I.; Raissi-Kerboua, D.; Djeraba, Z.; Touil-Boukoffa, C. Beneficial role of the probiotic mixture Ultrabiotique on maintaining the integrity of intestinal mucosal barrier in DSS-induced experimental colitis. Immunopharmacol. Immunotoxicol. 2013, 35, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Gudadappanavar, A.M.; Hombal, P.R.; Timashetti, S.S.; Javali, S.B. Influence of Lactobacillus acidophilus and Lactobacillus plantarum on wound healing in male Wistar rats - an experimental study. Int. J. Appl. Basic. Med. Res. 2017, 7, 233–238. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Filidou, E.; Kandilogiannakis, L.; Tarapatzi, G.; Spathakis, M.; Steiropoulos, P.; Mikroulis, D.; Arvanitidis, K.; Paspaliaris, V.; Kolios, G. Anti-Inflammatory and Anti-Fibrotic Effect of Immortalized Mesenchymal-Stem-Cell-Derived Conditioned Medium on Human Lung Myofibroblasts and Epithelial Cells. Int. J. Mol. Sci. 2022, 23, 4570. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


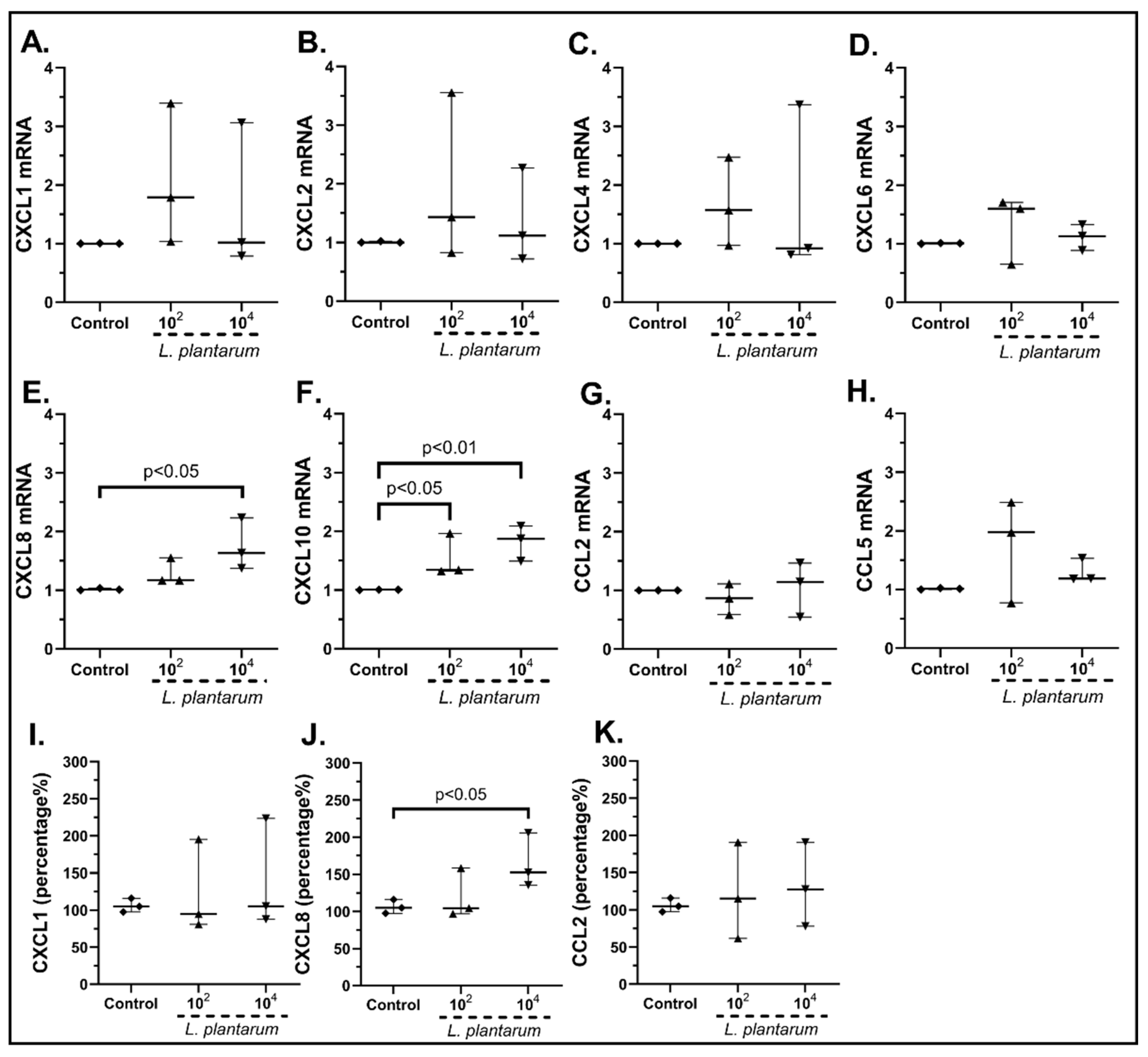
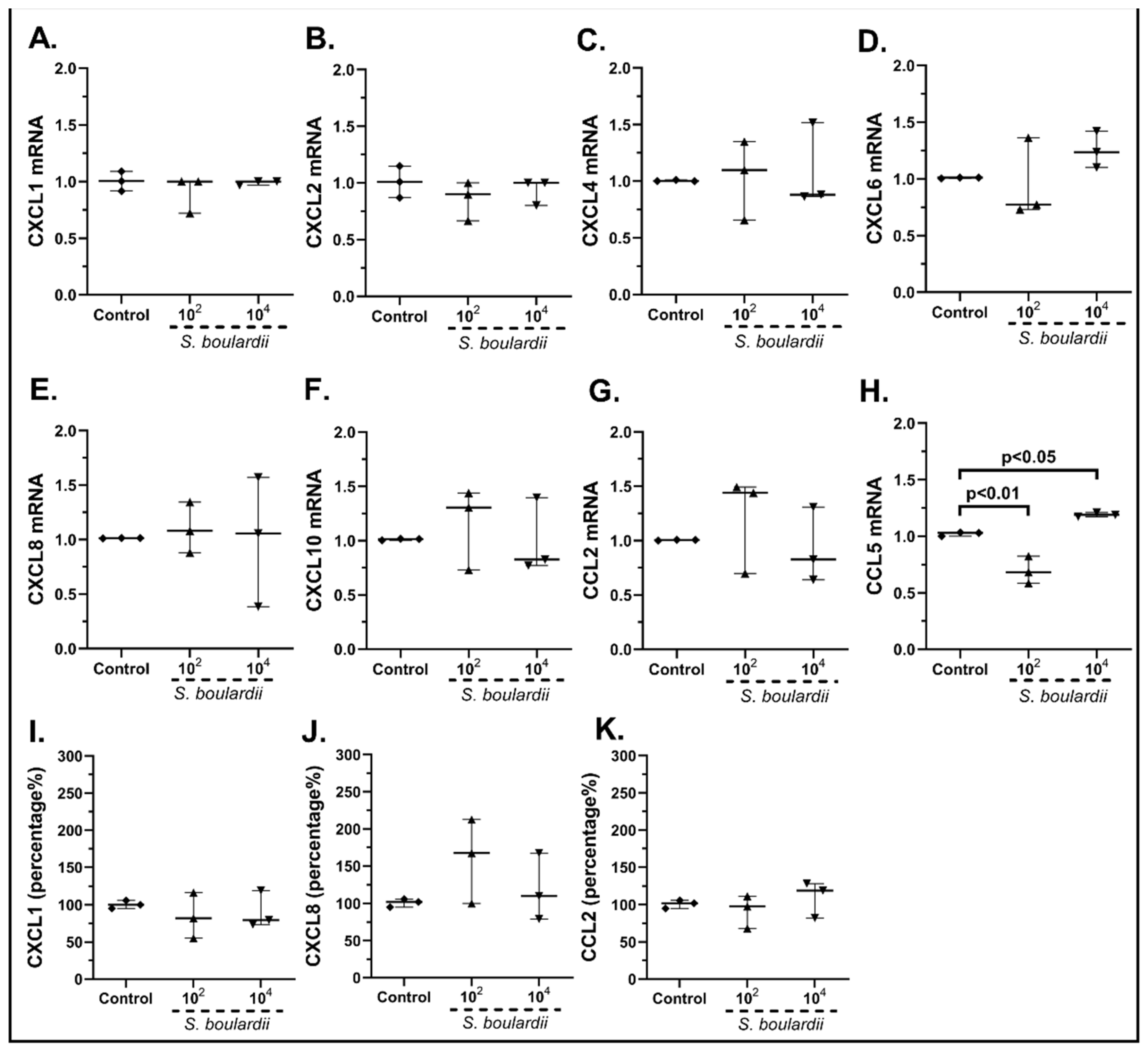
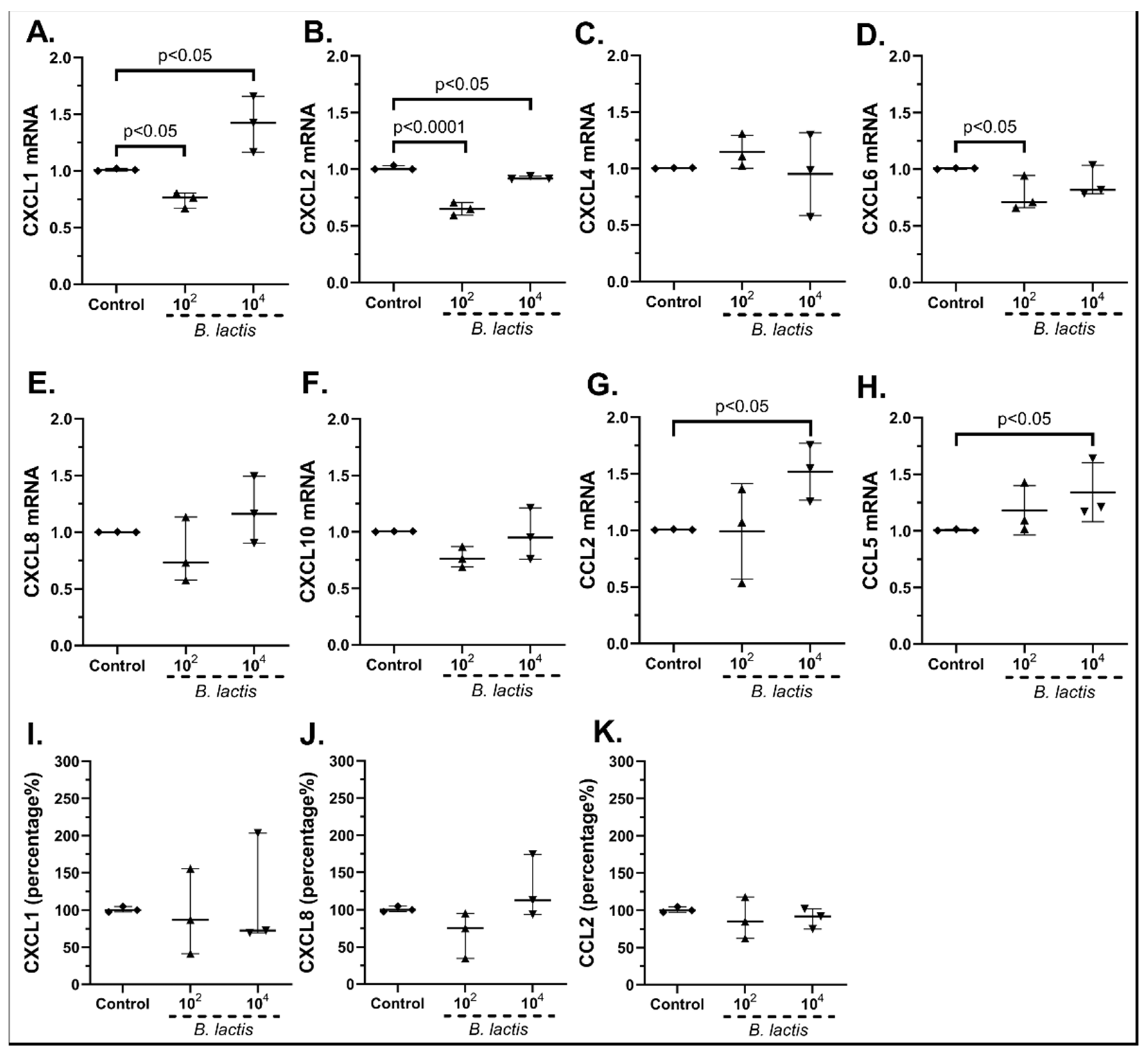

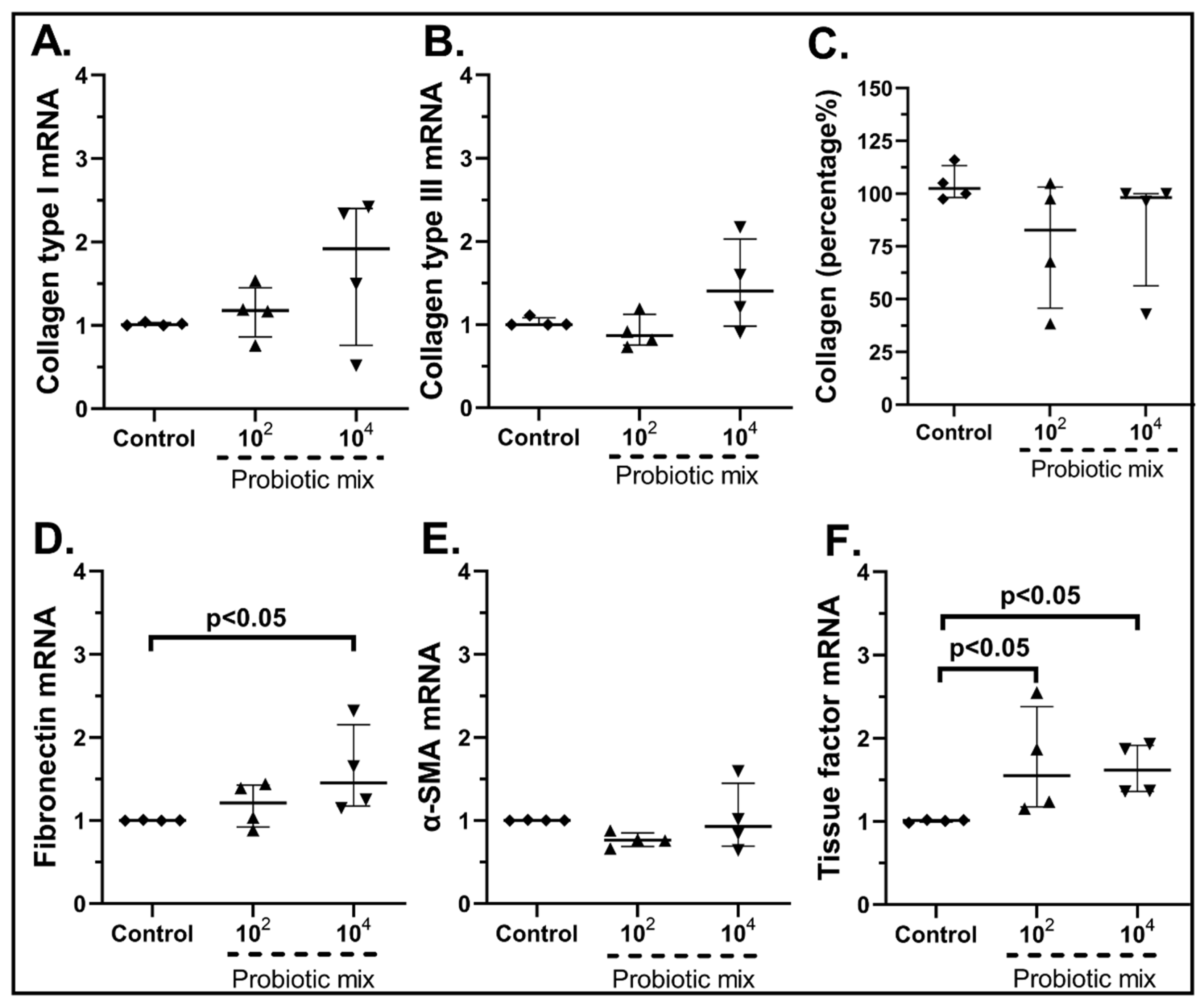



| Gene | Forward | Reverse | Tm (°C) |
|---|---|---|---|
| CCL2 | AGGAAGATCTCAGTGCAGAGG | AGTCTTCGGAGTTTGGGTTTG | 60 |
| CCL5 | CTCGCTGTCATCCTCATTGCT | TGTGGTGTCCGAGGAATATGG | 60 |
| CCL20 | GCTGCTTTGATGTCAGTGC | GCAGTCAAAGTTGCTTGCTTC | 56 |
| Collagen Type I | CCCTGGAAAGAATGGAGATGAT | ACTGAAACCTCTGTGTCCCTTCA | 60 |
| Collagen Type III | GCTCTGCTTCATCCCACTATTA | TGCGAGTCCTCCTACTGCTAC | 60 |
| CXCL1 | GCCCAAACCGAAGTCATAGCC | ATCCGCCAGCCTCTATCACA | 60 |
| CXCL2 | GCTTGTCTCAACCCCGCATC | TGGATTTGCCATTTTTCAGCATCTT | 60 |
| CXCL3 | CGCCCAAACCGAAGTCAT | GTGCTCCCCTTGTTCAGTATCT | 60 |
| CXCL4 | GTCCAGTGGCACCCTCCTGA | AATTGACATTTAGGCAGCTGA | 60 |
| CXCL5 | AGCTGCGTTGCGTTTGTTTAC | TGGCGAACACTTGCAGATTAC | 60 |
