Potential and Limits of Kidney Cells for Evaluation of Renal Excretion
Abstract
:1. Introduction
2. Results
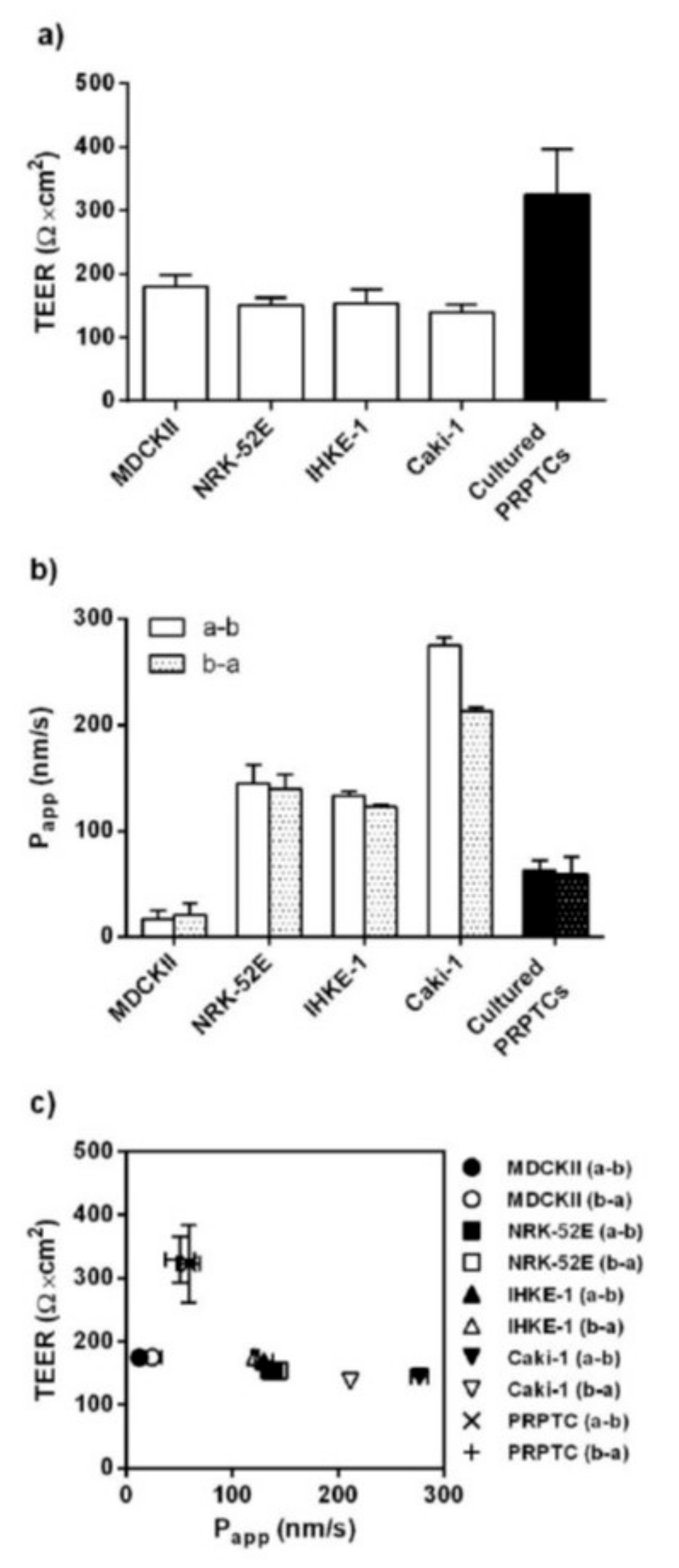
2.1. Phenotypic Characterization, Cell Morphology and Monolayer Integrity
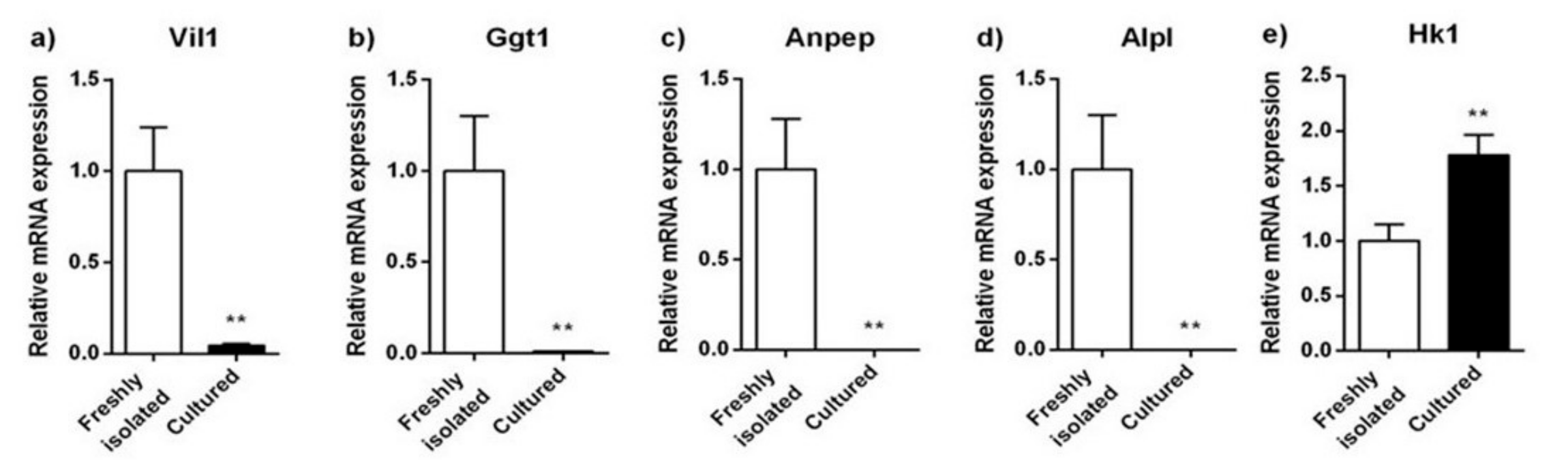
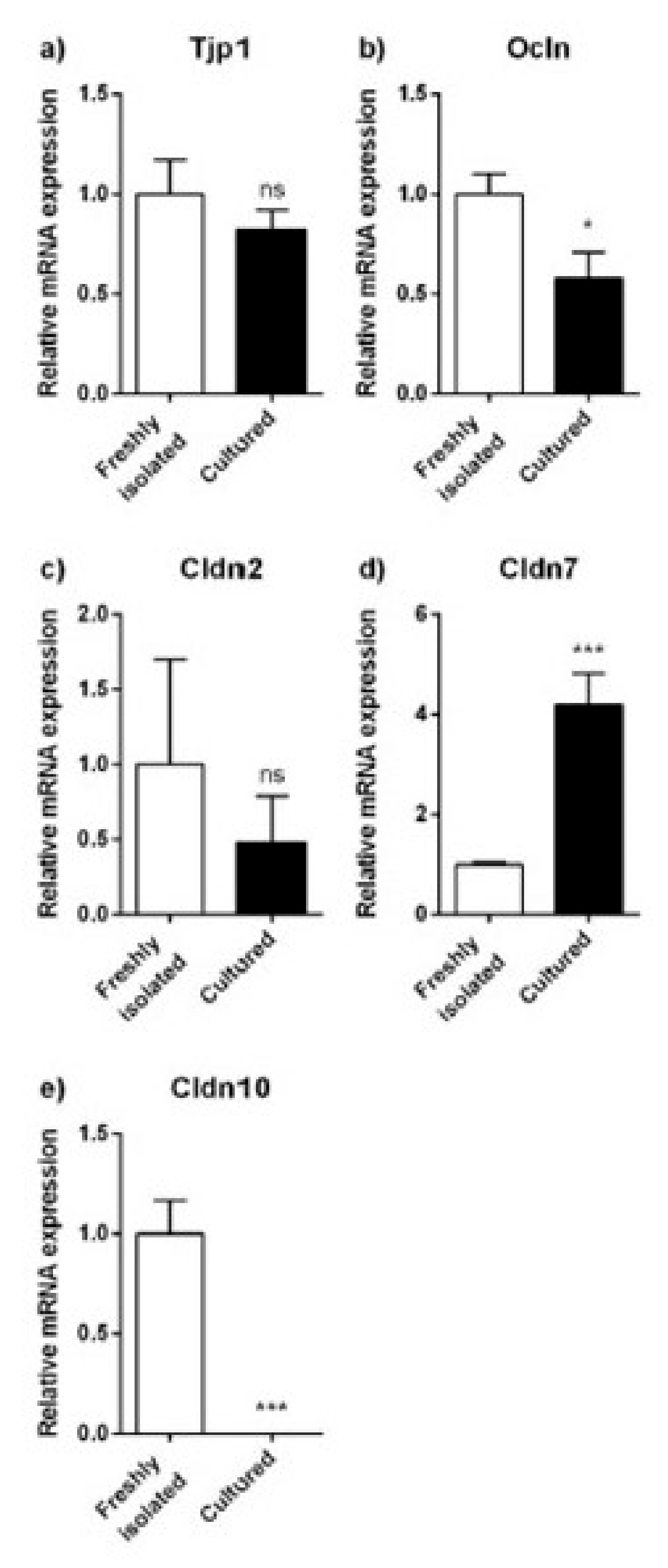
2.2. Expression Analyses of Proximal and Distal Tubule Markers
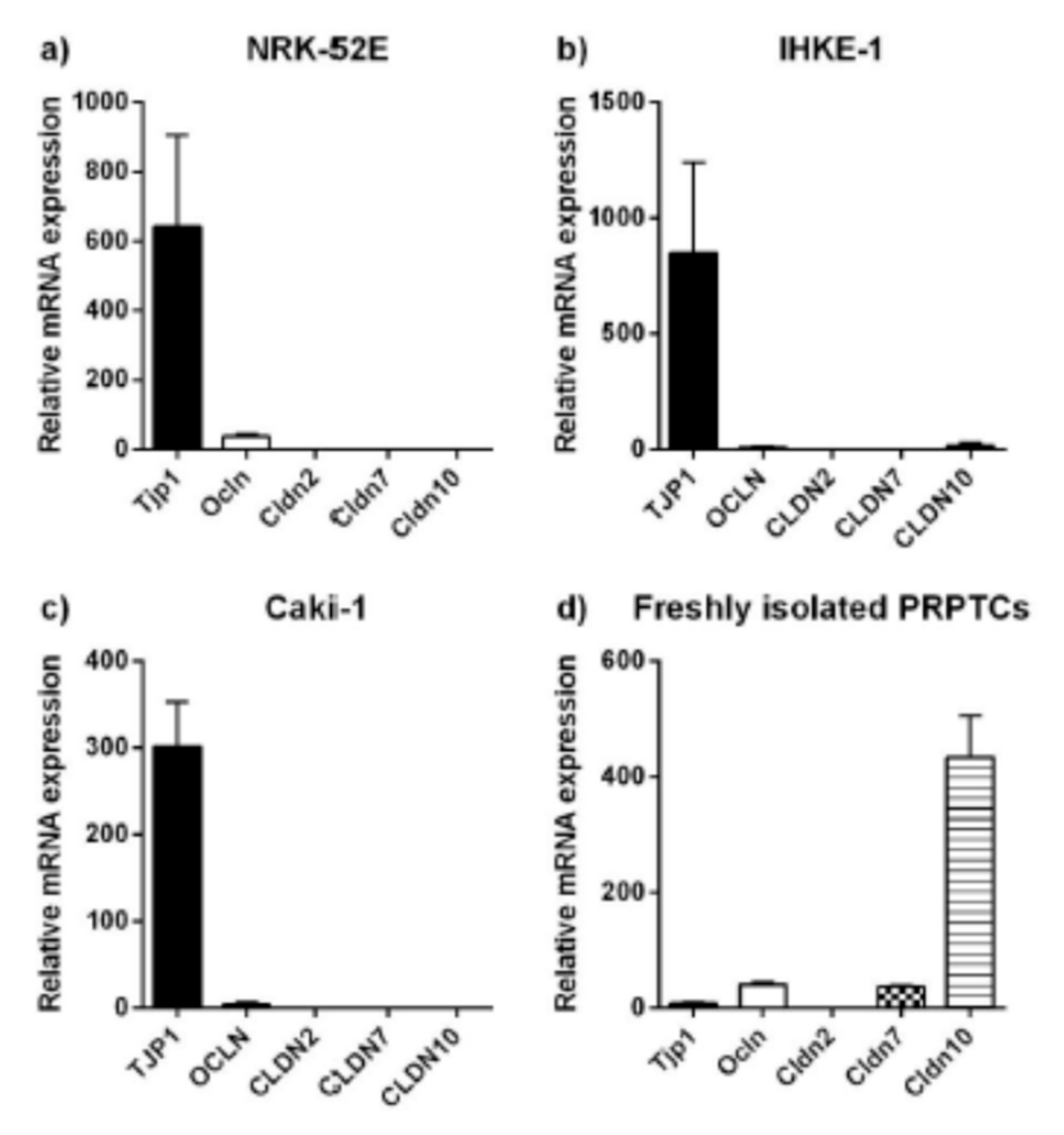
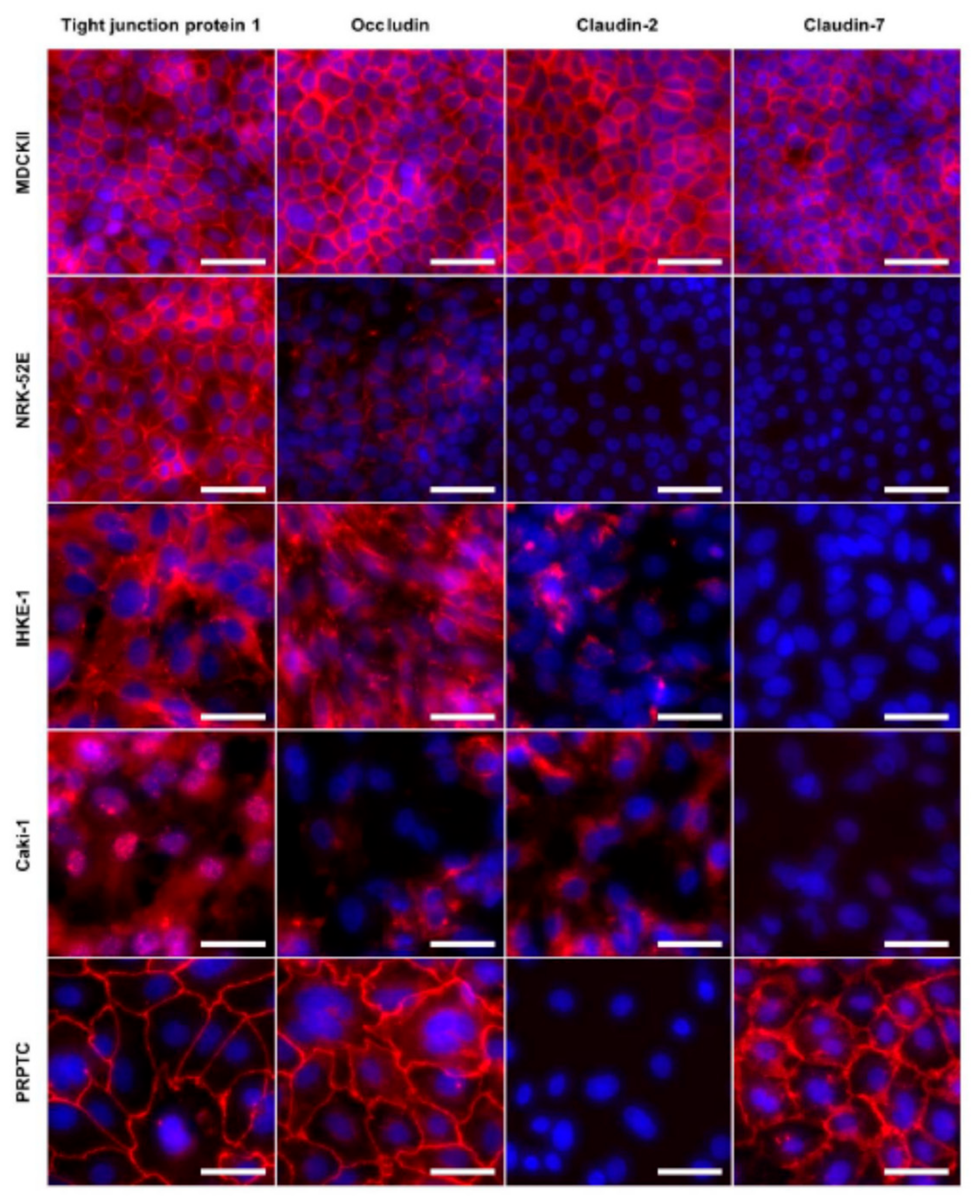
2.3. Expression Analyses of Tight Junction Proteins and Paracellular Transport
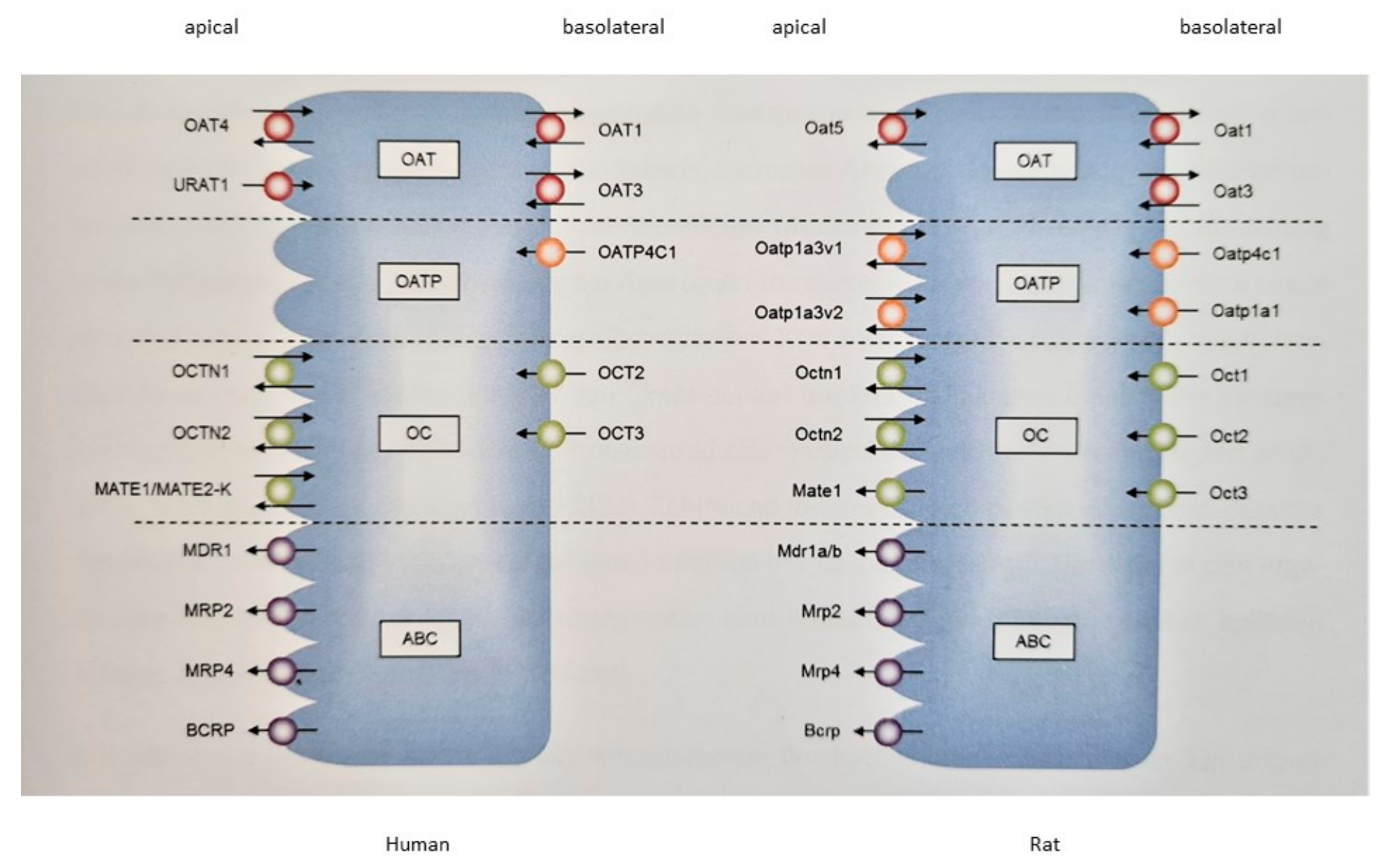
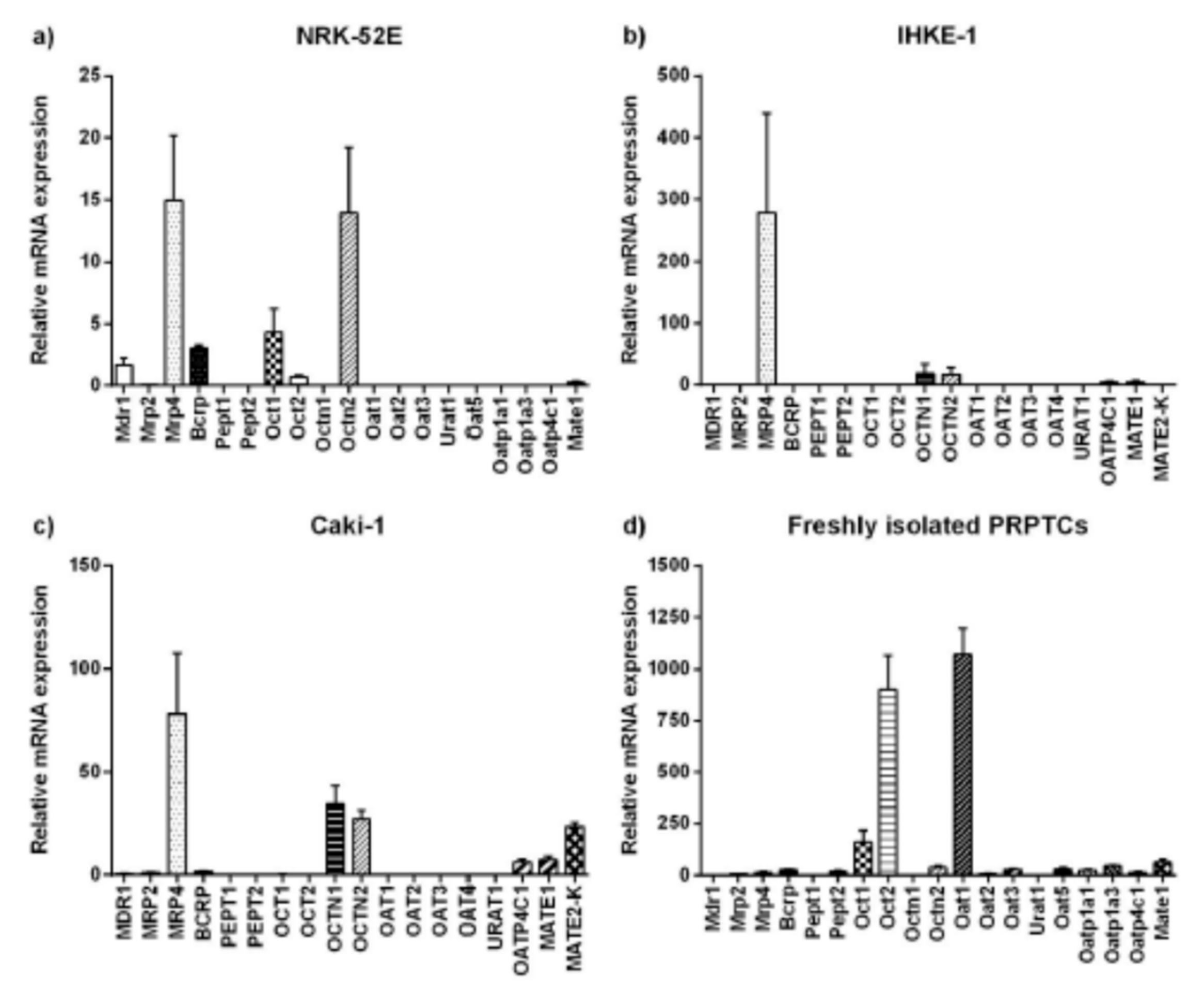
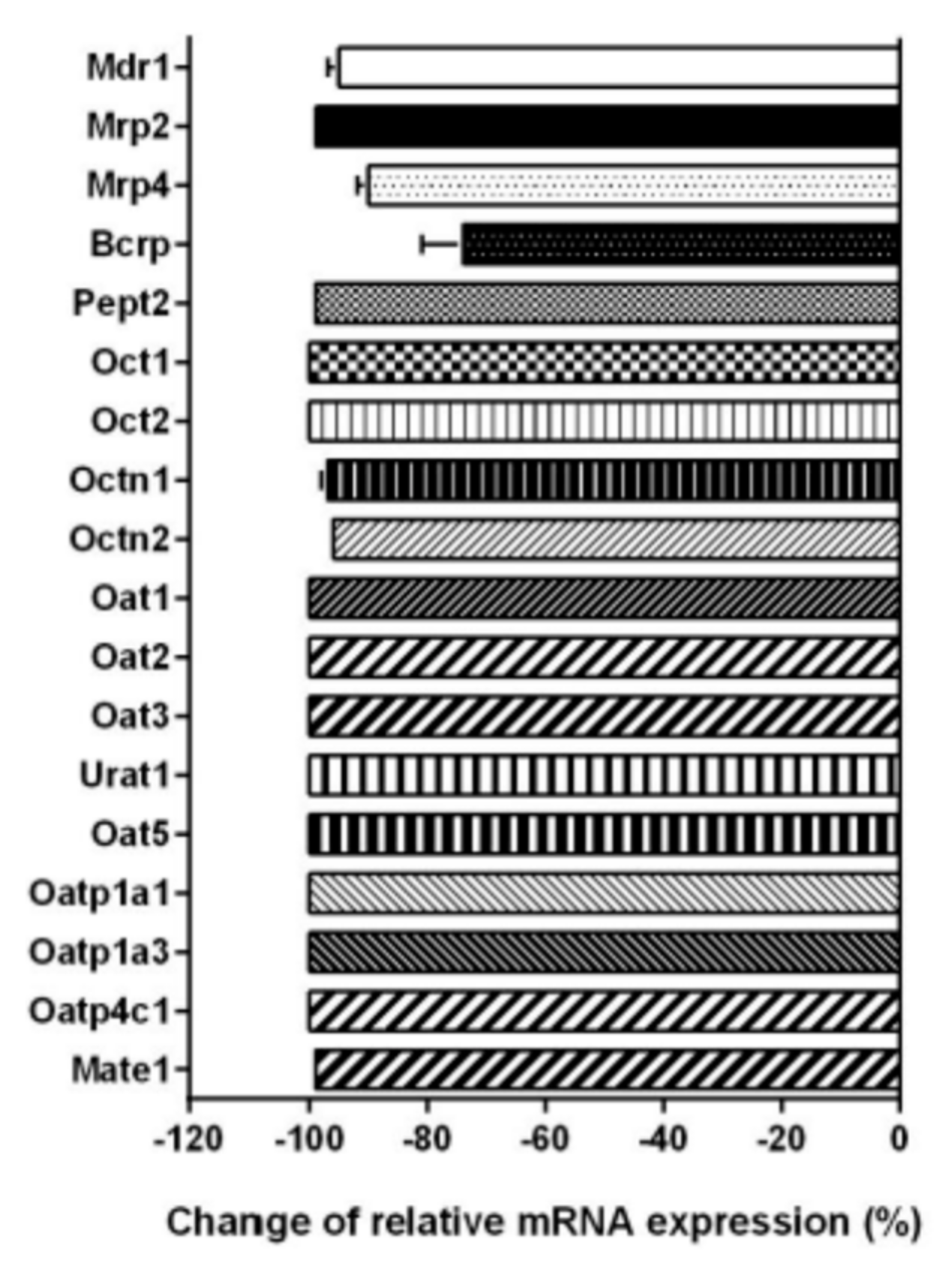
2.4. Expression of Transport Proteins
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Animals
4.3. Isolation of Proximal Tubular Cells from Rats
4.4. RNA Expression Analyses
4.5. Immunofluorescent Staining
