In Silico-Based Discovery of Natural Anthraquinones with Potential against Multidrug-Resistant E. coli
Abstract
:1. Introduction
2. Results and Discussion
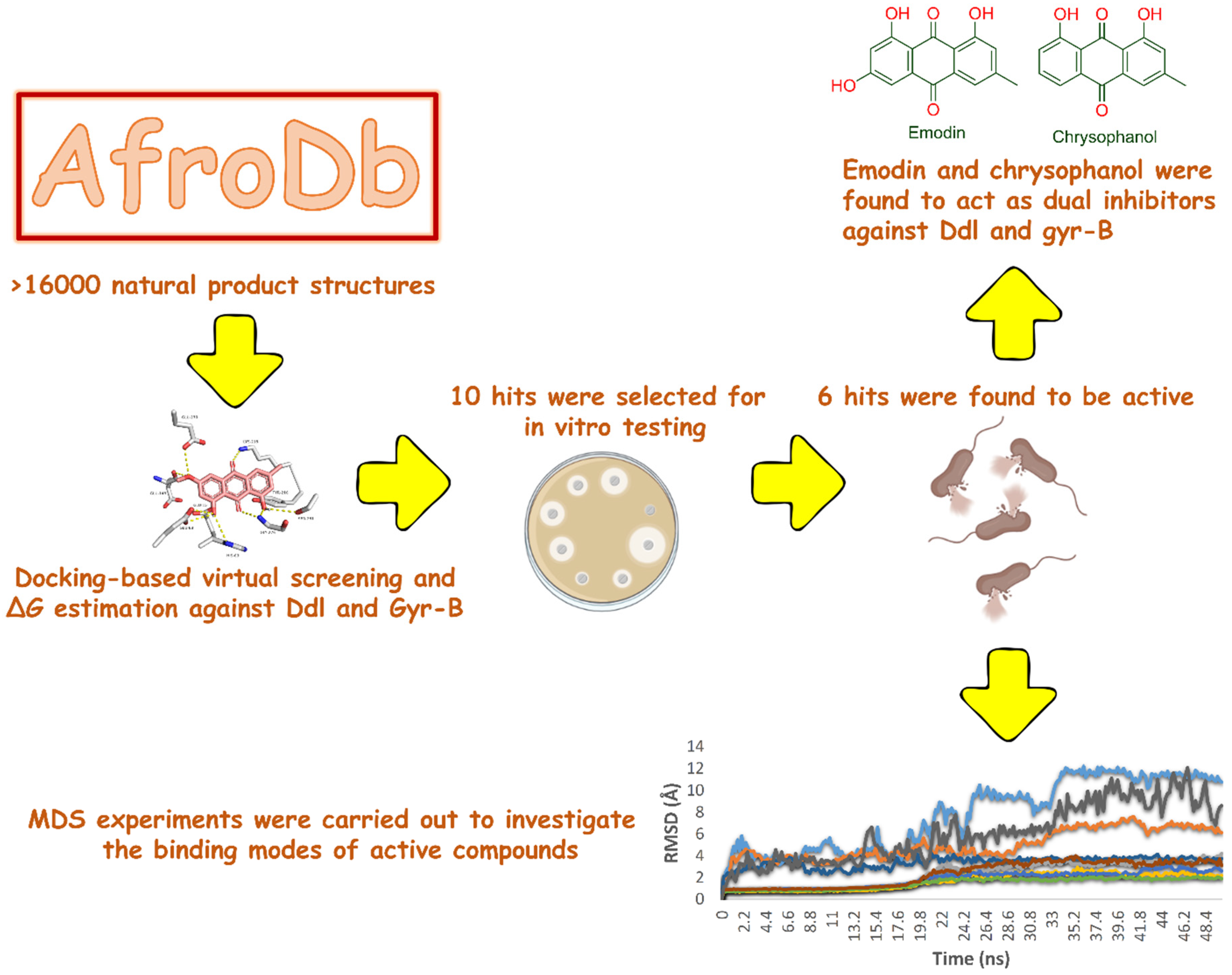
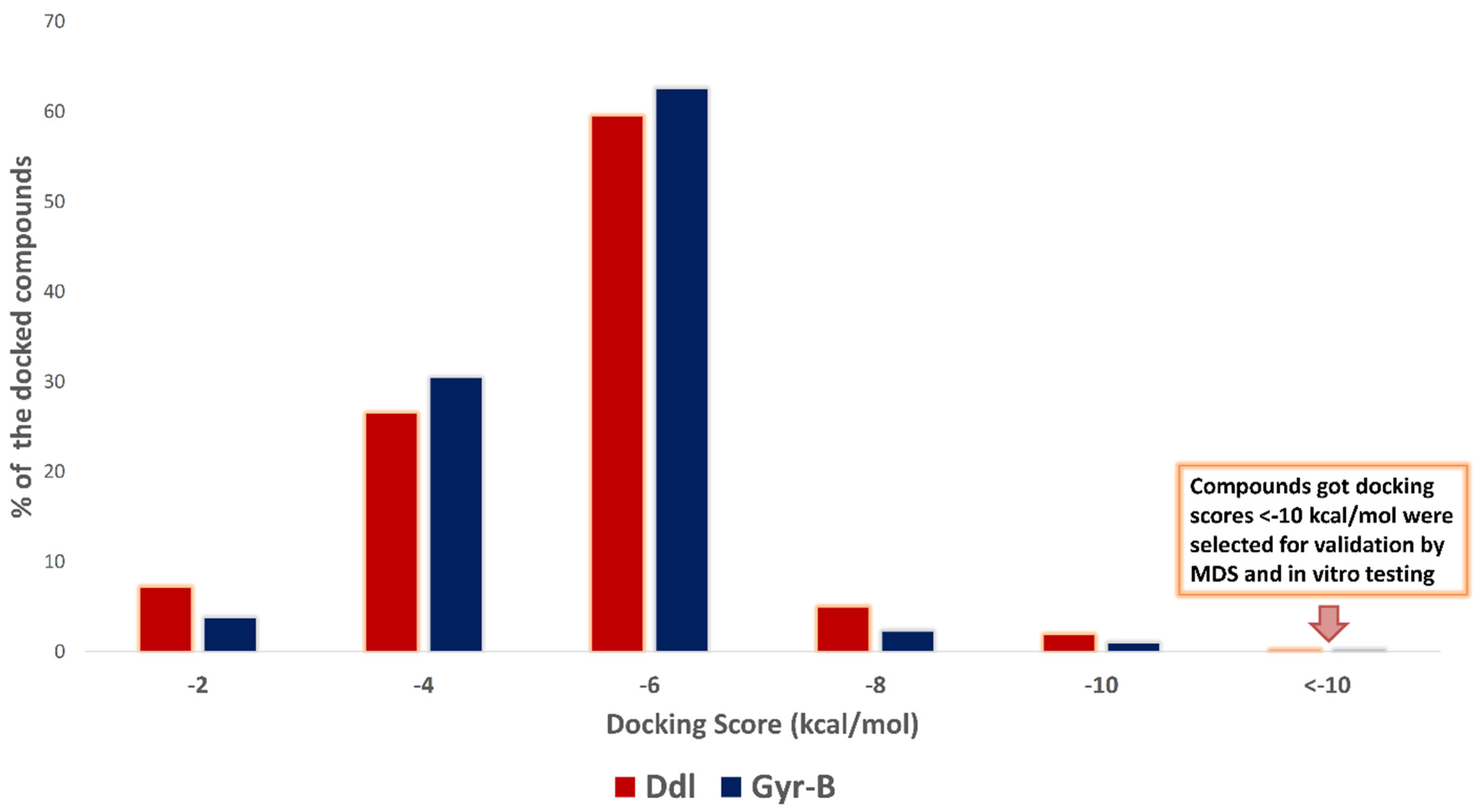
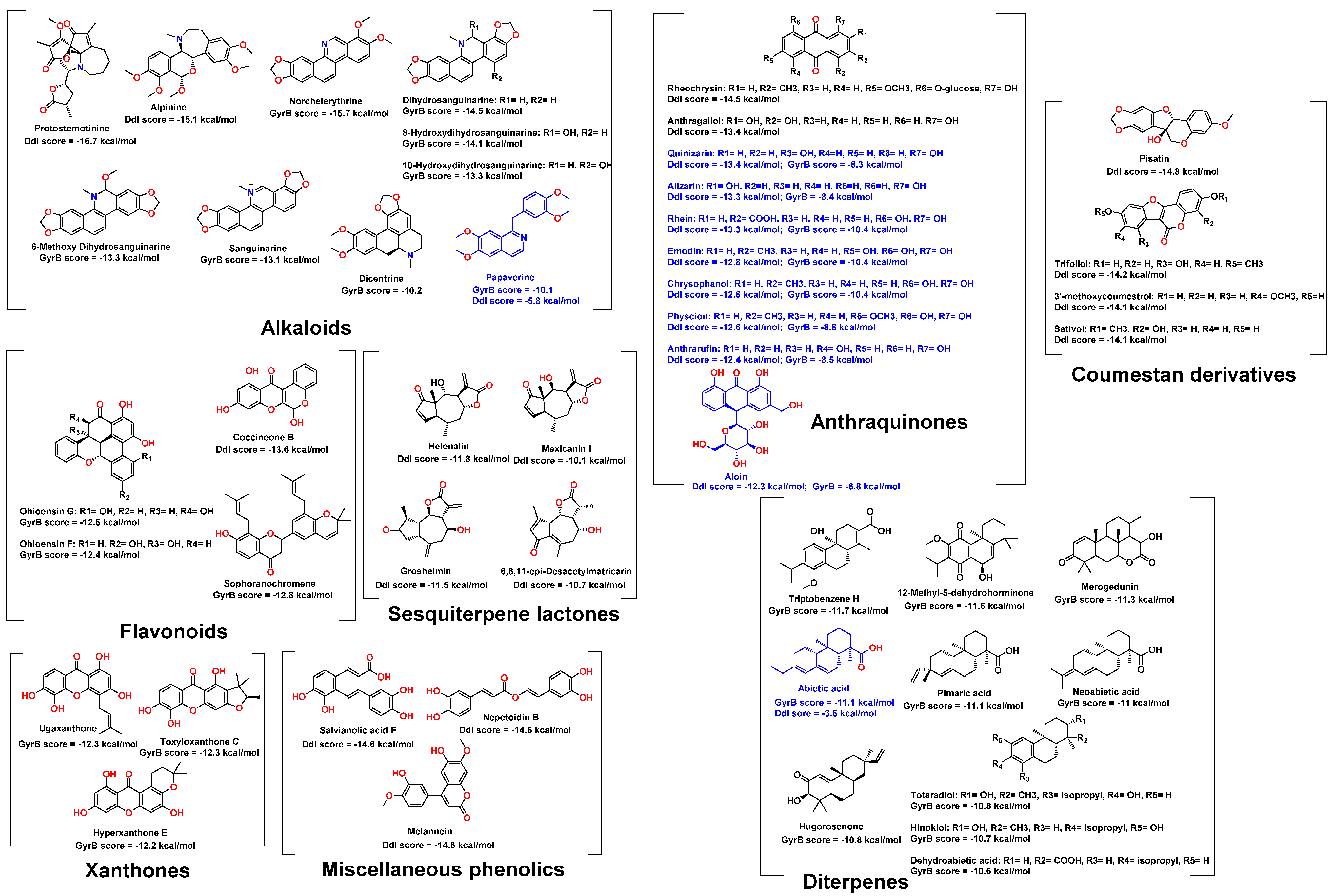
2.1. Molecular Docking-Based Virtual Screening
2.2. In Vitro Validation
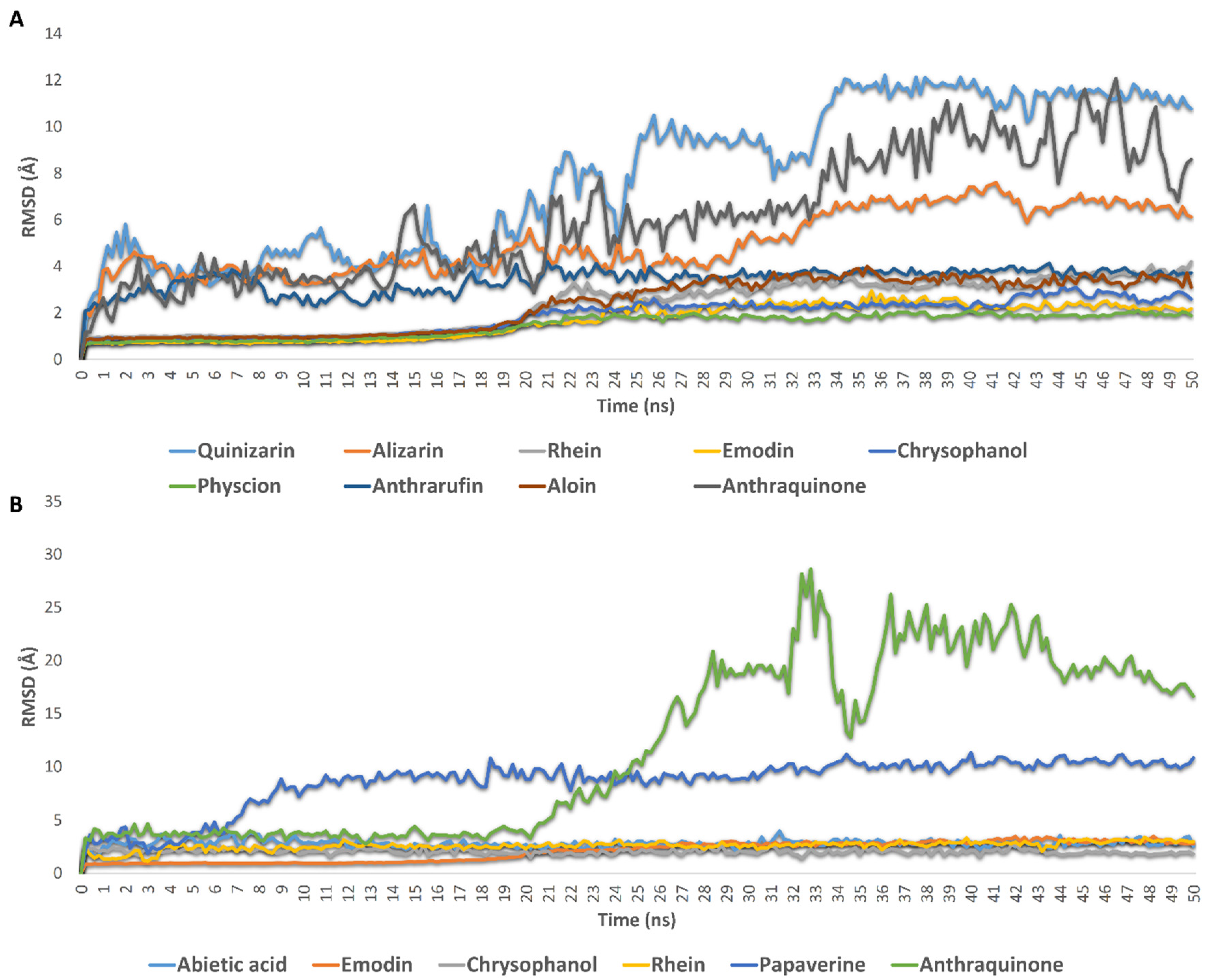
2.3. Molecular Modeling
3. Materials and Methods
3.1. Preparation of E. coli Target Proteins and the Compounds Dataset
3.2. Docking-Based Virtual Screening
3.3. Drug-Likeness Analysis
3.4. Molecular Dynamic Simulation and Binding Free Energy Estimation
3.5. Compounds Preparation
3.6. In Vitro Testing
3.6.1. Enzyme Assay
3.6.2. Antibacterial Evaluation
3.6.3. Cellular Toxicity against Normal Fibroblasts WI-38
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sayed, A.M.; Alhadrami, H.A.; El-Hawary, S.S.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Abdelmohsen, U.R.; Bakeer, W. Discovery of Two Brominated Oxindole Alkaloids as Staphylococcal DNA Gyrase and Pyruvate Kinase Inhibitors via Inverse Virtual Screening. Microorganisms 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culp, E.J.; Waglechner, N.; Wang, W.; Fiebig-Comyn, A.A.; Hsu, Y.-P.; Koteva, K.; Sychantha, D.; Coombes, B.K.; Van Nieuwenhze, M.S.; Brun, Y.V.; et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 2020, 578, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Kuru, E.; Radkov, A.; Meng, X.; Egan, A.; Alvarez, L.; Dowson, A.; Booher, G.; Breukink, E.; Roper, D.I.; Cava, F.; et al. Mechanisms of Incorporation for D-Amino Acid Probes That Target Peptidoglycan Biosynthesis. ACS Chem. Biol. 2019, 14, 