Nano Differential Scanning Fluorimetry-Based Thermal Stability Screening and Optimal Buffer Selection for Immunoglobulin G
Abstract
:1. Introduction
2. Results
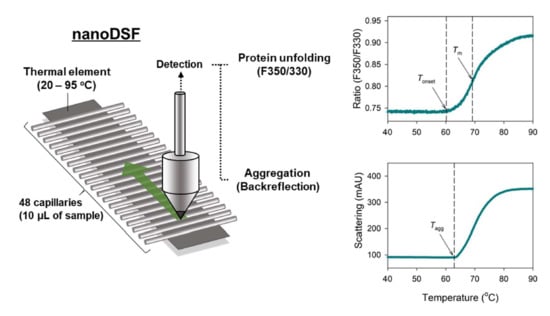
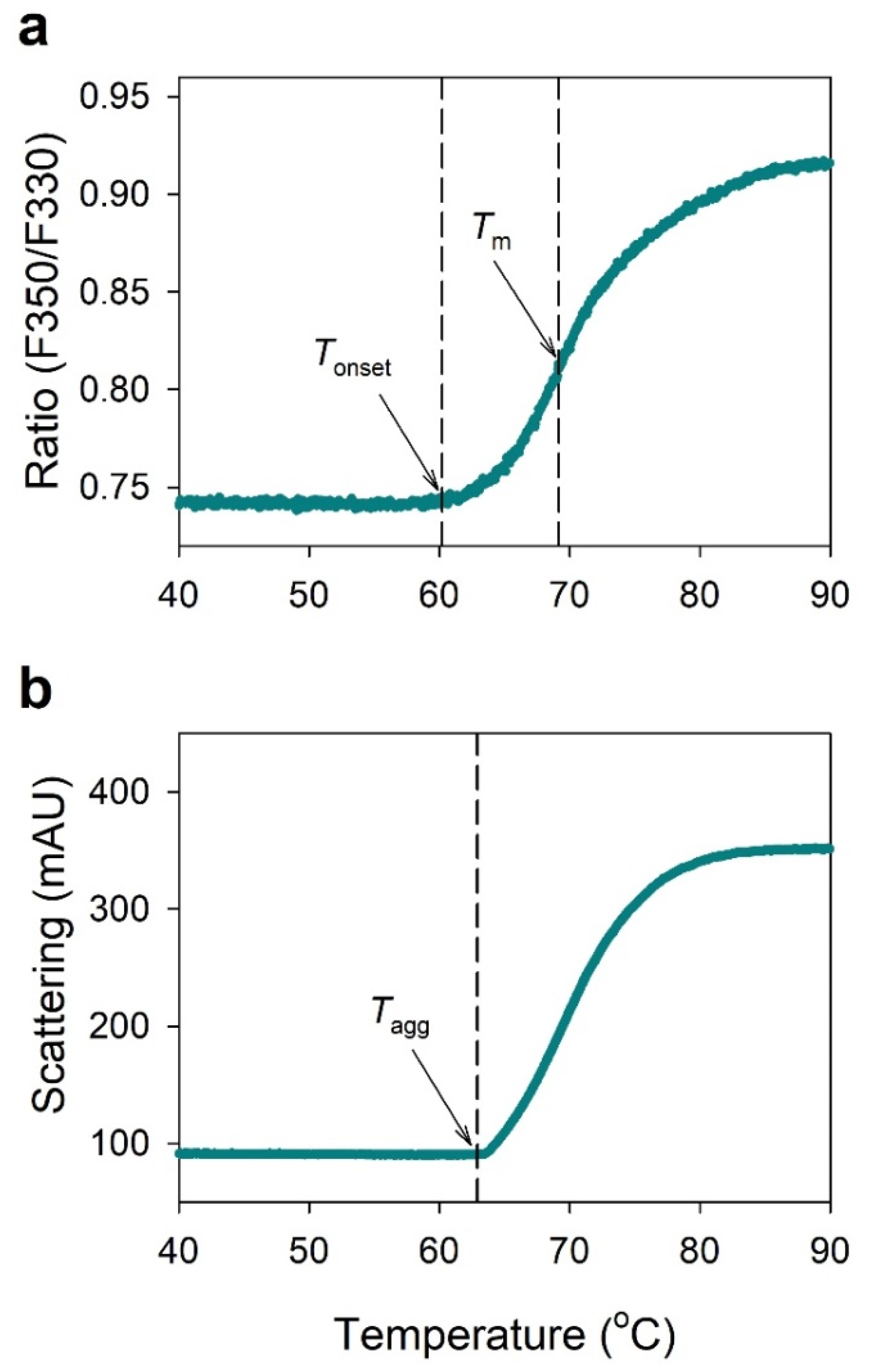
2.1. NanoDSF Analysis of IgG
2.2. Buffer Screening by NanoDSF
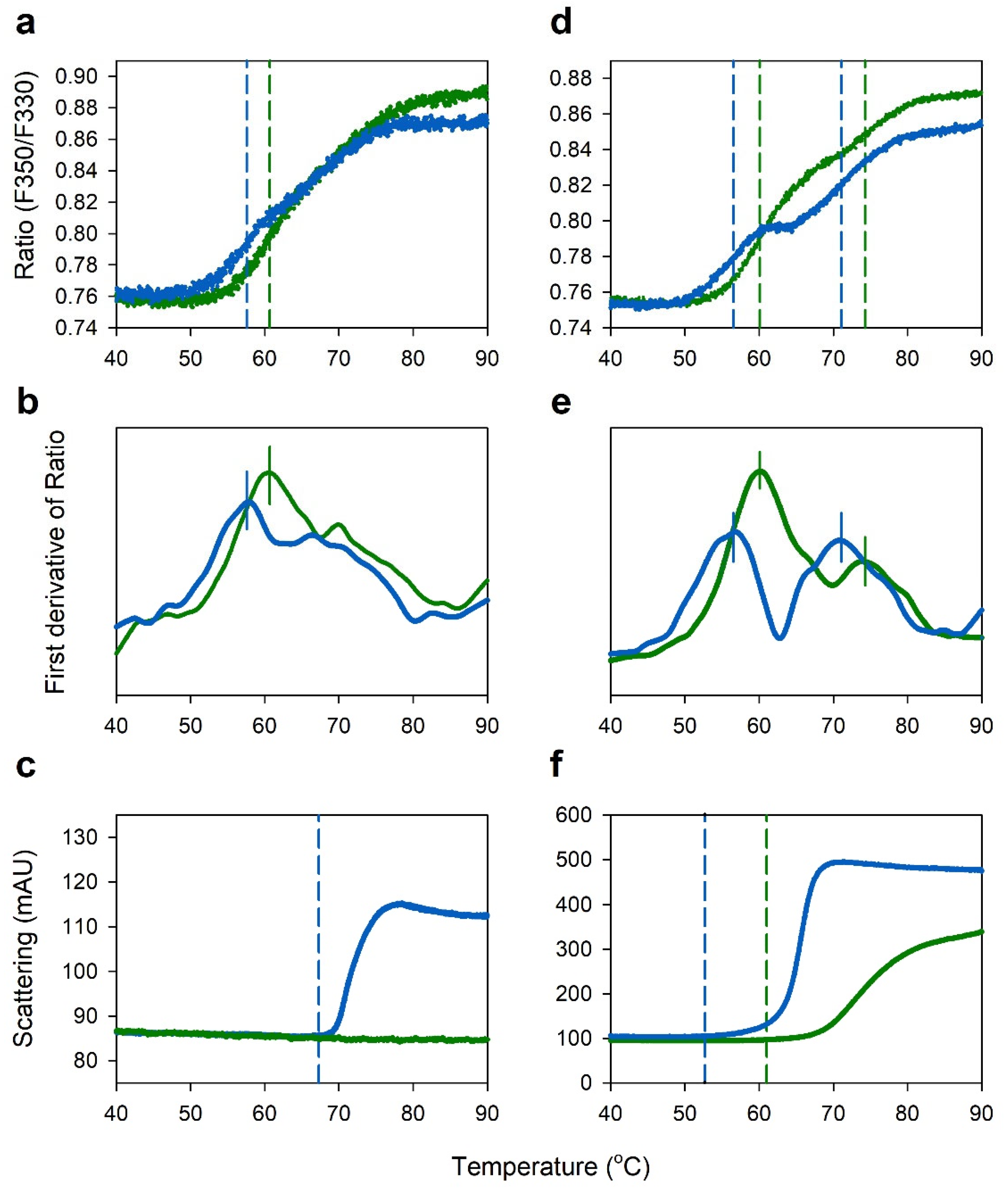
2.3. Comparison of Thermal Stability between Acetate and Citrate Buffers
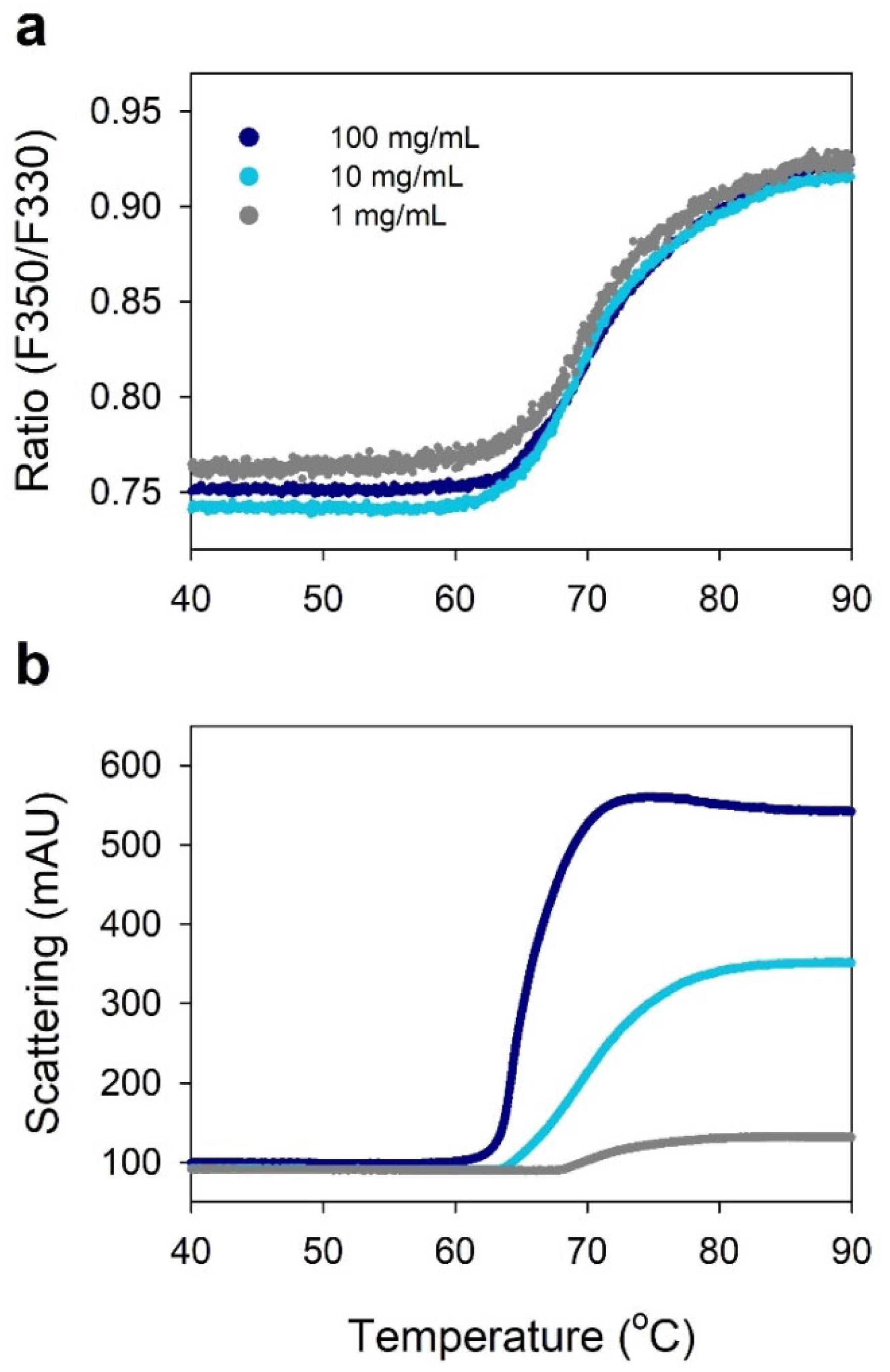
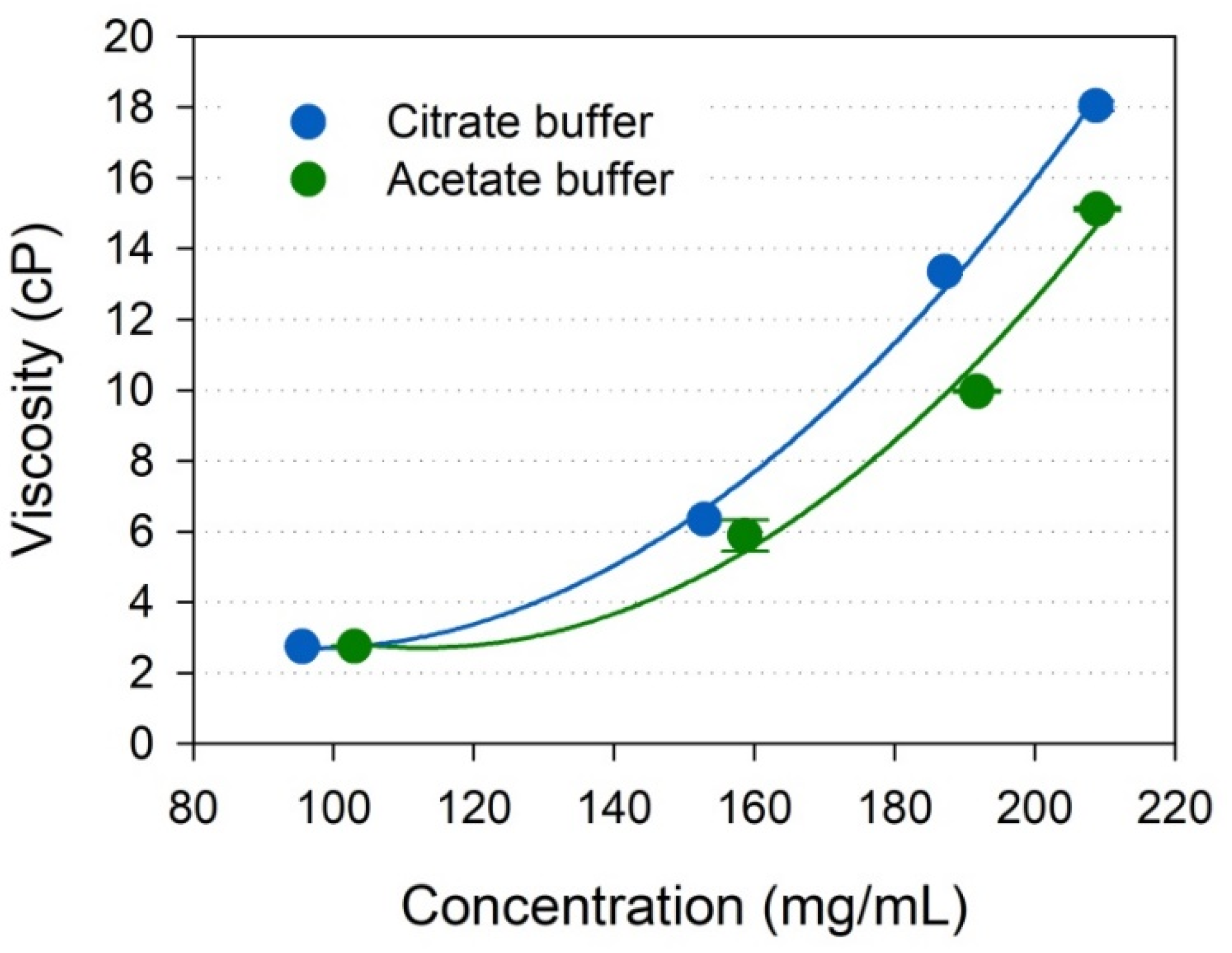
2.4. Viscosity of Highly Concentrated IgG
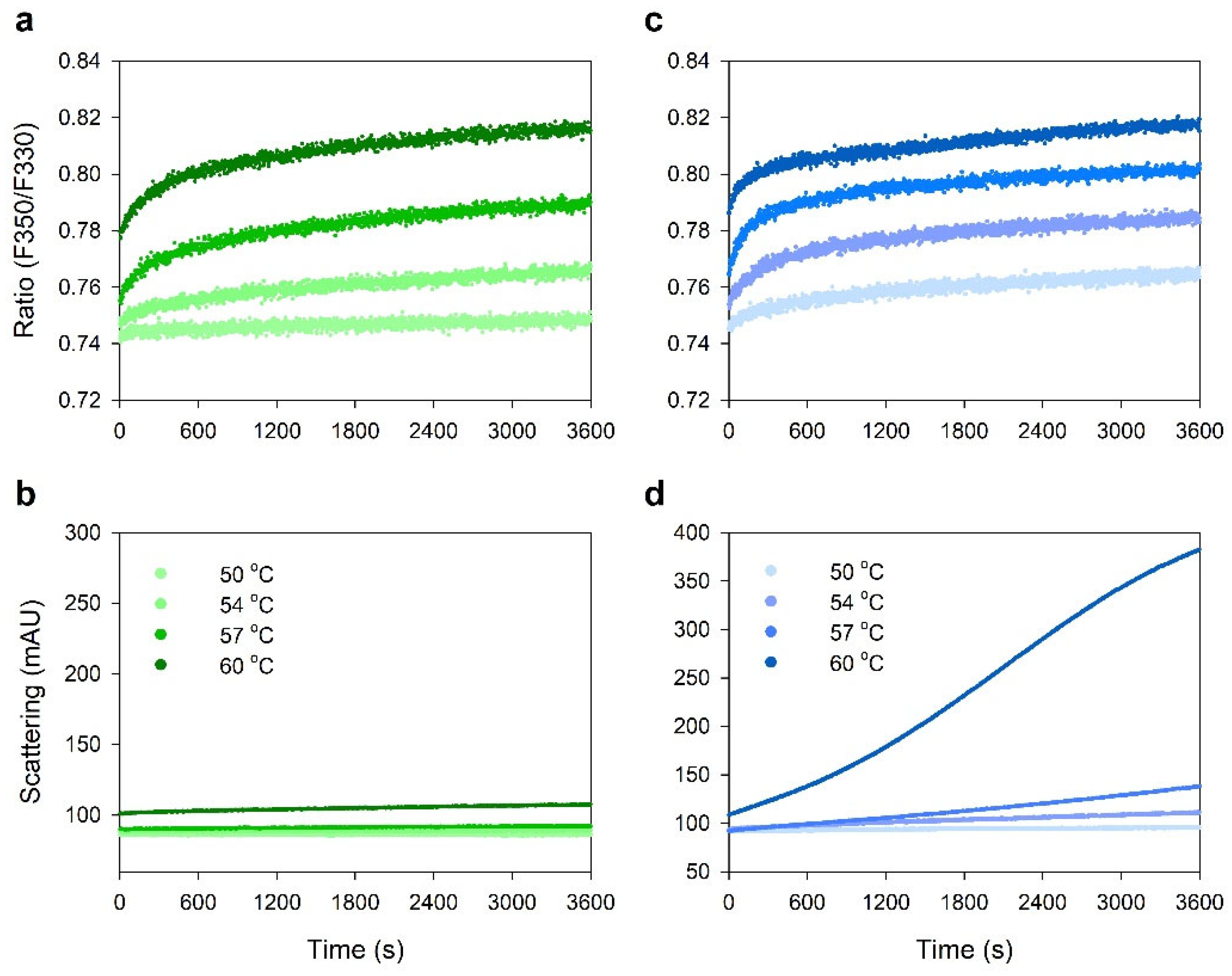
2.5. Protein–Protein Interactions
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Buffer Preparation
4.3. Buffer Exchange and Centrifugal Concentration
4.4. Nano Differential Scanning Fluorimetry
4.5. Viscosity Measurement
4.6. Dynamic and Static Light Scattering
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, S.; Kumar, N.K.; Dwiwedi, P.; Charan, J.; Kaur, R.; Sidhu, P.; Chugh, V.K. Monoclonal antibodies: A review. Curr. Clin. Pharmacol. 2018, 13, 85–99. [Google Scholar] [CrossRef]
- Mullard, A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 2021, 20, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Reichert, J.M. Antibodies to watch in 2021. mAbs 2021, 13, 1860476. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, E.J.; Na, D.H. Antibody-drug conjugates for targeted anticancer drug delivery. J. Pharm. Investig. 2016, 46, 341–349. [Google Scholar] [CrossRef]
- Shim, H. Bispecific antibodies and antibody-drug conjugates for cancer therapy: Technological considerations. Biomolecules 2020, 10, 360. [Google Scholar] [CrossRef] [Green Version]
