Testing Mitochondrial-Targeted Drugs in iPSC-RPE from Patients with Age-Related Macular Degeneration
Abstract
:1. Introduction
2. Results
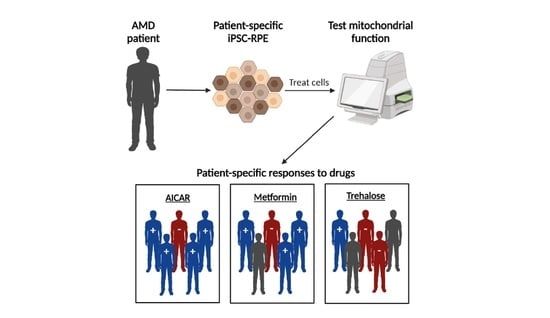
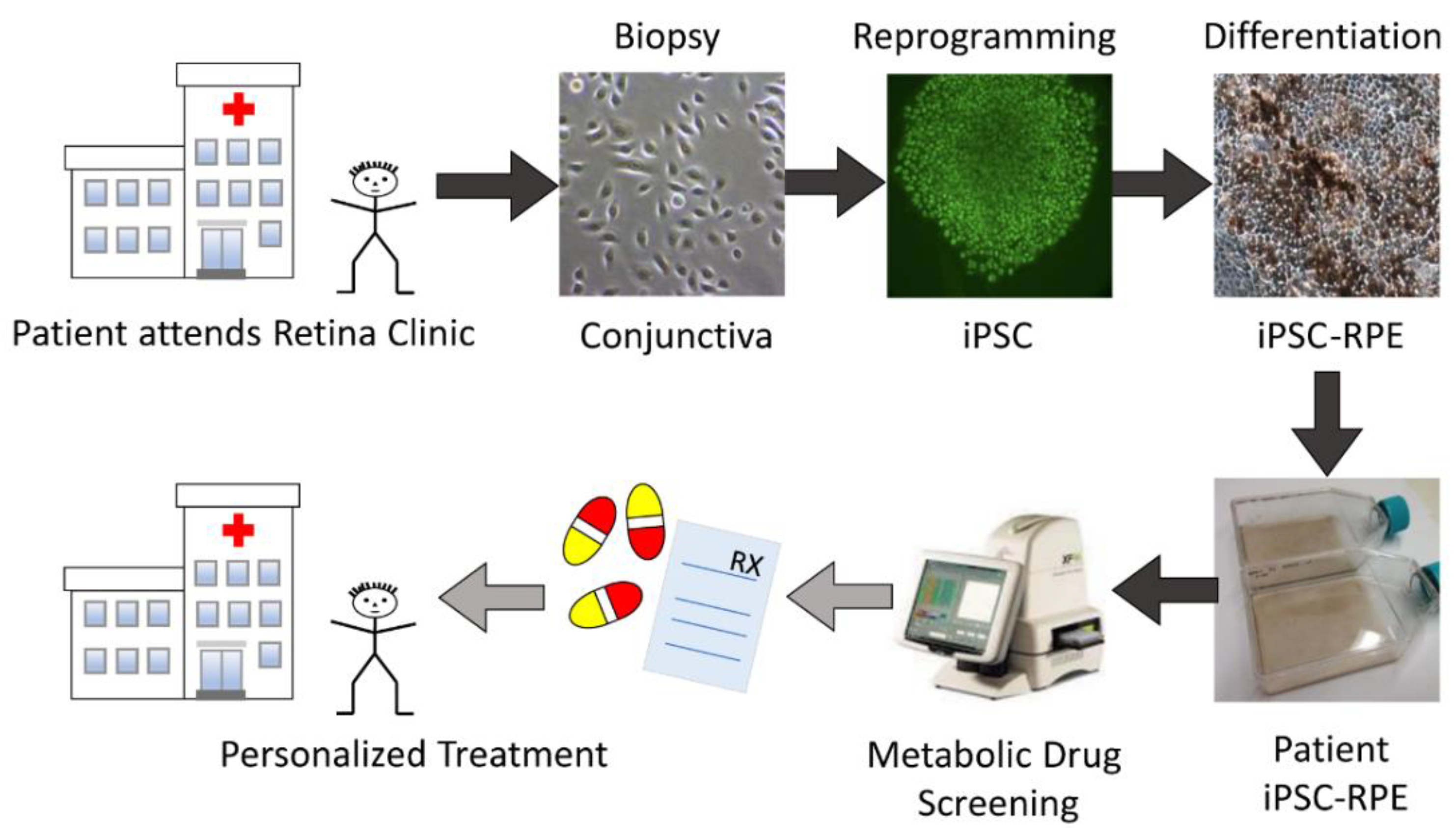
2.1. Proposed Drug Screening Approach to Determine Patient-Specific Treatment for AMD
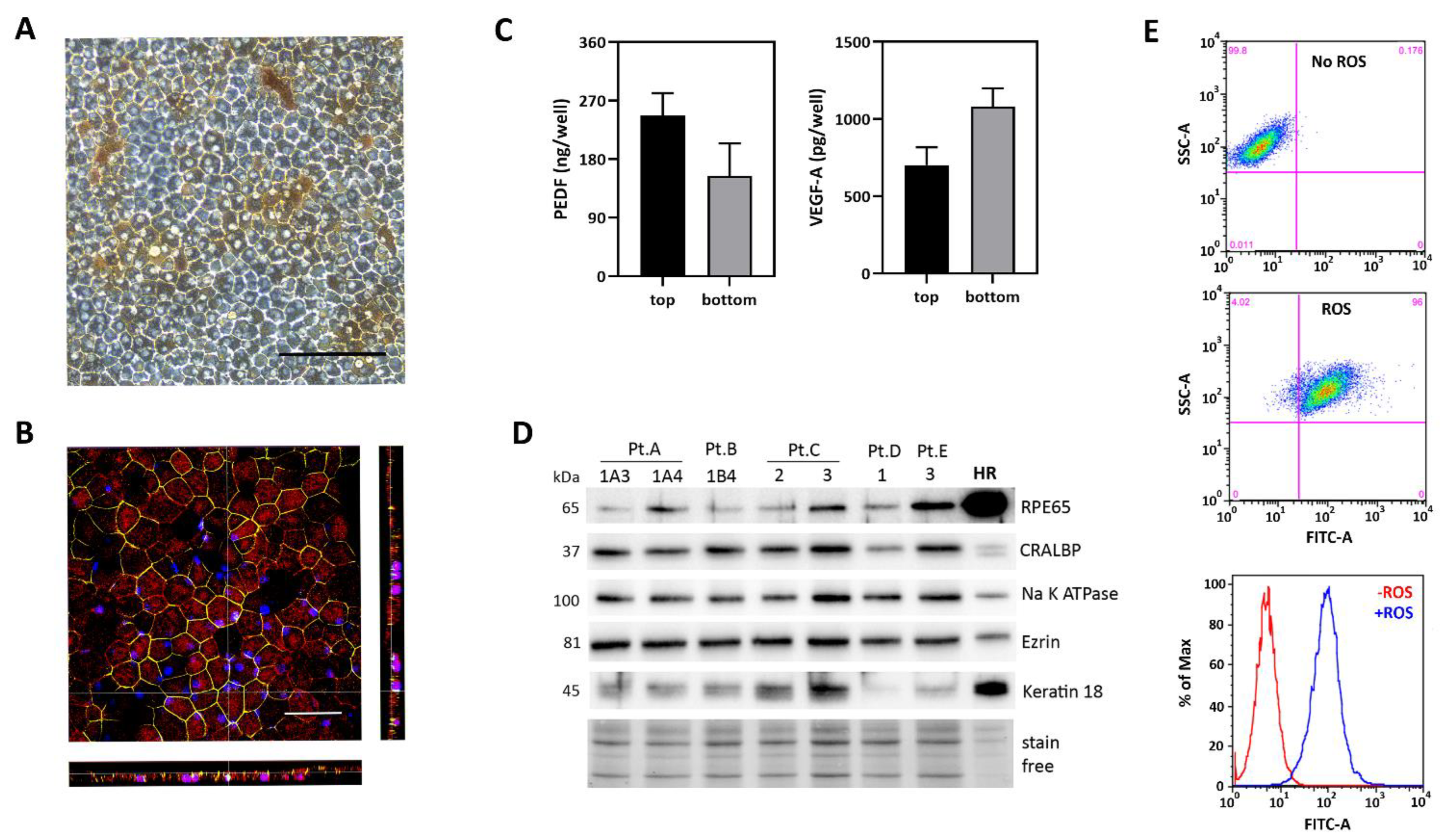
2.2. Characterization of iPSC-RPE Cultures Derived from AMD Patients
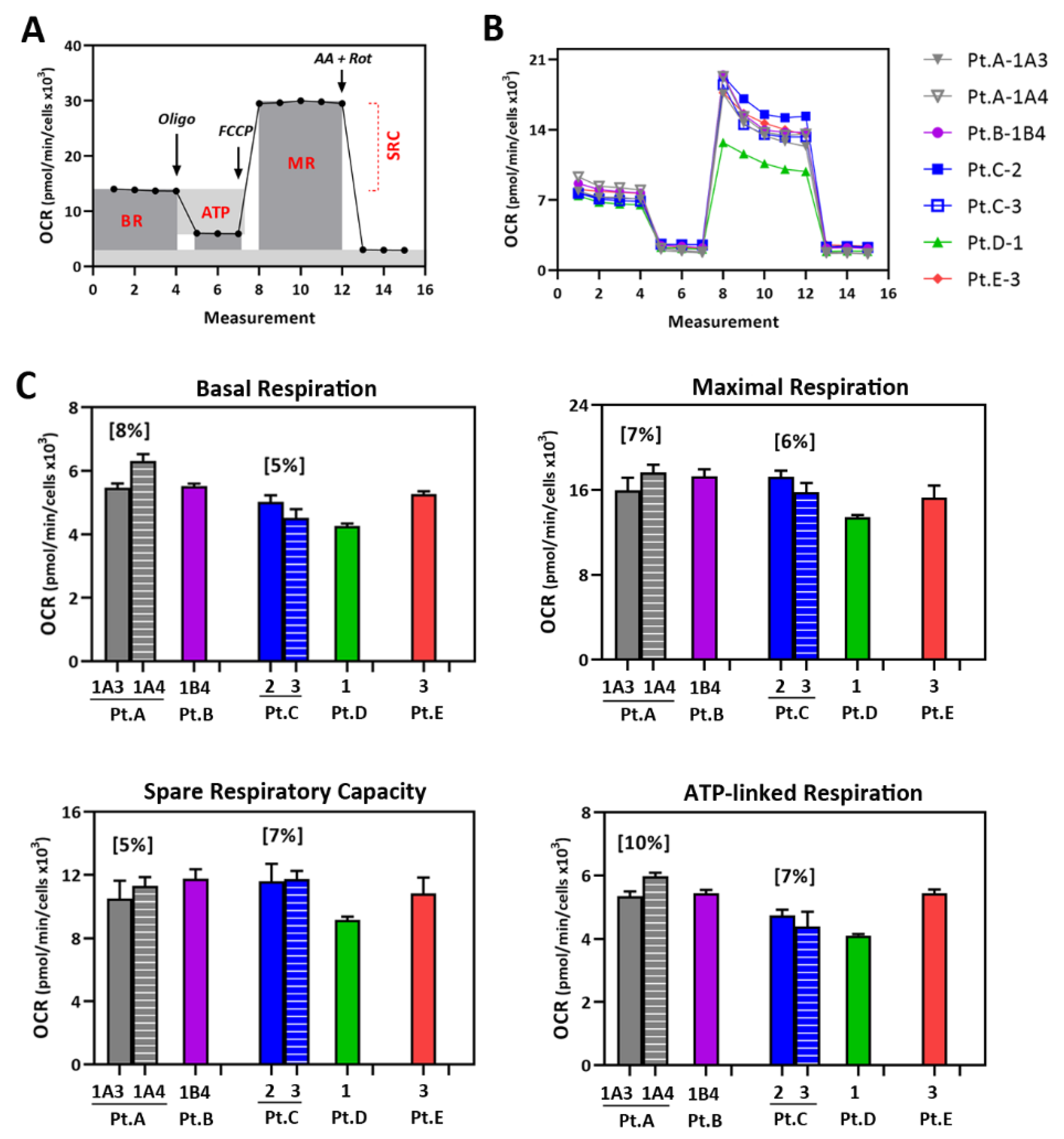
2.3. Mt Testing of iPSC-RPE from Individual AMD Patients
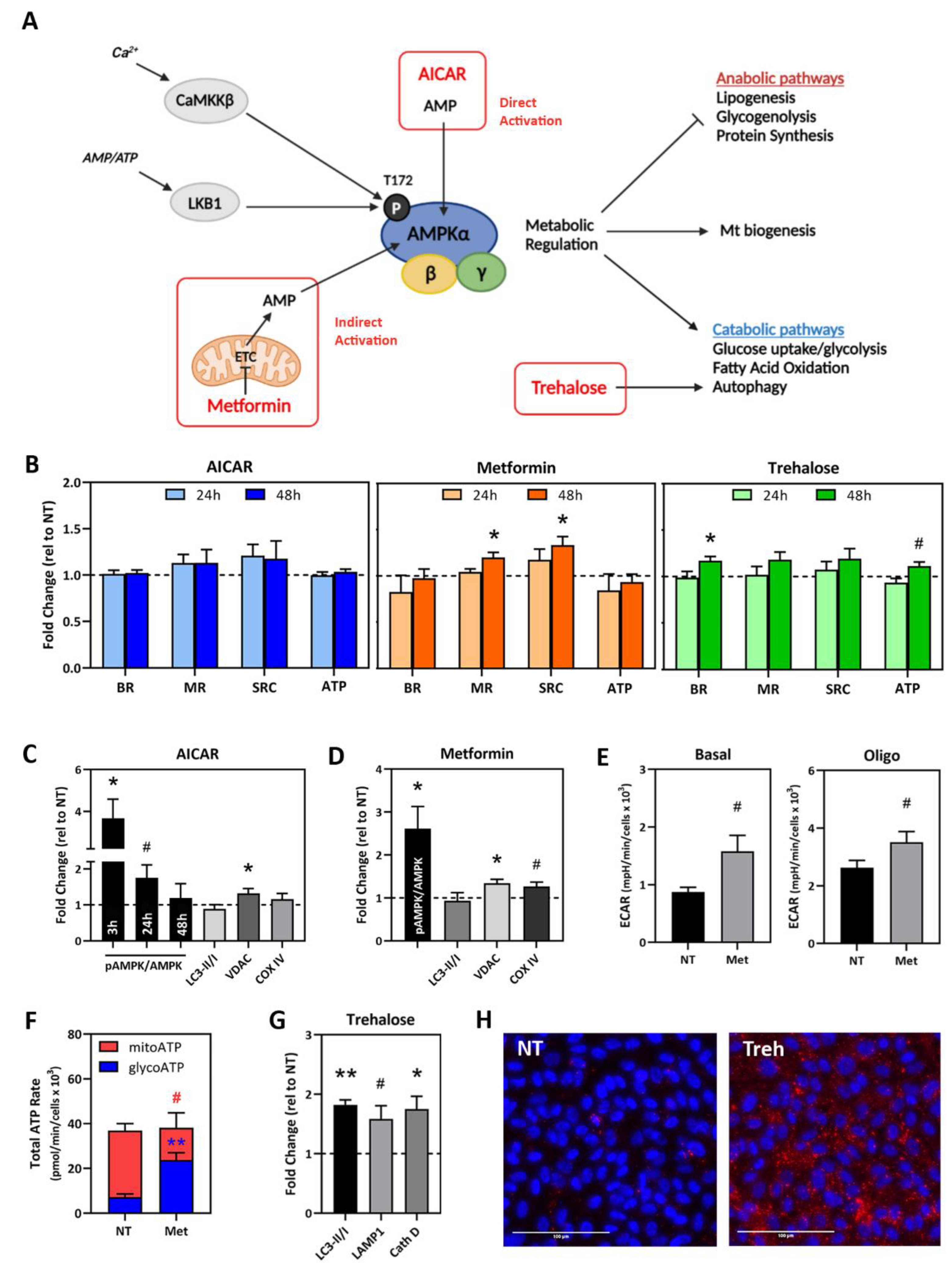
2.4. Testing Compounds That Target Mt Activity and Homeostasis in AMD Donor iPSC-RPE
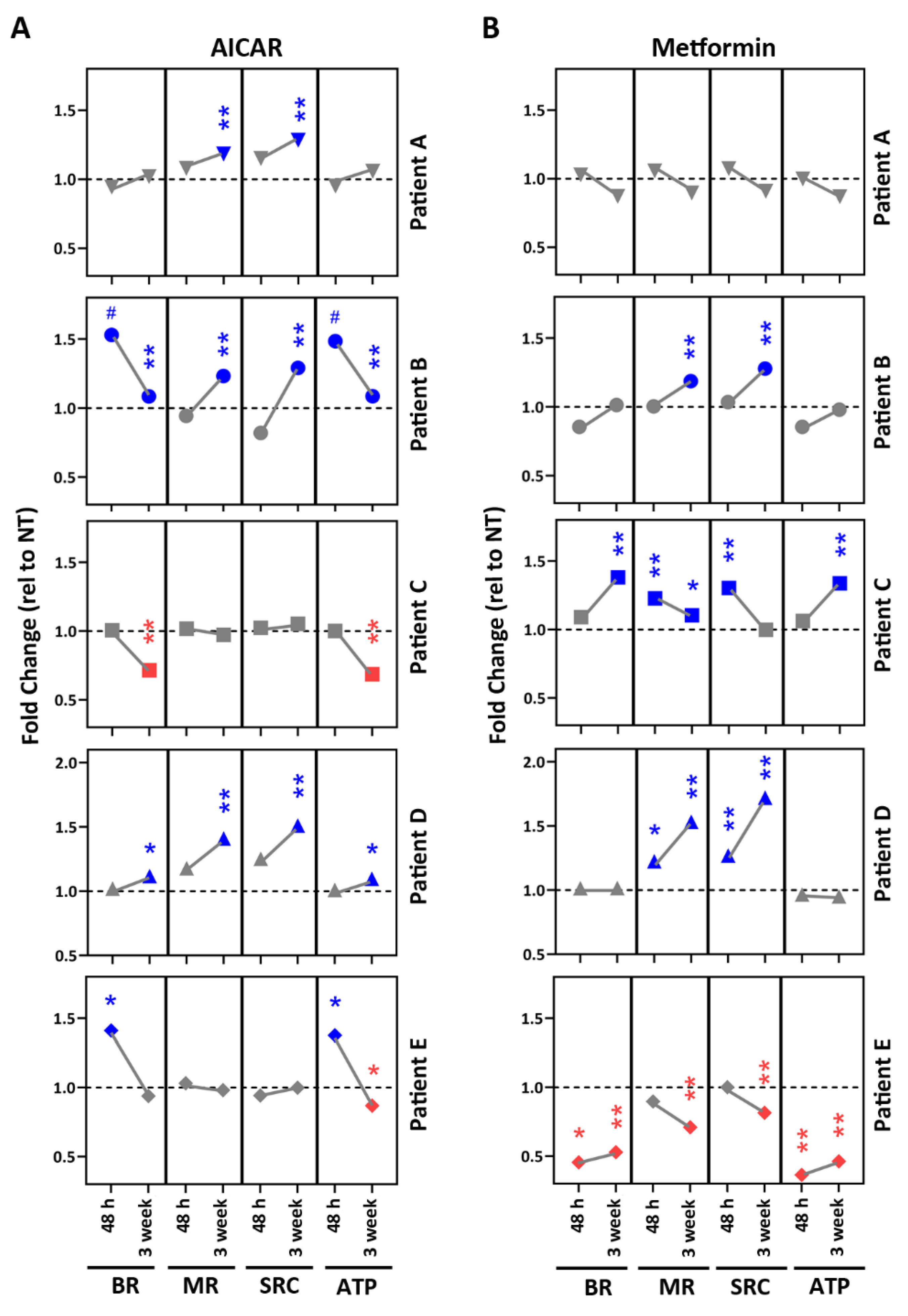
2.5. Drug Testing in Patient-Specific iPSC-RPE Using AMPK Activators
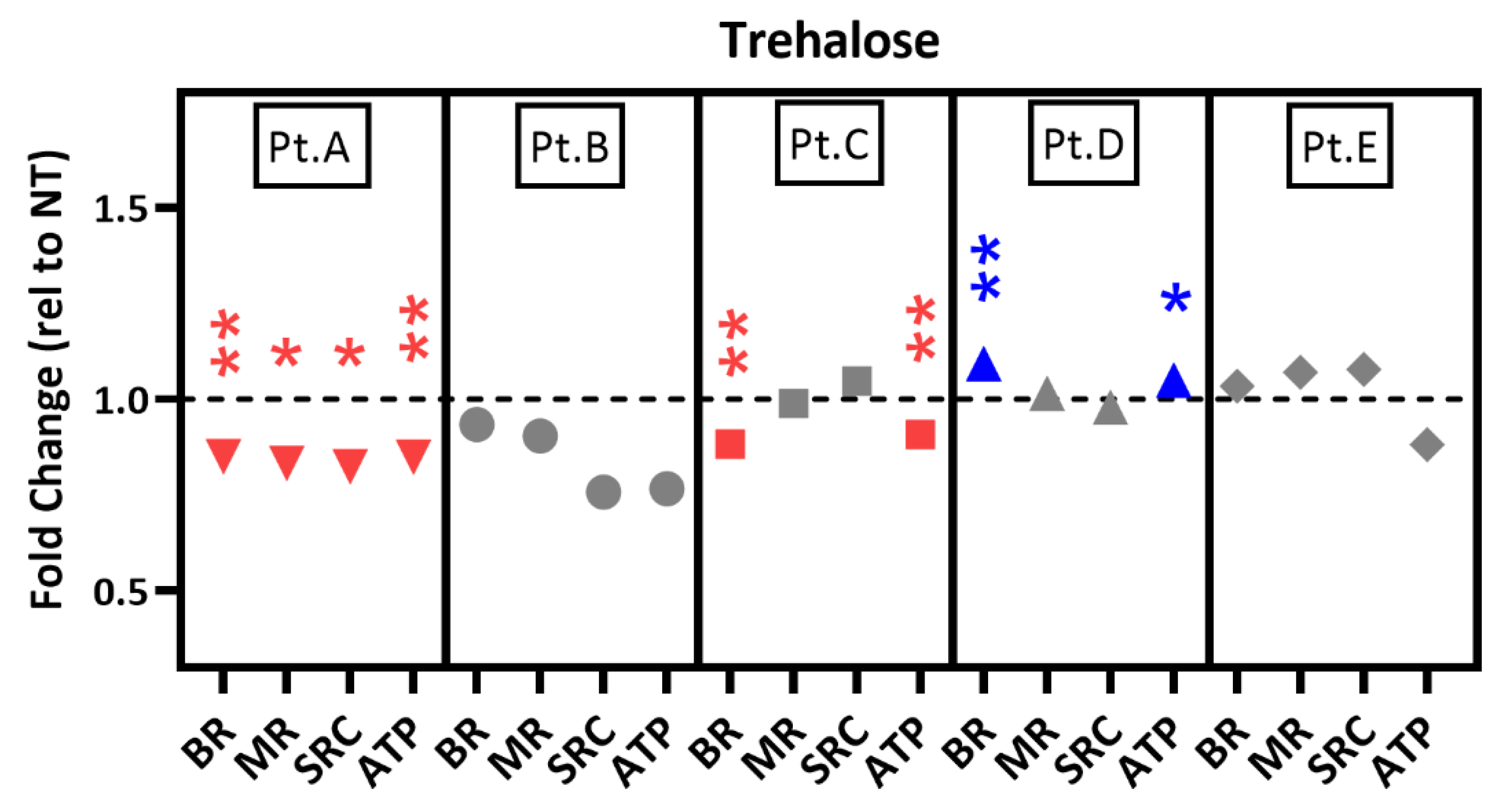
2.6. Drug Testing in Patient-Specific iPSC-RPE Using an Autophagy Inducer
3. Discussion
4. Materials and Methods
4.1. Conjunctiva Biopsies from AMD Patients
4.2. Culturing iPSC-RPE Cells
4.3. Genotyping
4.4. Immunofluorescence
4.5. Enzyme-Linked Immunosorbent Assay (ELISA)
4.6. Western Blotting
4.7. Phagocytosis of Outer Segments (OS)
4.8. Measuring RPE Mt Function Using the Cell Mito Stress Test (CMST)
4.9. Measuring ATP Production Rates Using Real-Time ATP Rate Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [Green Version]
- Feher, J.; Kovacs, I.; Artico, M.; Cavallotti, C.; Papale, A.; Balacco Gabrieli, C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging 2006, 27, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Nordgaard, C.L.; Karunadharma, P.P.; Feng, X.; Olsen, T.W.; Ferrington, D.A. Mitochondrial proteomics of the retinal pigment epithelium in age-related macular degeneration. investig. Ophthalmol. Vis. Sci. 2008, 49, 2848–2855. [Google Scholar] [CrossRef] [PubMed]
- Karunadharma, P.P.; Nordgaard, C.L.; Olsen, T.W.; Ferrington, D.A. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. investig. Ophthalmol. Vis. Sci. 2010, 51, 5470–5479. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Xu, H.; Liang, F.Q.; Liang, H.; Gupta, P.; Havey, A.N.; Boulton, M.E.; Godley, B.F. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. investig. Ophthalmol. Vis. Sci. 2011, 52, 3251–3259. [Google Scholar] [CrossRef] [PubMed]