4.6. Transepithelial Electrical Resistance and Bidirectional Transport Studies with [14C]-Mannitol
4.7. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ALPL | Alkaline phosphatase liver/bone/kidney |
| ANPEP | Alanine aminopeptidase |
| BCRP | Breast cancer resistance protein |
| Caki-1 | Carcinoma kidney-1 |
| CLDN | Claudin |
| Cy3 | Cyanin 3 |
| DAPI | 4′,6-diamidino-2-phenylindole |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| GGT1 | γ-glutamyl transferase 1 |
| HK1 | Hexokinase 1 |
| IHKE-1 | Immortalized human kidney epithelial-1 |
| MATE | Multidrug and toxin extrusion |
| MDCK | Madin-Darby canine kidney |
| MDR1 | Multidrug resistance 1 |
| MRP | Multidrug resistance |
| NRK-52E | Normal rat kidney-52E |
| OAT | Organic anion transporter |
| OATP | Organic anion transporting polypeptide |
| OCLN | Occludin |
| OCT | Organic cation transporter |
| OCTN | Organic cation transporter, novel |
| Papp | Apparent permeability coefficient |
| PEPT | Peptide transporter |
| PRPTC | Primary rat proximal tubule cell |
| qPCR | Quantitative real-time PCR |
| SLC | Solute carrier |
| SLCO | Solute carrier organic anion |
| TEER | Transepithelial electrical resistance |
| TJP1 | Tight junction protein 1 |
| URAT | Urate transporter |
| VIL1 | Villin 1 |
References
- Courjault-Gautier, J.C. Consecutive use of hormonally defined serum-free media to establish highly differentiated human renal proximal tubule cells in primary culture. J. Am. Soc. Nephrol. JASN 1995, 5, 1949–1963. [Google Scholar] [CrossRef]
- Gildea, J.J.; Shah, I.; Weiss, R.; Casscells, N.D.; McGrath, H.E.; Zhang, J.; Jones, J.E.; Felder, R.A. HK-2 Human Renal Proximal Tubule Cells as a Model for G Protein–Coupled Receptor Kinase Type 4–Mediated Dopamine 1 Receptor Uncoupling. Hypertension 2010, 56, 505–511. [Google Scholar] [CrossRef] [Green Version]