2745–2756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovac, A.; Konc, J.; Vehar, B.; Bostock, J.M.; Chopra, I.; Janezic, D.; Gobec, S. Discovery of New Inhibitors ofd-Alanine:d-Alanine Ligase by Structure-Based Virtual Screening. J. Med. Chem. 2008, 51, 7442–7448. [Google Scholar] [CrossRef] [PubMed]
- Pujol, M.; Miró, J.-M.; Shaw, E.; Aguado, J.-M.; San-Juan, R.; Puig-Asensio, M.; Pigrau, C.; Calbo, E.; Montejo, M.; Rodriguez-Álvarez, R.; et al. Daptomycin Plus Fosfomycin Versus Daptomycin Alone for Methicillin-resistant Staphylococcus aureus Bacteremia and Endocarditis: A Randomized Clinical Trial. Clin. Infect. Dis. 2020, 72, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef] [Green Version]
- Kouidmi, I.; Levesque, R.C.; Paradis-Bleau, C. The biology of Mur ligases as an antibacterial target. Mol. Microbiol. 2014, 94, 242–253. [Google Scholar] [CrossRef]
- Janupally, R.; Medepi, B.; Brindha, P.; Suryadevara, P.; Jeankumar, V.U.; Kulkarni, P.; Yogeeswari, P.; Sriram, D. Design and Biological Evaluation of Furan/Pyrrole/Thiophene-2-carboxamide Derivatives as Efficient DNA GyraseB Inhibitors of Staphylococcus aureus. Chem. Biol. Drug Des. 2015, 86, 918–925. [Google Scholar] [CrossRef]
- Dighe, S.N.; Collet, T.A. Recent advances in DNA gyrase-targeted antimicrobial agents. Eur. J. Med. Chem. 2020, 199, 112326. [Google Scholar] [CrossRef] [PubMed]
- Tari, L.W.; Trzoss, M.; Bensen, D.C.; Li, X.; Chen, Z.; Lam, T.; Zhang, J.; Creighton, C.J.; Cunningham, M.L.; Kwan, B.; et al. Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorg. Med. Chem. Lett. 2012, 23, 1529–1536. [Google Scholar] [CrossRef]