- Terluk, M.R.; Kapphahn, R.J.; Soukup, L.M.; Gong, H.; Gallardo, C.; Montezuma, S.R.; Ferrington, D.A. Investigating mitochondria as a target for treating age-related macular degeneration. J. Neurosci. 2015, 35, 7304–7311. [Google Scholar] [CrossRef] [Green Version]
- Ferrington, D.A.; Ebeling, M.C.; Kapphahn, R.J.; Terluk, M.R.; Fisher, C.R.; Polanco, J.R.; Roehrich, H.; Leary, M.M.; Geng, Z.; Dutton, J.R.; et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age-related macular degeneration. Redox Biol. 2017, 13, 255–265. [Google Scholar] [CrossRef]
- Golestaneh, N.; Chu, Y.; Xiao, Y.-Y.; Stoleru, G.L.; Theos, A.C. Dysfunctional autophagy in RPE, a contributing factor in age-related macular degeneration. Cell Death Dis. 2017, 8, e2537. [Google Scholar] [CrossRef]
- Kanow, M.A.; Giarmarco, M.M.; Jankowski, C.S.; Tsantilas, K.; Engel, A.L.; Du, J.; Linton, J.D.; Farnsworth, C.C.; Sloat, S.R.; Rountree, A.; et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife 2017, 6, e28899. [Google Scholar] [CrossRef]
- Age-Related Eye Disease Study 2 Research Group. Lutein + zeaxanthin and omega-3 fatty acids for age-related maular degeneration: The Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA 2013, 309, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Chew, E.Y.; Clemons, T.E.; Agron, E.; Sperduto, R.D.; Sangiovanni, J.P.; Kurinij, N.; Davis, M.D.; Age-Related Eye Disease Study Research Group. Long-Term effects of vitamins C and E, β-carotene, and zinc on age-related macular degeneration: AREDS report no. 35. Ophthalmology 2013, 120, 1604–1611.e4. [Google Scholar] [CrossRef] [Green Version]
- Ebeling, M.C.; Polanco, J.R.; Qu, J.; Tu, C.; Montezuma, S.R.; Ferrington, D.A. Improving retinal mitochondrial function as a treatment for age-related macular degeneration. Redox Biol. 2020, 34, 101552. [Google Scholar] [CrossRef] [PubMed]
- Schafer, N.; Wolf, H.N.; Enzbrenner, A.; Schikora, J.; Reichenthaler, M.; Enzmann, V.; Pauly, D. Properdin modulates complement component production in stressed human primary retinal pigment epithelium cells. Antioxidants 2020, 9, 793. [Google Scholar] [CrossRef]