| CXCL6 | AGAGCTGCGTTGCACTTGTT | GCAGTTTACCAATCGTTTTGGGG | 60 |
| CXCL7 | TGAGACAGAATGAAACAC | AGGTGATGAATCTGCTG | 60 |
| CXCL8 | TGGGTGCAGAGGGTTGTG | CAGACTAGGGTTGCCAGATTTA | 60 |
| CXCL9 | AAGAAGCACGTGGTAAAACA | TCTCGGTGGCTATCTTGTTA | 56 |
| CXCL10 | CCTGCTTCAAATATTTCCCT | CCTTCCTGTATGTGTTTGGA | 56 |
| CXCL11 | GACGCTGTCTTTGCATAGGC | GGATTTAGGCATCGTTGTCCTTT | 60 |
| CXCL12 | AGAGATGAAAGGGCAAAGAC | CGTATGCTATAAATGCAGGG | 60 |
| CXCL14 | TCCGGTCAGCATGAGGCTCC | CACCCTATTCTTCGTAGACC | 60 |
| Fibronectin | CCAGTCCACAGCTATTCCTG | ACAACCACGGATGAGCTG | 60 |
| GapdH | GACATCAAGAAGGTGGTGAA | TGTCATACCAGGAAATGAGC | 60 |
| Tissue Factor | TTCAGTGTTCAAGCAGTGATTCC | ATGATGACCACAAATACCACAGC | 51 |
| α-SMA | AATGCAGAAGGAGATCACGG | TCCTGTTTGCTGATCCACATC | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarapatzi, G.; Filidou, E.; Kandilogiannakis, L.; Spathakis, M.; Gaitanidou, M.; Arvanitidis, K.; Drygiannakis, I.; Valatas, V.; Kotzampassi, K.; Manolopoulos, V.G.; et