- Van der Hauwaert, C.; Savary, G.; Gnemmi, V.; Glowacki, F.; Pottier, N.; Bouillez, A.; Maboudou, P.; Zini, L.; Leroy, X.; Cauffiez, C.; et al. Isolation and Characterization of a Primary Proximal Tubular Epithelial Cell Model from Human Kidney by CD10/CD13 Double Labeling. PLoS ONE 2013, 8, e66750. [Google Scholar] [CrossRef] [Green Version]
- Kiuchi-Saishin, Y.; Gotoh, S.; Furuse, M.; Takasuga, A.; Tano, Y.; Tsukita, S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J. Am. Soc. Nephrol. 2002, 13, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Prozialeck, W.C.; Edwards, J.R.; Lamar, P.C.; Smith, C.S. Epithelial barrier characteristics and expression of cell adhesion molecules in proximal tubule-derived cell lines commonly used for in vitro toxicity studies. Toxicol. In Vitro 2006, 20, 942–953. [Google Scholar] [CrossRef]
- Reyes, J.L.; Lamas, M.; Martin, D.; Namorado, M.D.C.; Islas, S.; Luna, J.; Tauc, M.; González-Mariscal, L. The renal segmental distribution of claudins changes with development. Kidney Int. 2002, 62, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Mariscal, L.; Namorado, M.C.; Martin, D.; Luna, J.; Alarcon, L.; Islas, S.; Valencia, L.; Muriel, P.; Ponce, L.; Reyes, J.L. Tight junction proteins ZO-1, ZO-2, and occludin along isolated renal tubules11See Editorial by Brown, p. 2652. Kidney Int. 2000, 57, 2386–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Vapurcuyan, A.; Shahidullah, M.; Aleksunes, L.; Pelis, R.M. Expression of Organic Anion Transporter 2 in the Human Kidney and Its Potential Role in the Tubular Secretion of Guanine-Containing Antiviral Drugs. Drug Metab. Dispos. 2012, 40, 617–624. [Google Scholar] [CrossRef] [Green Version]