- Pham, N.B.; Meng, W.S. Protein aggregation and immunogenicity of biotherapeutics. Int. J. Pharm. 2020, 585, 119523. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ohtake, S. Science and art of protein formulation development. Int. J. Pharm. 2019, 568, 118505. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, L.; Hostrup, S.; Moeller, E.H.; Grohganz, H. Recent trends in stabilising peptides and proteins in pharmaceutical formulation—considerations in the choice of excipients. Expert Opin. Drug Deliv. 2009, 6, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Maharana, C.; Rathore, A.S. An application of nano differential scanning fluorimetry for higher order structure assessment between mAb originator and biosimilars: Trastuzumab and Rituximab as case studies. J. Pharm. Biomed. Anal. 2020, 186, 113270. [Google Scholar] [CrossRef]
- Emami, F.; Vatanara, A.; Vakhshiteh, F.; Kim, Y.; Kim, T.W.; Na, D.H. Amino acid-based stable adalimumab formulation in spray freeze-dried microparticles for pulmonary delivery. J. Drug Deliv. Sci. Technol. 2019, 54, 101249. [Google Scholar] [CrossRef]
- Wang, W. Protein aggregation and its inhibition in biopharmaceutics. Int. J. Pharm. 2005, 289, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lord, H.; Knutson, N.; Wikström, M. Nano differential scanning fluorimetry for comparability studies of therapeutic proteins. Anal. Biochem. 2020, 593, 113581. [Google Scholar] [CrossRef]
- Johnson, C.M. Differential scanning calorimetry as a tool for protein folding and stability. Arch. Biochem. Biophys. 2013, 531, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, A.O.; Szekrenyi, A.; Joosten, H.J.; Finnigan, J.; Charnock, S.; Fessner, W.D. nanoDSF as screening tool for enzyme libraries and biotechnology development. FEBS J. 2019, 286, 184–204. [Google Scholar] [CrossRef] [Green Version]
