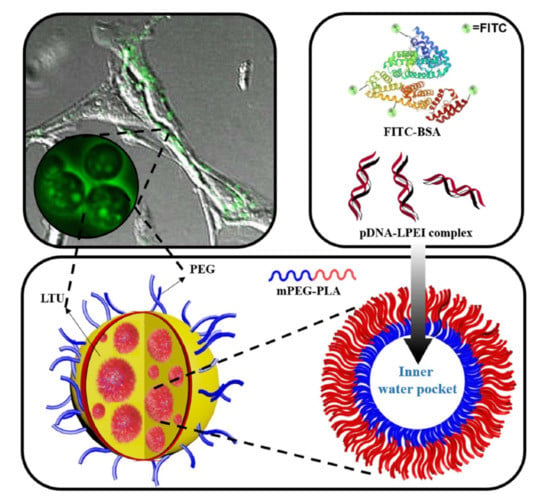
Fabrication and Biological Activities of Plasmid DNA Gene Carrier Nanoparticles Based on Biodegradable l-Tyrosine Polyurethane
Abstract
:1. Introduction
2. Results and Discussion
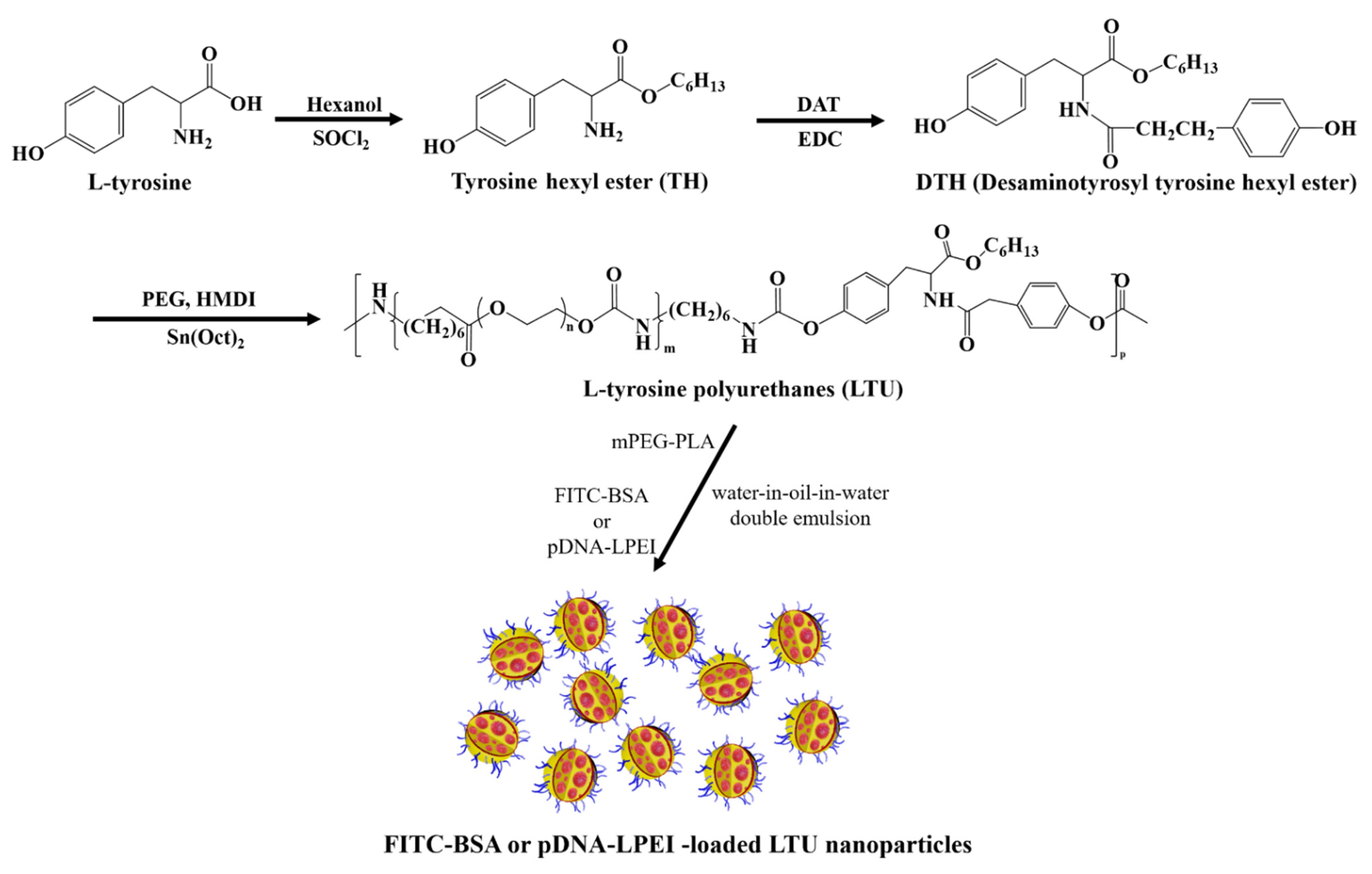
2.1. Synthesis and Characterization of LTU and mPEG-PLA
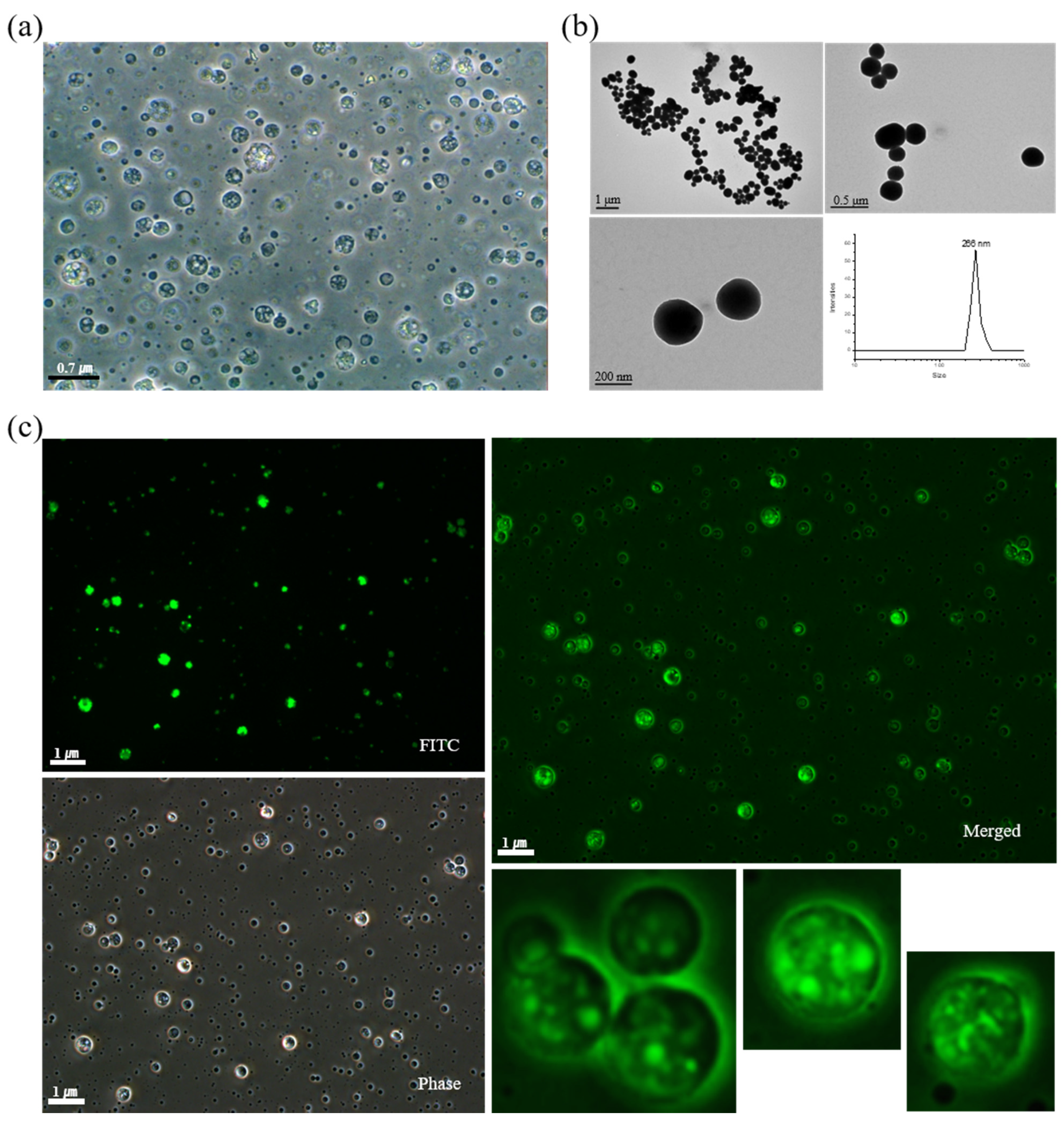
2.2. Characterization and Cellular Uptake Ability of FITC-BSA LTU NPs
2.3. Characterization of pDNA-LPEI LTU NPs
2.4. Transfection Efficiency of pDNA-LPEI LTU NPs
3. Materials and Methods
3.1. Materials
3.2. Characterization
3.3. Synthesis of LTU
3.4. Synthesis of mPEG-PLA
3.5. Fabrication of FITC-BSA LTU Nanoparticles (NPs)
3.6. Preparation of pDNA-LPEI LTU NPs
3.7. Cellular Uptake of FITC-BSA LTU NPs
3.8. Loading Confirmation and Release Behavior Study
3.9. Transfection of pDNA-LPEI LTU NPs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, S.; Huang, L. Nonviral gene therapy: Promises and challenges. Gene Ther. 2000, 7, 31–34. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, R.C. The basic science of gene therapy. Science 1993, 260, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-O.; Mahato, R.I.; Sung, Y.K.; Kim, S.W. Development of Biomaterials for Gene Therapy. Mol. Ther. 2000, 2, 302–317. [Google Scholar] [CrossRef]
- Sethuraman, V.A.; Na, K.; Bae, Y.H. pH-Responsive Sulfonamide/PEI System for Tumor Specific Gene Delivery: An in Vitro Study. Biomacromolecules 2006, 7, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Non-viral gene delivery systems. Curr. Opin. Biotechnol. 2002, 13, 128–131. [Google Scholar] [CrossRef]
- Abbasi, S.; Uchida, S. Multifunctional Immunoadjuvants for Use in Minimalist Nucleic Acid Vaccines. Pharmaceutics 2021, 13, 644. [Google Scholar] [CrossRef]
- Al-Dosari, M.S.; Gao, X. Nonviral gene delivery: Principle, limitations, and recent progress. AAPS J. 2009, 11, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramamoorth, M.; Narvekar, A. Nonviral vectors in gene therapy-An overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Kohn, J.; Langer, R. Polymerization reactions involving the side chains of alpha-L-amino acids. J. Am. Chem. Soc. 1987, 109, 817–820. [Google Scholar] [CrossRef]