- Tomasšičc, T.; Katsamakas, S.; Hodnik, Z.; Ilasš, J.; Brvar, M.; Solmajer, T.; Montalvão, S.; Paãvi, T.; Banjanac, M.; Ergovi´c, G.; et al. Discovery of 4, 5, 6, 7-tetrahydrobenzo [1, 2-d] thiazoles as novel DNA gyrase inhibitors targeting the ATP-binding site. J. Med. Chem. 2015, 58, 5501–5521. [Google Scholar] [CrossRef] [PubMed]
- Polshettiwar, V.; Varma, R.S. Aqueous microwave chemistry: A clean and green synthetic tool for rapid drug discovery. Chem. Soc. Rev. 2008, 37, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Shahzad, A.; Sobia, F.; Sahai, A.; Tripathi, T.; Singh, A.; Khan, H.M. Plant natural products as a potential source for antibacterial agents: Recent trends. Anti-Infect. Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Infect. Agents) 2009, 8, 211–225. [Google Scholar] [CrossRef]
- Porras, G.; Chassagne, F.; Lyles, J.T.; Marquez, L.; Dettweiler, M.; Salam, A.M.; Samarakoon, T.; Shabih, S.; Farrokhi, D.R.; Quave, C.L. Ethnobotany and the Role of Plant Natural Products in Antibiotic Drug Discovery. Chem. Rev. 2020, 121, 3495–3560. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Zofou, D.; Babiaka, S.B.; Meudom, R.; Scharfe, M.; Lifongo, L.L.; Mbah, J.A.; Mbaze, L.M.; Sippl, W.; Efange, S.M.N. AfroDb: A Select Highly Potent and Diverse Natural Product Library from African Medicinal Plants. PLoS ONE 2013, 8, e78085. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Moews, P.C.; Walsh, C.T.; Knox, J.R. Vancomycin resistance: Structure of D-alanine: D-alanine ligase at 2.3 Å resolution. Science 1994, 266, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Ushiyama, F.; Amada, H.; Takeuchi, T.; Tanaka-Yamamoto, N.; Kanazawa, H.; Nakano, K.; Mima, M.; Masuko, A.; Takata, I.; Hitaka, K.; et al. Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation. ACS Omega 2020, 5, 10145–10159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, S.T.; Tam, N.M.; Quan, P.M.; Nguyen, T.H. Benchmark of Popular Free Energy Approaches Revealing the Inhibitors Binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021, 61, 2302–2312. [Google Scholar] [CrossRef] [PubMed]
- Nong, W.J.; Chen, X.P.; Liang, J.Z.; Wang, L.L.; Tong, Z.F.; Huang, K.L.; Wu, R.; Xie, Q.R.; Jia, Y.H.; Li, K.X. Isolation and Characterization of Abietic Acid. In Advanced Materials Research; Trans Tech Publications Ltd.: Freienbach, Switzerland, 2014; Volume 887, pp. 551–556. [Google Scholar]
- Pi, L.; Hu, F.; Shi, Z. Determination of papaverine in seeds of Papaver somniferum L. and soup of chafing dish by high performance liquid chromatography-fluorescence detection. Chin. J. Chromatogr. 2005, 23, 639–641. [Google Scholar]
- Chalothorn, T.; Rukachaisirikul, V.; Phongpaichit, S.; Pannara, S.; Tansakul, C. Synthesis and antibacterial activity of emodin and its derivatives against methicillin-resistant Staphylococcus aureus. Tetrahedron Lett. 2019, 60, 151004. [Google Scholar] [CrossRef]
- Ubbink-Kok, T.R.E.E.E.S.; Anderson, J.A.; Konings, W.N. Inhibition of electron transfer and uncoupling effects by emodin and emodinanthrone in Escherichia coli. Antimicrob. Agents Chemother. 1986, 30, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Soomro, N.A.; Wu, Q.; Amur, S.A.; Liang, H.; Rahman, A.U.; Yuan, Q.; Wei, Y. Natural drug physcion encapsulated zeolitic imidazolate framework, and their application as antimicrobial agent. Colloids Surfaces B Biointerfaces 2019, 182, 110364. [Google Scholar] [CrossRef] [PubMed]