- Gao, K.; Oerlemans, R.; Groves, M.R. Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 2020, 12, 85–104. [Google Scholar] [CrossRef] [Green Version]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef]
- Chattopadhyay, G.; Varadarajan, R. Facile measurement of protein stability and folding kinetics using a nano differential scanning fluorimeter. Protein Sci. 2019, 28, 1127–1134. [Google Scholar] [CrossRef]
- Ghisaidoobe, A.B.; Chung. S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- McClure, S.M.; Ahl, P.L.; Blue, J.T. High throughput differential scanning fluorimetry (DSF) formulation screening with complementary dyes to assess protein unfolding and aggregation in presence of surfactants. Pharm. Res. 2018, 35, 81. [Google Scholar] [CrossRef] [PubMed]
- Zbacnik, T.J.; Holcomb, R.E.; Katayama, D.S.; Murphy, B.M.; Payne, R.W.; Coccaro, R.C.; Evans, G.J.; Matsuura, J.E.; Henry, C.S.; Manning, M.C. Role of buffers in protein formulations. J. Pharm. Sci. 2017, 106, 713–733. [Google Scholar] [CrossRef]
- Hamuro, Y.; Derebe, M.G.; Venkataramani, S.; Nemeth, J.F. The effects of intramolecular and intermolecular electrostatic repulsions on the stability and aggregation of NISTmAb revealed by HDX-MS, DSC, and nanoDSF. Protein Sci. 2021, 30, 1686–1700. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.A.; Heo, B.; Jeong, S.H. Rapid methodology for basal system selection of therapeutic proteins during the early stage biopharmaceutical development. J. Pharm. Investig. 2020, 50, 363–372. [Google Scholar] [CrossRef]