- Park, S.-Y.; Kang, J.; Yoon, J.-Y.; Chung, I. Synthesis and Characterization of Polyfumarateurethane Nanoparticles for Sustained Release of Bupivacaine. Pharmaceutics 2020, 12, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, D. Development and Characterization of l-Tyrosine Based Polyurethanes for Tissue Engineering Applications. Ph.D. Thesis, The University of Akron, Akron, OH, USA, August 2007. [Google Scholar]
- Sarkar, D.; Yang, J.-C.; Klettlinger, N.; Lopina, S.T. Blends of l-tyrosine polyurethanes for biomaterial applications. Exp. Polym. Lett. 2007, 1, 724–733. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, S.-Y.; Kim, T.; Ahn, H.; Chung, I. Syntheses of biodegradable polymer networks based on polycaprolactone and glutamic acid. Polym. Adv. Technol. 2019, 30, 872–878. [Google Scholar] [CrossRef]
- Kim, T.; Park, S.Y.; Lee, M.-H.; Kim, D.-H.; Chung, I. Syntheses of polyrotaxane conjugated with 5-fluorouracil and vitamins with improved antitumor activities. J. Bioact. Compat. Polym. 2019, 34, 25–38. [Google Scholar] [CrossRef]
- Kim, T.; Mays, J.; Chung, I. Porous poly(ε-caprolactone) microspheres via UV photodegradation of block copolymers prepared by RAFT polymerization. Polymer 2018, 158, 198–203. [Google Scholar] [CrossRef]
- Hyun, J.; Wang, S.; Kim, J.; Rao, K.M.; Park, S.Y.; Chung, I.; Ha, C.-S.; Kim, S.-W.; Yun, Y.H.; Jung, Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat. Commun. 2016, 7, 10993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.; Ji, W.; Wang, C. Synthesis and characterization of new poly(ortho ester amidine) copolymers for non-viral gene delivery. Polymer 2011, 52, 921–932. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-M.; Bala, Y.S.; Lee, W.-K.; Jo, N.-J.; Chung, I. Antitumor and antiangiogenic active dendrimer/5-fluorouracil conjugates. J. Biomed. Mater. Res. Part A 2013, 101A, 2306–2312. [Google Scholar] [CrossRef]
- Amani, A.; Kabiri, T.; Shafiee, S.; Hamidi, A. Preparation and characterization of PLA-PEG-PLA/PEI/DNA nanoparticles for improvement of transfection efficiency and controlled release of DNA in gene delivery systems. Iran. J. Pharm. Res. 2019, 18, 125–141. [Google Scholar]
- Abebe, D.G.; Kandil, R.; Kraus, T.; Elsayed, M.; Merkel, O.M. Three-layered biodegradable micelles prepared by two-step self-assembly of PLA-PEI-PLA and PLA-PEG-PLA triblock copolymers as efficient gene delivery system. Micromol. Biosci. 2015, 15, 698–711. [Google Scholar] [CrossRef]