- Prateeksha, P.; Bajpai, R.; Rao, C.V.; Upreti, D.K.; Barik, S.K.; Singh, B.N. Chrysophanol-Functionalized Silver Nanoparticles for Anti-Adhesive and Anti-Biofouling Coatings to Prevent Urinary Catheter-Associated Infections. ACS Appl. Nano Mater. 2021, 4, 1512–1528. [Google Scholar] [CrossRef]
- Ito, Y.; Ito, T.; Yamashiro, K.; Mineshiba, F.; Hirai, K.; Omori, K.; Yamamoto, T.; Takashiba, S. Antimicrobial and antibiofilm effects of abietic acid on cariogenic Streptococcus mutans. Odontology 2019, 108, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Helfenstein, A.; Vahermo, M.; Nawrot, D.A.; Demirci, F.; Iscan, G.; Krogerus, S.; Yli-Kauhaluoma, J.; Moreira, V.M.; Tammela, P. Antibacterial profiling of abietane-type diterpenoids. Bioorg. Med. Chem. 2017, 25, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Batson, S.; De Chiara, C.; Majce, V.; Lloyd, A.J.; Gobec, S.; Rea, D.; Fülöp, V.; Thoroughgood, C.W.; Simmons, K.; Dowson, C.; et al. Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine. Nat. Commun. 2017, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology; Humana Press: New York, NY, USA, 2015; pp. 243–250. [Google Scholar]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tutone, M.; Perricone, U.; Almerico, A.M. Conf-VLKA: A structure-based revisitation of the Virtual Lock-and-key Approach. J. Mol. Graph. Model. 2017, 71, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Alhadrami, H.A.; Sayed, A.M.; Melebari, S.A.; Khogeer, A.A.; Abdulaal, W.H.; Al-Fageeh, M.B.; Rateb, M.E. Targeting allosteric sites of human aromatase: A comprehensive in-silico and in-vitro workflow to find potential plant-based anti-breast cancer therapeutics. J. Enzym. Inhib. Med. Chem. 2021, 36, 1334–1345. [Google Scholar] [CrossRef]