- Kameoka, D.; Masuzaki, E.; Ueda, T.; Imoto, T. Effect of buffer species on the unfolding and the aggregation of humanized IgG. J. Biochem. 2007, 142, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Barnett, G.V.; Razinkov, V.I.; Kerwin, B.A.; Laue, T.M.; Woodka, A.H.; Butler, P.D.; Perevozchikova, T.; Roberts, C.J. Specific-ion effects on the aggregation mechanisms and protein-protein interactions for anti-streptavidin immunoglobulin gamma-1. J. Phys. Chem. B 2015, 119, 5793–5804. [Google Scholar] [CrossRef]
- Barnett, G.V.; Razinkov, V.I.; Kerwin, B.A.; Hillsley, A.; Roberts, C.J. Acetate- and citrate-specific ion effects on unfolding and temperature-dependent aggregation rates of anti-streptavidin IgG1. J. Pharm. Sci. 2016, 105, 1066–1073. [Google Scholar] [CrossRef] [Green Version]
- Oyama, H.; Koga, H.; Tadokoro, T.; Maenaka, K.; Shiota, A.; Yokoyama, M.; Noda, M.; Torisu, T.; Uchiyama, S. Relation of colloidal and conformational stabilities to aggregate formation in a monoclonal antibody. J. Pharm. Sci. 2020, 109, 308–315. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.W.; Kim, K.Y.; Song, H.-J.; Han, K.; Jee, S.; Kim, J. Standardization and validation of molecular size distribution test for human immunoglobulin products in Korea. Yakhak Hoeji 2021, 65, 23–30. [Google Scholar] [CrossRef]
- Emami, F.; Vatanara, A.; Najafabadi, A.R.; Kim, Y.; Park, E.J.; Sardari, S.; Na, D.H. Effect of amino acids on the stability of spray freeze-dried immunoglobulin G in sugar-based matrices. Eur. J. Pharm. Sci. 2018, 119, 39–48. [Google Scholar] [CrossRef]
- Galeotti, C.; Kaveri, S.V.; Bayry, J. IVIG-mediated effector functions in autoimmune and inflammatory diseases. Int. Immunol. 2017, 29, 491–498. [Google Scholar] [CrossRef]