- Saini, J.S.; Corneo, B.; Miller, J.D.; Kiehl, T.R.; Wang, Q.; Boles, N.C.; Blenkinsop, T.A.; Stern, J.H.; Temple, S. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell 2017, 20, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Chang, W.C.; Hung, K.H.; Yang, D.M.; Cheng, Y.H.; Liao, Y.W.; Woung, L.C.; Tsai, C.Y.; Hsu, C.C.; Lin, T.C.; et al. The generation of induced pluripotent stem cells for macular degeneration as a drug screening platform: Identification of curcumin as protective agent for retinal pigment epithelial cells against oxidative stress. Front. Aging Neurosci. 2014, 6, 191. [Google Scholar] [CrossRef] [Green Version]
- Troung, V.; Viken, K.; Geng, Z.; Barkan, S.; Johnson, B.; Ebeling, M.C.; Montezuma, S.R.; Ferrington, D.A.; Dutton, J.R. Automating human induced pluripotent stem cell culture and differentiation of iPSC-derived retinal pigment epithelium for personalized drug testing. SLAS Technol. 2021, 26, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Swaroop, A.; Chew, E.Y.; Rickman, C.B.; Abecasis, G.R. Unraveling a multifactorial late-onset disease: From genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu. Rev. Genom. Hum. Genet. 2009, 10, 19–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellit, M.S.; Barbar, L.; Tesar, P.J. Drug screening for human genetic diseases using iPSC models. Hum. Mol. Genet. 2018, 27, R89–R98. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Myers, R.; Li, Y.; Chen, Y.; Shen, X.; Fenyk-Melody, J.; Wu, M.; Ventre, J.; Doebber, T.; Fujii, N.; et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. investig. 2001, 108, 1167–1174. [Google Scholar] [CrossRef]
- Ghavami, S.; Shojaei, S.; Yeganeh, B.; Ande, S.R.; Jangamreddy, J.R.; Mehrpour, M.; Christoffersson, J.; Chaabane, W.; Moghadam, A.R.; Kashani, H.H.; et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog. Neurobiol. 2014, 112, 24–49. [Google Scholar] [CrossRef] [Green Version]
- Geng, Z.; Walsh, P.J.; Troung, V.; Hill, C.; Ebeling, M.; Kapphahn, R.J.; Montezuma, S.R.; Yuan, C.; Roehrich, H.; Ferrington, D.A.; et al. Generation of retinal pigmented epithelium from iPSCs derived from the conjunctiva of donors with and without age-related macular degeneration. PLoS ONE 2017, 12, e0173575. [Google Scholar] [CrossRef]