- Ribeiro, J.V.; Bernardi, R.C.; Rudack, T.; Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Schulten, K. QwikMD—Integrative molecu-lar dynamics toolkit for novices and experts. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Caflisch, A. Molecular dynamics in drug design. Eur. J. Med. Chem. 2015, 91, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Gecibesler, I.H.; Disli, F.; Bayindir, S.; Toprak, M.; Tufekci, A.R.; Yaglıoglu, A.S.; Altun, M.; Kocak, A.; Demirtas, I.; Adem, S. The isolation of secondary metabolites from Rheum ribes L. and the synthesis of new semi-synthetic anthraquinones: Isolation, synthesis and biological activity. Food Chem. 2021, 342, 128378. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, Y.K.; Lee, D.-S.; Yoo, J.-E.; Shin, M.-S.; Yamabe, N.; Kim, S.-N.; Lee, S.; Kim, K.H.; Lee, H.-J.; et al. Abietic acid isolated from pine resin (Resina Pini) enhances angiogenesis in HUVECs and accelerates cutaneous wound healing in mice. J. Ethnopharmacol. 2017, 203, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
| Compound | Docking Score | ΔG | IC50 (µM) | Ki (µM) | MIC (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ddl-B | Gyr-B | Ddl-B | Gyr-B | Ddl-B | Gyr-B | Ddl-B | Gyr-B | E. coli (MDR) a | E. coli (MDR) b | MRSA | VRSA | |
| Quinizarin | −13.4 | −8.3 | −9.1 | −5.5 | 515 ± 8.3 | 38.8 ± 2.3 | 230 ± 8.3 | 17.9 ± 1.4 | 64 | 64 | 64 | 16 |
| Alizarin | −13.3 | −8.4 | −8.5 | −6.5 | 468 ± 6.5 | 17.9 ± 1.2 | 220 ± 4.3 | 10.4 ± 1.1 | 64 | >64 | 64 | >64 |
| Rhein | −13.3 | −10.4 | −8.4 | −9.1 | 281 ± 6.1 | 11.5 ± 1.2 | 133 ± 2.3 | 7.33 ± 0.9 | 8 | 2 | >64 | 8 |
| Emodin | −12.8 | −10.4 | −9.6 | −9.9 | 216 ± 5.6 | 0.81 ± 0.3 | 102 ± 4.8 | 0.22 ± 0.1 | 2 | 2 | 2 | 2 |
| Chrysophanol | −12.6 | −10.4 | −9.1 | −9.3 | 236 ± 8.9 | 1.5 ± 0.5 | 105 ± 4.9 | 0.83 ± 0.2 | 4 | 4 | >64 | 4 |