- Chowdhury, A.; Guruprasad, G.; Chen, A.T.; Karouta, C.A.; Blanco, M.A.; Truskett, T.M.; Johnston, K.P. Protein-protein interactions, clustering, and rheology for bovine IgG up to high concentrations characterized by small angle X-ray scattering and molecular dynamics simulations. J. Pharm. Sci. 2020, 109, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Dear, B.J.; Hung, J.J.; Laber, J.R.; Wilks, L.R.; Sharma, A.; Truskett, T.M.; Johnston, K.P. Enhancing stability and reducing viscosity of a monoclonal antibody with cosolutes by weakening protein-protein interactions. J. Pharm. Sci. 2019, 108, 2517–2526. [Google Scholar] [CrossRef]
- Hung, J.J.; Dear, B.J.; Karouta, C.A.; Chowdhury, A.A.; Godfrin, P.D.; Bollinger, J.A.; Nieto, M.P.; Wilks, L.R.; Shay, T.Y.; Ramachandran, K.; et al. Protein-protein interactions of highly concentrated monoclonal antibody solutions via static light scattering and influence on the viscosity. J. Phys. Chem. B 2019, 123, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Shire, S.J.; Kalonia, D.S. Viscosity behavior of high-concentration monoclonal antibody solutions: Correlation with interaction parameter and electroviscous effects. J. Pharm. Sci. 2012, 101, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Holm, L.; Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 1994, 242, 309–320. [Google Scholar] [CrossRef]
- Nezlin, R. Dynamic aspects of the immunoglobulin structure. Immunol. Investig. 2019, 48, 771–780. [Google Scholar] [CrossRef]
- Wu, H.; Kroe-Barrett, R.; Singh, S.; Robinson, A.S.; Roberts, C.J. Competing aggregation pathways for monoclonal antibodies. FEBS Lett. 2014, 588, 936–941. [Google Scholar] [CrossRef]
- Wurster, U.; Haas, J. Passage of intravenous immunoglobulin and interaction with the CNS. J. Neurol. Neurosurg. Psychiatry 1994, 57, 21–25. [Google Scholar] [CrossRef] [Green Version]