- Ebeling, M.C.; Geng, Z.; Kapphahn, R.J.; Roehrich, H.; Montezuma, S.R.; Dutton, J.R.; Ferrington, D.A. Impaired mitochondrial function in iPSC-retinal pigment epithelium with complement factor H polymorphism for age-related macular degeneration. Cells 2021, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Hallam, D.; Collin, J.; Bojic, S.; Chichagova, V.; Buskin, A.; Xu, Y.; Lafage, L.; Otten, E.G.; Anyfantis, G.; Mellough, C.; et al. An induced pluripotent stem cell patient specific model of complement factor H (Y402H) polymorphism displays characteristic features of age-related macular degeneration and indicates a beneficial role for UV light exposure. Stem Cells 2017, 35, 2305–2320. [Google Scholar] [CrossRef] [Green Version]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrington, D.A.; Kenney, M.C.; Atilano, S.R.; Hurley, J.B.; Brown, E.E.; Ash, J.D. Mitochondria: The retina’s Achilles’ heel in AMD. Adv. Exp. Med. Biol. 2021, 1256, 237–264. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, S.J.; Jo, D.H.; Park, K.S.; Kim, J.H. Blockade of mTORC-1-NOX signaling pathway inhibits TGF-Beta1-mediated senescence-like structural alterations of the retinal pigment epithelium. FASEB J. 2021, 353, e21403. [Google Scholar] [CrossRef]
- Matoba, R.; Morizane, Y.; Shiode, Y.; Hirano, M.; Doi, S.; Toshima, S.; Araki, R.; Hosogi, M.; Yonezawa, T.; Kanzaki, Y.; et al. Suppressive effect of AMP-activated protein kinase on the epithelial-mesenchymal transition in retinal pigment epithelial cells. PLoS ONE 2017, 12, e0181481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Liu, L.; Jiang, Y.; Silva, M.; Zhen, X.; Zheng, W. Protective effect of Metformin against hydrogen peroxide-induced oxidative damage in human retinal pigment epithelial (RPE) cells by enhancing autophagy through activation of AMPK pathway. Oxid. Med. Cell. Longev. 2020, 2020, 2524174. [Google Scholar] [CrossRef]
- Karim, M.R.; Fisher, C.R.; Kapphahn, R.J.; Polanco, J.R.; Ferrington, D.A. Investigating AKT activation and autophagy in immunoproteasome-deficient retinal cells. PLoS ONE 2020, 15, e0231212. [Google Scholar] [CrossRef] [PubMed]