| Physcion | −12.6 | −8.8 | −8.3 | −6.9 | 244 ± 7.2 | 10.9 ± 0.5 | 119 ± 9.1 | 6.6 ± 0.9 | 2 | 2 | 2 | 2 |
| Anthrarufin | −12.4 | −8.5 | −8.1 | −7.7 | 350 ± 7.4 | 6.93 ± 1.6 | 345 ± 10.1 | 4.1 ± 1.6 | 2 | 2 | 2 | >64 |
| Aloin | −12.3 | −6.8 | −8.7 | −5.2 | 927 ± 7.4 | 22.4 ± 2.1 | 529 ± 12.3 | 10.48 ± 1.6 | >64 | >64 | >64 | >64 |
| Anthraquinone | −9.3 | −7.8 | −5.1 | −5.6 | >1000 | >100 | >1000 | >1000 | >64 | >64 | >64 | 64 |
| Abietic acid | −3.6 | −11.1 | −1.7 | −8.8 | >1000 | 1.4 ± 0.4 | >1000 | 0.77 ± 0.1 | 64 | 16 | >64 | 16 |
| Papaverine | −5.8 | −10.1 | −2.2 | −6.2 | >1000 | >100 | >1000 | >1000 | >64 | >64 | >64 | >64 |
| D-cycloserine * | −7.3 | - | −7.8 | - | 362 | - | 118 ± 11.7 | - | 16 | 16 | 32 | 32 |
| Novobiocin ** | - | −15.3 | - | −10.4 | - | 0.47 | - | - | 64 | 64 | 1 | 1 |
| Compound | CC50 (µM) |
|---|---|
| Quinizarin | 20.11 ± 0.56 |
| Alizarin | 25.89 ± 0.69 |
| Rhein | >50 |
| Emodin | 49.23 ± 1.39 |
| Chrysophanol | 48.15 ± 1.39 |
| Physcion | >50 |
| Anthrarufin | 29.67 ± 0.76 |
| Aloin | 34.74 ± 0.94 |
| Abietic acid | >50 |
| Papaverine | 36.83 ± 0.93 |
| Doxorubicin | 26.18 ± 0.61 |
| Compound | Interaction | |
|---|---|---|
| H-Bonding | Hydrophobic | |
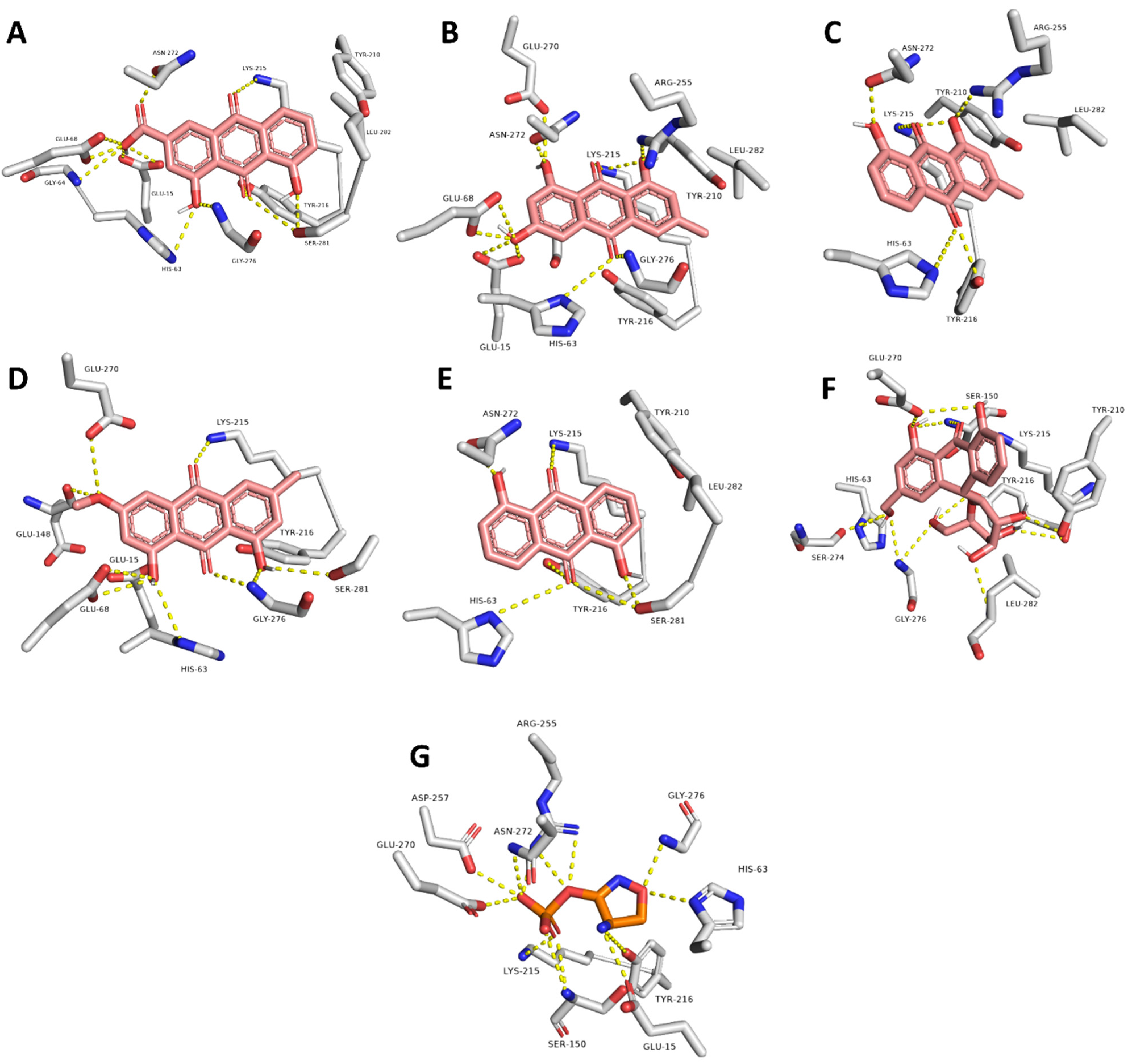
| Rhein | GLU-15, HIS-63, GLY-64, GLU-68, LYS-215, TYR-216, ASN-272, SER-281 | LYS-215, LEU-282 |
| Emodin | GLU-15, HIS-63, GLY-64, LYS-215, ARG-255, GLU-270, ASN-272, GLY-276 | LYS-215, LEU-282 |
| Chrysophanol | HIS-63, LYS-215, TYR-216, ARG-255 | LYS-215, LEU-282 |
| Physcion | GLU-15, HIS-63, GLY-64, GLU-68, GLU-148, GLY-276, SER-281 | LYS-215 |
| Anthrarufin | HIS-63, LYS-215, TYR-216, ASN-272, SER-281 | LYS-215 |