- Prin, C.; Bene, M.C.; Gobert, B.; Montagne, P.; Faure, G.C. Isoelectric restriction of human immunoglobulin isotypes. Biochim. Biophys. Acta. 1995, 1243, 287–289. [Google Scholar] [CrossRef]
- Mark, S.M. Comparison of intravenous immunoglobulin formulations: Product formulary, and cost considerations. Hosp. Pharm. 2011, 46, 668–676. [Google Scholar] [CrossRef]
- Strickley, R.G.; Lambert, W.J. A review of formulations of commercially available antibodies. J. Pharm. Sci. 2021, 110, 2590–2608. [Google Scholar] [CrossRef]
- Schmidt, A.; Koulov, A.; Huwyler, J.; Mahler, H.C.; Jahn, M. Stabilizing polysorbate 20 and 80 against oxidative degradation. J. Pharm. Sci. 2020, 109, 1924–1932. [Google Scholar] [CrossRef]
- Burckbuchler, V.; Mekhloufi, G.; Giteau, A.P.; Grossiord, J.L.; Huille, S.; Agnely, F. Rheological and syringeability properties of highly concentrated human polyclonal immunoglobulin solutions. Eur. J. Pharm. Biopharm. 2010, 76, 351–356. [Google Scholar] [CrossRef]
- Perez, E.E.; Orange, J.S.; Bonilla, F.; Chinen, J.; Chinn, I.K.; Dorsey, M.; El-Gamal, Y.; Harville, T.O.; Hossny, E.; Mazer, B.; et al. Update on the use of immunoglobulin in human disease: A review of evidence. J. Allergy Clin. Immunol. 2017, 139, S1–S46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jezek, J.; Rides, M.; Derham, B.; Moore, J.; Cerasoli, E.; Simler, R.; Perez-Ramirez, B. Viscosity of concentrated therapeutic protein compositions. Adv. Drug Deliv. Rev. 2011, 63, 1107–1117. [Google Scholar] [CrossRef]