- Golestaneh, N.; Chu, Y.; Cheng, S.K.; Cao, H.; Poliakov, E.; Berinstein, D.M. Repressed SIRT1/PGC-1α pathway and mitochondrial disintegration in iPSC-derived RPE disease model of age-related macular degeneration. J. Trans. Med. 2016, 14, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagishima, K.J.; Wan, Q.; Corneo, B.; Sharma, R.; Lotfi, M.R.; Boles, N.C.; Hua, F.; Maminishkis, A.; Zhang, C.; Blenkinsop, T.; et al. In pursuit of authenticity: Induced pluripotent stem cell-derived retinal pigment epithelium for clinical applications. Stem Cells Trans. Med. 2016, 5, 1562–1574. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shen, W.; Kuai, D.; Martin, J.M.; Guo, X.; Smith, M.A.; Perez, E.T.; Phillips, M.J.; Simonett, J.M.; Wallace, K.A.; et al. iPS cell modeling of Best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 2013, 22, 593–607. [Google Scholar] [CrossRef] [Green Version]
- Polinati, P.P.; Ilmarinen, T.; Trokovic, R.; Hyotylainen, T.; Otonkoski, T.; Suomalainen, A.; Skottman, H.; Tyni, T. Patient-Specific induced pluripotent stem cell-derived RPE cells: Understanding the pathogenesis of retinopathy in Long-Chain 3-Hydroxyacyl-CoA Dehydrogenase deficiency. investig. Ophthalmol. Vis. Sci. 2015, 56, 3371–3382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galloway, C.A.; Dalvi, S.; Hung, S.S.C.; MacDonald, L.A.; Latchney, L.R.; Wong, R.C.B.; Guymer, R.H.; Mackey, D.A.; Williams, D.S.; Chung, M.M.; et al. Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc. Natl. Acad. Sci. USA 2017, 114, E8214–E8223. [Google Scholar] [CrossRef] [Green Version]
- Duong, T.T.; Vasireddy, V.; Ramachandran, P.; Herrera, P.S.; Leo, L.; Merkel, C.; Bennett, J.; Mills, J.A. Use of induced pluripotent stem cell models to probe the pathogenesis of Choroideremia and to develop a potiental treatment. Stem Cell Res 2018, 27, 140–150. [Google Scholar] [CrossRef]
- Lee, J.H.; Oh, J.O.; Lee, C.S. Induced pluripotent stem cell modeling of Best disease and autosomal recessive Bestrophinopathy. Yonsei Med. J. 2020, 61, 816–825. [Google Scholar] [CrossRef]