| Aloin | GLU-15, HIS-63, GLY-64, GLU-68, SER-150, LYS-215, TYR-216, ASN-272, LEU-282 | - |
| Co-crystalized ligand (D-cycloserine) | GLU-15, HIS-63, GLY-64, SER-150, ARG-255, GLU-270, ASN-272, GLY-276 | - |
| Compound | Interaction | |
|---|---|---|
| H-Bonding | Hydrophobic | |
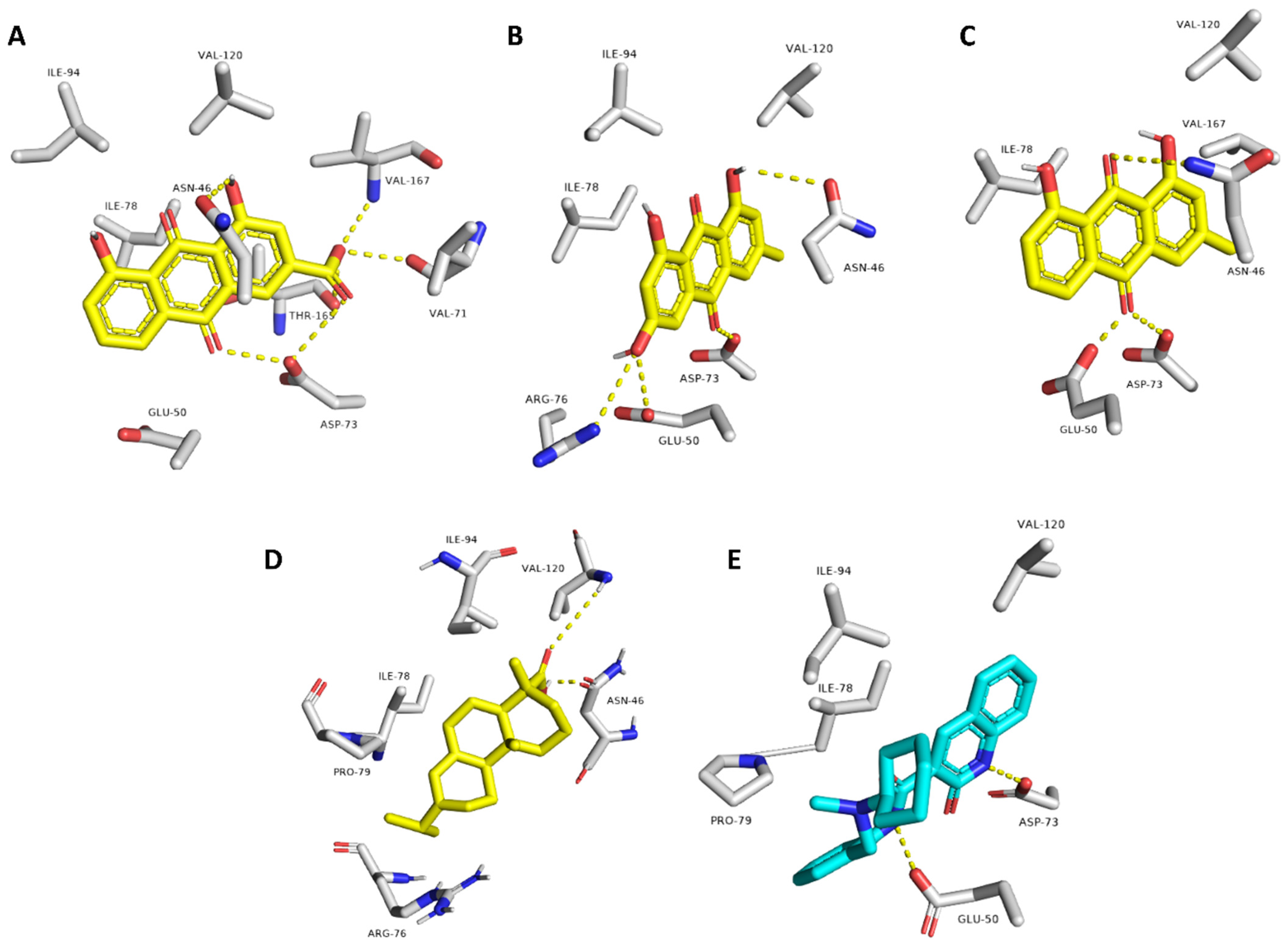
| Rhein | ASN-46, VAL-71, ASP-73, VAL-167 | ILE-78 |
| Emodin | ASN-46, GLU-50, ASP-73, ARG-76 | ILE-78 |
| Chrysophanol | ASN-46, GLU-50, ASP-73 | ILE-78 |
| Abietic acid | ASN-46, VAL-120 | ILE-78, PRO-79, ILE-94 |
| Co-crystalized ligand (2-oxo-1,2-dihydroquinoline) | GLU-50, ASP-73 | VAL-120, ILE-78, PRO-79 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhadrami, H.A.; Abdulaal, W.H.; Hassan, H.M.; Alhakamy, N.A.; Sayed, A.M. In Silico-Based Discovery of Natural Anthraquinones with Potential against Multidrug-Resistant E. coli. Pharmaceuticals 2022, 15, 86. https://doi.org/10.3390/ph15010086
Alhadrami HA, Abdulaal WH, Hassan HM, Alhakamy NA, Sayed AM. In Silico-Based Discovery of Natural Anthraquinones with Potential against Multidrug-Resistant E. coli. Pharmaceuticals. 2022; 15(1):86. https://doi.org/10.3390/ph15010086
Chicago/Turabian StyleAlhadrami, Hani A., Wesam H. Abdulaal, Hossam M. Hassan, Nabil A. Alhakamy, and Ahmed M. Sayed. 2022. "In Silico-Based Discovery of Natural Anthraquinones with Potential against Multidrug-Resistant E. coli" Pharmaceuticals 15, no. 1: 86. https://doi.org/10.3390/ph15010086
APA StyleAlhadrami, H. A., Abdulaal, W. H., Hassan, H. M., Alhakamy, N. A., & Sayed, A. M. (2022). In Silico-Based Discovery of Natural Anthraquinones with Potential against Multidrug-Resistant E. coli. Pharmaceuticals, 15(1), 86. https://doi.org/10.3390/ph15010086