- Shah, M.; Corbett, D.; Lanzaro, A.; Roche, A.; Sibanda, N.; Davis, P.; Uddin, S.; van der Walle, C.F.; Curtis, R.; Pluen, A. Micro- and macro-viscosity relations in high concentration antibody solutions. Eur. J. Pharm. Biopharm. 2020, 153, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Maeder, W.; Lieby, P.; Sebald, A.; Spycher, M.; Pedrussio, R.; Bolli, R. Local tolerance and stability up to 24 months of a new 20% proline-stabilized polyclonal immunoglobulin for subcutaneous administration. Biologicals 2011, 39, 43–49. [Google Scholar] [CrossRef] [PubMed]
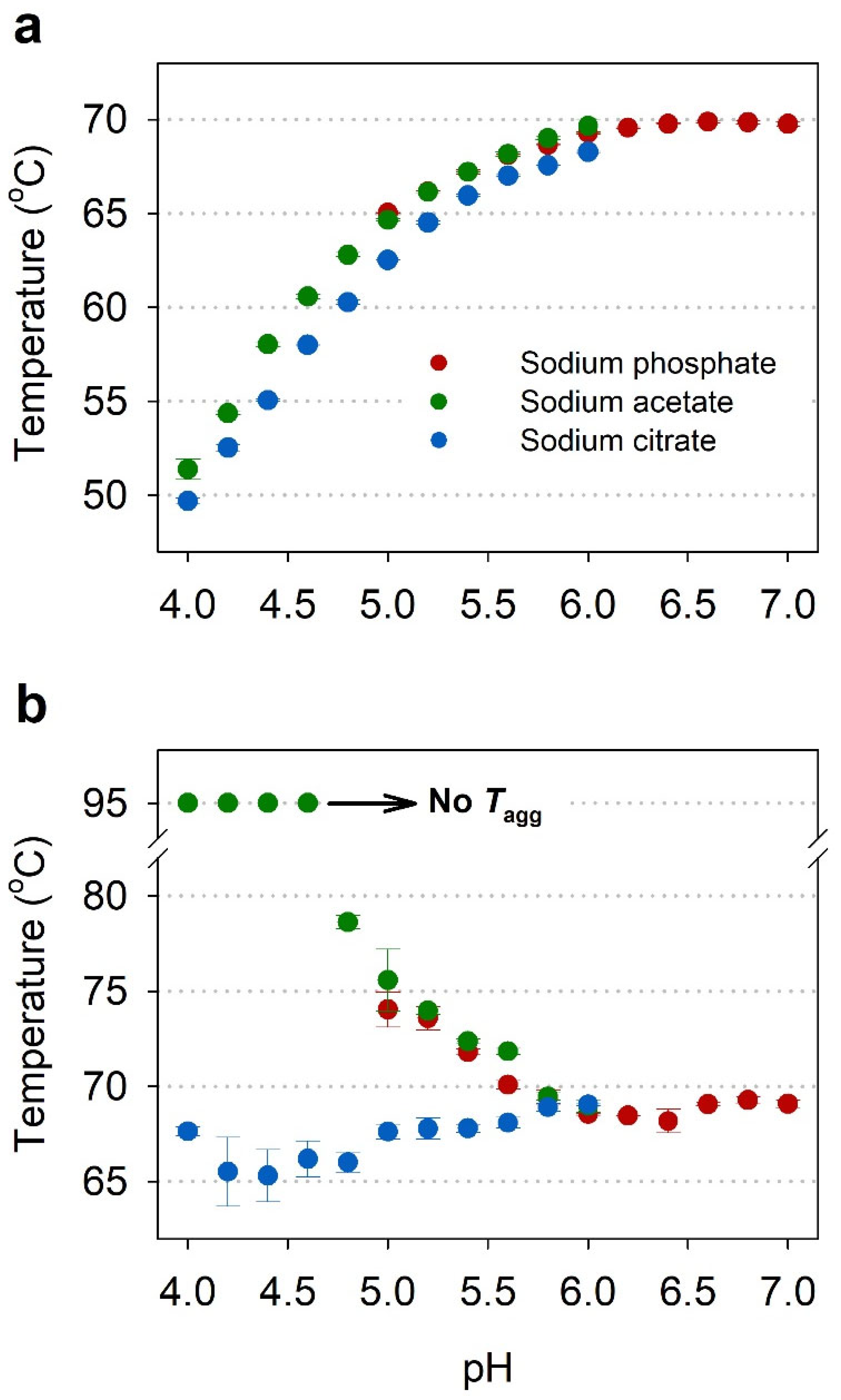

| Buffers | pH | Thermal Stability Parameters (°C) | ||
|---|---|---|---|---|
| Tonset | Tm | Tagg | ||
| Sodium phosphate | 5.0 | 57.9 ± 0.5 | 65.0 ± 0.0 | 74.0 ± 0.9 |
| 5.2 | 58.7 ± 0.8 | 66.1 ± 0.0 | 73.6 ± 0.6 | |
| 5.4 | 59.9 ± 0.4 | 67.2 ± 0.1 | 71.8 ± 0.2 | |
| 5.6 | 60.7 ± 0.9 | 68.1 ± 0.0 | 70.1 ± 0.2 | |
| 5.8 | 61.2 ± 0.3 | 68.7 ± 0.0 | 69.4 ± 0.4 | |
| 6.0 | 61.4 ± 0.1 | 69.3 ± 0.1 | 68.5 ± 0.0 | |
| 6.2 | 61.6 ± 0.3 | 69.6 ± 0.0 | 68.5 ± 0.0 | |
| 6.4 | 61.8 ± 0.4 | 69.8 ± 0.0 | 68.2 ± 0.6 | |
| 6.6 | 61.4 ± 0.0 | 69.9 ± 0.1 | 69.1 ± 0.1 | |
| 6.8 | 60.1 ± 0.3 | 69.8 ± 0.1 | 69.3 ± 0.2 | |
| 7.0 | 60.8 ± 0.1 | 69.8 ± 0.1 | 69.1 ± 0.2 | |
| 7.3 | 60.4 ± 0.5 | 69.5 ± 0.1 | 69.5 ± 0.2 | |
| 7.5 | 60.2 ± 0.2 | 69.4 ± 0.1 | 70.1 ± 0.1 | |
| 7.7 | 59.7 ± 0.9 | 69.2 ± 0.0 | 69.2 ± 0.6 | |
| 8.0 | 58.9 ± 0.2 | 69.0 ± 0.0 | 70.5 ± 0.5 | |
| Sodium acetate | 4.0 | 42.3 ± 0.1 | 51.4 ± 0.5 | No aggregation |
| 4.2 | 45.0 ± 0.0 | 54.4 ± 0.1 | No aggregation | |
| 4.4 | 50.1 ± 0.5 | 58.0 ± 0.1 | No aggregation | |
| 4.6 | 53.2 ± 0.1 | 60.6 ± 0.1 | No aggregation | |
| 4.8 | 55.2 ± 0.2 | 62.8 ± 0.1 | 78.6 ± 0.4 | |
| 5.0 | 57.0 ± 0.3 | 64.6 ± 0.1 | 75.6 ± 1.6 | |
| 5.2 | 58.6 ± 0.1 | 66.1 ± 0.0 | 74.0 ± 0.2 | |
| 5.4 | 59.9 ± 0.1 | 67.2 ± 0.0 | 72.4 ± 0.1 | |