- Gong, J.; Cai, H.; NYSCF Global Stem Cell Array Team; Noggle, S.; Paull, D.; Rizzolo, L.J.; Del Priore, L.V.; Fields, M.A. Stem cell-derived retinal pigment epithelium from patients with age-related macular degeneration exhibit reduced metabolism and matrix interactions. Stem Cells Transl. Med. 2020, 9, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Cerniauskas, E.; Kurwana-Akanbi, M.; Xie, L.; Hallam, D.; Moya-Molina, M.; White, K.; Steel, D.; Doherty, M.; Whitfield, P.; Al-Aama, J.; et al. Complement modulation reverses pathology in Y402H-retinal pigment epithelium cell model of age-related macular degeneration by restoring lysosomal function. Stem Cells Transl. Med. 2020, 9, 1585–1603. [Google Scholar] [CrossRef]
- Cai, H.; Gong, J.; Abriola, L.; Hoyer, D.; NYSCF Global Stem Cell Array Team; Noggle, S.; Paull, D.; Del Priore, L.V.; Fields, M.A. High-Throughput screening identifies compounds that protect RPE cells from physiological stressors present in AMD. Exp. Eye Res. 2019, 185, 107641. [Google Scholar] [CrossRef]
- Beatty, S.; Koh, H.; Phil, D.; Henson, D.; Boulton, M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv. Ophthalmol. 2000, 45, 115–134. [Google Scholar] [CrossRef] [Green Version]
- Kauppinen, A.; Paterno, J.J.; Blasiak, J.; Salminen, A.; Kaarniranta, K. Inflammation and its role in age-related macular degeneration. Cell. Mol. Life Sci. 2016, 73, 1765–1786. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Jiang, N.; Chu, Y.; Postnikova, O.; Varghese, R.; Horvath, A.; Cheema, A.K.; Golestaneh, N. Dysregulated metabolic pathways in age-related macular degeneration. Sci. Rep. 2020, 10, 2464. [Google Scholar] [CrossRef] [Green Version]
- Vial, G.; Detaille, D.; Guigas, B. Role of mitochondria in the mechanism(s) of action of Metformin. Front. Endocrinol. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; An, H.; Liu, T.; Qin, C.; Sesaki, H.; Guo, S.; Radovick, S.; Hussain, M.; Maheshwari, A.; Wondisford, E.E.; et al. Metformin improves mitochondrial respiratory activity through activation of AMPK. Cell Rep. 2019, 29, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kong, L.; Wang, J.; Ash, J.D. Stimulation of AMPK prevents degeneration of photoreceptors and the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2018, 115, 10475–10480. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.E.; Ball, J.D.; Chen, Z.; Khurshid, G.S.; Prosperi, M.; Ash, J.D. The common antidiabetic drug Metformin reduces odds of developing age-related macular degeneration. investig. Ophthalmol. Vis. Sci. 2019, 60, 1470–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blitzer, A.L.; Ham, S.A.; Colby, K.A.; Skondra, D. Association of Metformin use with age-related macular degeneration: A case-control study. JAMA Ophthalmol. 2021, 139, 302–309. [Google Scholar] [CrossRef]
- Palmieri, M.; Pal, R.; Nelvagal, H.R.; Lotfi, P.; Stinnett, G.R.; Seymour, M.L.; Chaudhury, A.; Bajaj, L.; Bondar, V.V.; Bremner, L.; et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017, 8, 14338. [Google Scholar] [CrossRef]
- Kulkarni, A.S.; Gubbi, S.; Barzilai, N. Benefits of Metformin in attenuating the hallmarks of aging. Cell Metab. 2020, 32, 15–30. [Google Scholar] [CrossRef] [PubMed]
- McNeish, J.; Gardner, J.P.; Wainger, B.J.; Woolf, C.J.; Eggan, K. From Dish to bedside: Lessons learned while translating findings from a stem cell model of disease to a clinical trial. Cell Stem Cell 2015, 17, 8–10. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.D.; Gangnon, R.E.; Lee, L.Y.; Hubbard, L.D.; Klein, B.E.; Klein, R.; Ferris, F.L.; Bressler, S.B.; Milton, R.C.; Age-Related Eye Disease Study Research Group. The age-related eye disease study severity scale for age-related macular degeneration. Arch. Ophthalmol. 2005, 123, 1484–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crombie, D.E.; Danieszewski, M.; Liang, H.H.; Kulkarni, T.; Li, F.; Lidgerwood, G.E.; Conquest, A.; Hernandez, D.; Hung, S.S.; Gill, K.P.; et al. Development of a modular automated system for maintenance and differentiation of adherent human pluripotent stem cells. SLAS Discov. 2017, 22, 1016–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regent, F.; Morizur, L.; Lesueur, L.; Habeler, W.; Plancheron, A.; Ben M’Barek, K.; Monville, C. Automation of human pluripotent stem cell differentiation toward retinal pigment epithelial cells for large scale productions. Sci. Rep. 2019, 9, 10646. [Google Scholar] [CrossRef] [Green Version]