- Moret, I.; Peris, J.E.; Guillem, V.M.; Benet, M.; Revert, F.; Dasí, F.; Crespo, A.; Aliño, S.F. Stability of PEI-DNA and DOTAP-DNA complexes: Effect of alkaline pH, heparin and serum. J. Control. Release 2001, 76, 169–181. [Google Scholar] [CrossRef]
- Yue, Z.; Eccleston, M.E.; Slater, N.K.H. PEGylation and aqueous solution behavior of pH responsive poly(L-lysine iso-phthalamide). Polymer 2005, 46, 2497–2505. [Google Scholar] [CrossRef]
- Casettari, L.; Vllasaliu, D.; Mantovani, G.; Howdle, S.M.; Stolnik, S.; Illum, L. Effect of PEGylation on the toxicity and permeability enhancement of chitosan. Biomacromolecules 2010, 11, 2854–2865. [Google Scholar] [CrossRef] [PubMed]
- Amatya, R.; Hwang, S.; Park, T.; Min, K.A.; Shin, C.S. In Vitro and In Vivo Evaluation of PEGylated Starch-Coated Iron Oxide Nanoparticles for Enhanced Photothermal Cancer Therapy. Pharmaceutics 2021, 13, 871. [Google Scholar] [CrossRef] [PubMed]
- Calderon, F.L.; Bibette, J.; Schmitt, V. Double Emulsions; Springer: New York, NY, USA, 2007. [Google Scholar]
- Shum, H.C.; Lee, D.; Yoon, I.; Kodger, T.; Weitz, D.A. Double emulsion templated monodisperse phospholipid vesicles. Langmuir 2008, 24, 7651–7653. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Feng, S.-S. Methoxy poly(ethylene glycol)-poly(lactide) (MPEG-PLA) nanoparticles for controlled delivery of anticancer drugs. Biomaterials 2004, 25, 2843–2849. [Google Scholar] [CrossRef]
- Hans, M.; Shimoni, K.; Danino, D.; Siegel, S.J.; Lowman, A. Synthesis and characterization of mPEG-PLA prodrug micelles. Biomacromolecules 2005, 6, 2708–2717. [Google Scholar] [CrossRef]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Nguyen, J.; Steele, T.; Merkel, O.; Kissel, T. A new synthesis method and degradation of hyper-branched polyethylenimine grafted polycaprolactone block mono-methoxyl poly (ethylene glycol) copolymers (hy-PEI-g-PCL-b-mPEG) as potential DNA delivery vectors. Polymer 2009, 50, 3895–3904. [Google Scholar] [CrossRef]
- Richard, I.; Marc, T.; Crescenzo, G.D.; Buschmann, M.D.; Lavertu, M. Ionization behavior of chitosan and chitosan-DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as PEI. Biomacromolecules 2013, 14, 1732–1740. [Google Scholar] [CrossRef]