al. The Probiotic Strains Bifidοbacterium lactis, Lactobacillus acidophilus, Lactiplantibacillus plantarum and Saccharomyces boulardii Regulate Wound Healing and Chemokine Responses in Human Intestinal Subepithelial Myofibroblasts. Pharmaceuticals 2022, 15, 1293. https://doi.org/10.3390/ph15101293
Tarapatzi G, Filidou E, Kandilogiannakis L, Spathakis M, Gaitanidou M, Arvanitidis K, Drygiannakis I, Valatas V, Kotzampassi K, Manolopoulos VG, et al. The Probiotic Strains Bifidοbacterium lactis, Lactobacillus acidophilus, Lactiplantibacillus plantarum and Saccharomyces boulardii Regulate Wound Healing and Chemokine Responses in Human Intestinal Subepithelial Myofibroblasts. Pharmaceuticals. 2022; 15(10):1293. https://doi.org/10.3390/ph15101293
Chicago/Turabian StyleTarapatzi, Gesthimani, Eirini Filidou, Leonidas Kandilogiannakis, Michail Spathakis, Maria Gaitanidou, Konstantinos Arvanitidis, Ioannis Drygiannakis, Vassilis Valatas, Katerina Kotzampassi, Vangelis G. Manolopoulos, and et al. 2022. "The Probiotic Strains Bifidοbacterium lactis, Lactobacillus acidophilus, Lactiplantibacillus plantarum and Saccharomyces boulardii Regulate Wound Healing and Chemokine Responses in Human Intestinal Subepithelial Myofibroblasts" Pharmaceuticals 15, no. 10: 1293. https://doi.org/10.3390/ph15101293
APA StyleTarapatzi, G., Filidou, E., Kandilogiannakis, L., Spathakis, M., Gaitanidou, M., Arvanitidis, K., Drygiannakis, I., Valatas, V., Kotzampassi, K., Manolopoulos, V. G., Kolios, G., & Vradelis, S. (2022). The Probiotic Strains Bifidοbacterium lactis, Lactobacillus acidophilus, Lactiplantibacillus plantarum and Saccharomyces boulardii Regulate Wound Healing and Chemokine Responses in Human Intestinal Subepithelial Myofibroblasts. Pharmaceuticals, 15(10), 1293. https://doi.org/10.3390/ph15101293