- Elanzew, A.; Niessing, B.; Langendoerfer, D.; Rippel, O.; Piotrowski, T.; Schenk, F.; Kulik, M.; Peitz, M.; Breitkreuz, Y.; Jung, S.; et al. The StemCellFactory: A modular system integration for automated generation and expansion of human induced pluripotent stem cells. Front. Bioeng. Biotechnol. 2020, 8, 580352. [Google Scholar] [CrossRef]
- Andreux, P.A.; Houtkeeper, R.H.; Auwerx, J. Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 2013, 12, 465–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
|
iPSC-RPE Line ID | Age a/Gender b | Disease Stage c |
CFH d/ARMS2 e Genotype | Figures Using Data from Specific Lines |
|---|---|---|---|---|
| Pt.A-1A3 + | 84/F | AREDS3 | CT/GT | 2C-E, 3B-C, 5, 6 |
| Pt.A-1A4 + | 84/F | AREDS3 | CT/GT | 2C-D, 3B-C, 5, 6 |
| Pt.B-1B4 | 76/F | AREDS4 | CC/TT | 2B-D, 3B-C, 5, 6 |
| Pt.C-2 # | 65/M | AREDS3 | CC/TT | 2C-D, 3B-C, 5, 6 |
| Pt.C-3 # | 65/M | AREDS3 | CC/TT | 2A,C-D, 3B-C, 5, 6 |
| Pt.D-1 | 76/F | AREDS3 | CC/GT | 2C-D, 3B-C, 5, 6 |
| Pt.E-3 | 63/F | AREDS2 | TT/GT | 2C-D, 3B-C, 5, 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebeling, M.C.; Geng, Z.; Stahl, M.R.; Kapphahn, R.J.; Roehrich, H.; Montezuma, S.R.; Ferrington, D.A.; Dutton, J.R. Testing Mitochondrial-Targeted Drugs in iPSC-RPE from Patients with Age-Related Macular Degeneration. Pharmaceuticals 2022, 15, 62. https://doi.org/10.3390/ph15010062
Ebeling MC, Geng Z, Stahl MR, Kapphahn RJ, Roehrich H, Montezuma SR, Ferrington DA, Dutton JR. Testing Mitochondrial-Targeted Drugs in iPSC-RPE from Patients with Age-Related Macular Degeneration. Pharmaceuticals. 2022; 15(1):62. https://doi.org/10.3390/ph15010062
Chicago/Turabian StyleEbeling, Mara C., Zhaohui Geng, Madilyn R. Stahl, Rebecca J. Kapphahn, Heidi Roehrich, Sandra R. Montezuma, Deborah A. Ferrington, and James R. Dutton. 2022. "Testing Mitochondrial-Targeted Drugs in iPSC-RPE from Patients with Age-Related Macular Degeneration" Pharmaceuticals 15, no. 1: 62. https://doi.org/10.3390/ph15010062
APA StyleEbeling, M. C., Geng, Z., Stahl, M. R., Kapphahn, R. J., Roehrich, H., Montezuma, S. R., Ferrington, D. A., & Dutton, J. R. (2022). Testing Mitochondrial-Targeted Drugs in iPSC-RPE from Patients with Age-Related Macular Degeneration. Pharmaceuticals, 15(1), 62. https://doi.org/10.3390/ph15010062