- Akinc, A.; Thomas, M.; Klibanov, A.M.; Langer, R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005, 7, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Florea, B.I.; Meaney, C.; Junginger, C.M.; Borchard, G. Transfection efficiency and toxicity of polyethylenimine in differentiated Calu-3 and nondifferentiated COS-1 cell cultures. AAPS Pharm. Sci. 2002, 4, E12. [Google Scholar] [CrossRef] [PubMed]
- Nair, L.S.; Laurencin, C.T. Biodegradable polymers as biomaterials. Prog. Polym. Sci. 2007, 32, 762–798. [Google Scholar] [CrossRef]
- Aoki, K.; Saito, N. Biodegradable Polymers as Drug Delivery Systems for Bone Regeneration. Pharmaceutics 2020, 12, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.K.; Goyal, A.K.; Gupta, P.N.; Khatri, K.; Mishra, N.; Mangal, S.; Vyas, S.P. Synthesis, characterization and evaluation of novel triblock copolymer based nanoparticles for vaccine delivery against hepatitis B. J. Control Release 2009, 136, 161–169. [Google Scholar] [CrossRef]
- Ghasemi, R.; Abdollahi, M.; Zadeh, E.E.; Khodabakhshi, K.; Badeli, A.; Bagheri, H.; Hosseinkhani, S. mPEG-PLA and PLA-PEG-PLA nanoparticles as new carriers for delivery of recombinant human Growth Hormone (rhGH). Sci. Rep. 2018, 8, 9584. [Google Scholar] [CrossRef]
- Hosseinkhani, H.; He, W.-J.; Chiang, C.-H.; Hong, P.-D.; Yu, D.-S.; Domb, A.J.; Ou, K.-L. Biodegradable nanoparticles for gene therapy technology. J. Nanopart. Res. 2013, 15, 1794. [Google Scholar] [CrossRef]
- Csaba, N.; Caamaño, P.; Sánchez, A.; Dominguez, F.; Alonso, M.J. PLGA:poloxamer and PLGA:poloxamine blend nanoparticles: New carriers for gene delivery. Biomacromolecules 2005, 6, 271–278. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, M.; Zhang, Z.; Hong, G.; Xiong, Q. Bovine serum albumin nanoparticles as controlled release carrier for local drug delivery to the inner ear. Nanoscale Res. Lett. 2014, 9, 343–349. [Google Scholar] [CrossRef]
- Maghsoudi, A.; Shojaosadati, S.A.; Farahani, E.V. 5-Fluorouracil-loaded BSA nanoparticles: Formulation optimization and in vitro release study. AAPS PharmSciTech. 2008, 9, 1092–1096. [Google Scholar] [CrossRef] [Green Version]
- Joanna, R.; Volker, O.; Inge, S.Z.; Dick, H. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar]