| 5.6 | 60.3 ± 0.5 | 68.2 ± 0.1 | 71.8 ± 0.2 | |
| 5.8 | 60.9 ± 0.3 | 69.0 ± 0.1 | 69.5 ± 0.2 | |
| 6.0 | 61.4 ± 0.3 | 69.7 ± 0.4 | 69.0 ± 0.3 | |
| Sodium citrate | 4.0 | 40.1 ± 0.2 | 49.7 ± 0.2 | 67.6 ± 0.2 |
| 4.2 | 44.3 ± 0.1 | 52.5 ± 0.2 | 65.5 ± 1.8 | |
| 4.4 | 47.6 ± 0.3 | 55.1 ± 0.1 | 65.3 ± 1.4 | |
| 4.6 | 50.5 ± 0.5 | 58.0 ± 0.1 | 66.2 ± 0.9 | |
| 4.8 | 53.0 ± 0.5 | 60.3 ± 0.1 | 66.0 ± 0.5 | |
| 5.0 | 55.4 ± 0.1 | 62.5 ± 0.0 | 67.6 ± 0.4 | |
| 5.2 | 57.7 ± 0.0 | 64.5 ± 0.1 | 67.8 ± 0.5 | |
| 5.4 | 58.4 ± 0.4 | 66.0 ± 0.1 | 67.8 ± 0.2 | |
| 5.6 | 59.7 ± 0.3 | 67.0 ± 0.0 | 68.1 ± 0.3 | |
| 5.8 | 60.2 ± 0.5 | 67.6 ± 0.0 | 68.9 ± 0.2 | |
| 6.0 | 60.6 ± 0.2 | 68.3 ± 0.0 | 69.0 ± 0.1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Yoo, H.J.; Park, E.J.; Na, D.H. Nano Differential Scanning Fluorimetry-Based Thermal Stability Screening and Optimal Buffer Selection for Immunoglobulin G. Pharmaceuticals 2022, 15, 29. https://doi.org/10.3390/ph15010029
Kim SH, Yoo HJ, Park EJ, Na DH. Nano Differential Scanning Fluorimetry-Based Thermal Stability Screening and Optimal Buffer Selection for Immunoglobulin G. Pharmaceuticals. 2022; 15(1):29. https://doi.org/10.3390/ph15010029
Chicago/Turabian StyleKim, Soo Hyun, Han Ju Yoo, Eun Ji Park, and Dong Hee Na. 2022. "Nano Differential Scanning Fluorimetry-Based Thermal Stability Screening and Optimal Buffer Selection for Immunoglobulin G" Pharmaceuticals 15, no. 1: 29. https://doi.org/10.3390/ph15010029
APA StyleKim, S. H., Yoo, H. J., Park, E. J., & Na, D. H. (2022). Nano Differential Scanning Fluorimetry-Based Thermal Stability Screening and Optimal Buffer Selection for Immunoglobulin G. Pharmaceuticals, 15(1), 29. https://doi.org/10.3390/ph15010029