- Ahn, S.J.; Costa, J.; Emanuel, J.R. PicoGreen quantitation of DNA: Effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996, 24, 2623–2625. [Google Scholar] [CrossRef] [PubMed]
- Kimpton, J.; Emerman, M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 1992, 66, 2232–2239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelenin, A.V.; Kolesnikov, V.A.; Tarasenko, O.A.; Shafei, R.A.; Zelenina, I.A.; Mikhailov, V.V.; Semenova, M.L.; Kovalenko, D.V.; Artemyeva, O.V.; Ivaschenko, T.E.; et al. Bacterial β-galactosidase and human dystrophin genes are expressed in mouse skeletal muscle fibers after ballistic transfection. FEBS Lett. 1997, 414, 319–322. [Google Scholar] [PubMed] [Green Version]
- Kalyane, D.; Raval, N.; Maheshwari, R.; Tambe, V.; Kalia, K.; Tekade, R.K. Empolyment of enhanced permeability and retention effect (EPR): Nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater. Sci. Eng. C 2019, 98, 1252–1276. [Google Scholar] [CrossRef]
- Abdalla, A.M.E.; Xiao, L.; Ulah, M.W.; Yu, M.; Ouyang, C.; Yang, G. Current challenges of cancer anti-angiogenic therapy and the promise of nanotherapeutics. Theranostics 2018, 8, 533–548. [Google Scholar] [CrossRef]
- Sarkar, D.; Yang, J.-C.; Lopina, S.T. Structure-property relationship of l-tyrosine-based polyurethanes for biomaterial applications. J. Appl. Polym. Sci. 2008, 108, 2345–2355. [Google Scholar] [CrossRef]
- Gupta, A.S.; Lopina, S.T. l-tyrosine-based backbone-modified poly(amino acids). J. Biomater. Sci. Polym. Ed. 2002, 13, 1093–1104. [Google Scholar] [CrossRef]
- Houssier, C.; Hardy, B.; Fredericq, E. Interaction of ethidium bromide with DNA. Optical and electrooptical study. Biopolymers 1974, 13, 1141–1160. [Google Scholar] [CrossRef]
- Ramalingam, K.; Castro, R.; Pires, P.; Shi, X.; Rodrigues, J.; Xiao, S.; Tomás, H. Gene delivery using dendrimer/pDNA complexes immobilized in electrospun fibers using the Layer-by-Layer technique pain. RSC Adv. 2016, 6, 97116–97128. [Google Scholar] [CrossRef]
- Helmy, I.M.; Azim, A.M.A. Efficacy of ImageJ in the assessment of apoptosis. Diagn. Pathol. 2012, 7, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venter, C.; Niesler, C.U. Rapid quantification of cellular proliferation and migration using imageJ. BioTechniques 2019, 66, 99–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, P.N.; Manthe, R.L.; Lopina, S.T.; Yun, Y.H. Electrospinning of l-tyrosine polyurethanes for potential biomedical applications. Polymer 2009, 50, 2281–2289. [Google Scholar] [CrossRef]





| Polymer | Conc. (mg/mL) | Mass (mg) | Vol (mL) | Mass % (w/w) | Vol % (v/v) |
|---|---|---|---|---|---|
| FITC-BSA LTU nanoparticles | |||||
| LTU | 32.3 | 291.0 | 9.0 | 97.0 | 8.0 |
| mPEG-PLA | 3.0 | 3.0 | 1.0 | 1.0 | 0.9 |
| FITC-BSA | 3.0 | 3.0 | 1.0 | 1.0 | 0.9 |
| LPEI | 3.0 | 3.0 | 1.0 | 1.0 | 0.9 |
| 5% PVA | 100.0 | 89.2 | |||
| Total | 300.0 | 112.0 | |||
| Blank LTU nanoparticles | |||||
| LTU | 32.6 | 294.0 | 9.0 | 98.0 | 8.1 |
| mPEG-PLA | 3.0 | 3.0 | 1.0 | 1.0 | 0.9 |
| LPEI | 3.0 | 3.0 | 1.0 | 1.0 | 0.9 |
| 5% PVA | 100.0 | 90.1 | |||
| total | 300.0 | 111.0 | |||
| Polymer | Conc. (mg/mL) | Mass (mg) | Vol (mL) | Mass % (w/w) | Vol % (v/v) |
|---|---|---|---|---|---|
| pDNA-LPEI LTU nanoparticles | |||||
| LTU | 21.6 | 194.0 | 9.0 | 73.0 | 8.1 |
| mPEG-PLA | 2.0 | 2.0 | 1.0 | 6.8 | 0.9 |
| pDNA-LPEI | 6.0 | 6.0 | 1.0 | 20.3 | 0.9 |
| 5% PVA | 100.0 | 90.1 | |||
| Total | 202.0 | 111.0 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-Y.; Yun, Y.H.; Park, B.-J.; Seo, H.-I.; Chung, I. Fabrication and Biological Activities of Plasmid DNA Gene Carrier Nanoparticles Based on Biodegradable l-Tyrosine Polyurethane. Pharmaceuticals 2022, 15, 17. https://doi.org/10.3390/ph15010017
Park S-Y, Yun YH, Park B-J, Seo H-I, Chung I. Fabrication and Biological Activities of Plasmid DNA Gene Carrier Nanoparticles Based on Biodegradable l-Tyrosine Polyurethane. Pharmaceuticals. 2022; 15(1):17. https://doi.org/10.3390/ph15010017
Chicago/Turabian StylePark, Soo-Yong, Yang H. Yun, Bum-Joon Park, Hyung-Il Seo, and Ildoo Chung. 2022. "Fabrication and Biological Activities of Plasmid DNA Gene Carrier Nanoparticles Based on Biodegradable l-Tyrosine Polyurethane" Pharmaceuticals 15, no. 1: 17. https://doi.org/10.3390/ph15010017
APA StylePark, S.-Y., Yun, Y. H., Park, B.-J., Seo, H.-I., & Chung, I. (2022). Fabrication and Biological Activities of Plasmid DNA Gene Carrier Nanoparticles Based on Biodegradable l-Tyrosine Polyurethane. Pharmaceuticals, 15(1), 17. https://doi.org/10.3390/ph15010017