- Kuo, K.-L.; Zhu, H.; McNamara, P.J.; Leggas, M. Localization and Functional Characterization of the Rat Oatp4c1 Transporter in an In Vitro Cell System and Rat Tissues. PLoS ONE 2012, 7, e39641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.; Kim, R.B. Transporters and renal drug elimination. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 137–166. [Google Scholar] [CrossRef]
- Giacomini, K.M.; Huang, S.-M.; Tweedie, D.J.; Benet, L.Z.; Brouwer, K.L.R.; Chu, X.; Dahlin, A.; Evers, R.; Fischer, V.; Hillgren, K.M.; et al. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010, 9, 215–236. [Google Scholar]
- Jennings, P.; Koppelstätter, C.; Lechner, J.; Pfaller, W. Renal cell culture models: Contribution to the understanding of nephrotoxic mechanisms. In Clinical Nephrotoxins; Springer: Berlin/Heidelberg, Germany, 2008; pp. 223–249. [Google Scholar]
- Markowska, M.; Oberle, R.; Juzwin, S.; Hsu, C.-P.; Gryszkiewicz, M.; Streeter, A.J. Optimizing Caco-2 cell monolayers to increase throughput in drug intestinal absorption analysis. J. Pharmacol. Toxicol. Methods 2001, 46, 51–55. [Google Scholar] [CrossRef]
- Anderberg, E.K.; Artursson, P. Epithelial Transport of Drugs in Cell Culture. VIII: Effects of Sodium Dodecyl Sulfate on Cell Membrane and Tight Junction Permeability in Human Intestinal Epithelial (Caco-2) Cells. J. Pharm. Sci. 1993, 82, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Gartzke, D.; Delzer, J.; Laplanche, L.; Uchida, Y.; Hoshi, Y.; Tachikawa, M.; Terasaki, T.; Sydor, J.; Fricker, G. Genomic Knockout of Endogenous Canine P-Glycoprotein in Wild-Type, Human P-Glycoprotein and Human BCRP Transfected MDCKII Cell Lines by Zinc Finger Nucleases. Pharm. Res. 2015, 32, 2060–2071. [Google Scholar] [CrossRef] [PubMed]
- Gartzke, D.; Fricker, G. Establishment of Optimized MDCK Cell Lines for Reliable Efflux Transport Studies. J. Pharm. Sci. 2014, 103, 1298–1304. [Google Scholar] [CrossRef]
- Di, L.; Whitney-Pickett, C.; Umland, J.P.; Zhang, H.; Zhang, X.; Gebhard, D.F.; Lai, Y.; Federico, J.J., 3rd; Davidson, R.E.; Smith, R.; et al. Development of a new permeability assay using low-efflux MDCKII cells. J. Pharm. Sci. 2011, 100, 4974–4985. [Google Scholar] [CrossRef] [PubMed]
- Madden, E.F.; Akkerman, M.; Fowler, B.A. A comparison of 60, 70, and 90 kDa stress protein expression in normal rat NRK-52 and human HK-2 kidney cell lines following in vitro exposure to arsenite and cadmium alone or in combination. J. Biochem. Mol. Toxicol. 2002, 16, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Yuan, Y.; Zheng, J.; Hu, N. Hydrogen sulfide alleviates uranium-induced kidney cell apoptosis mediated by ER stress via 20S proteasome involving in Akt/GSK-3β/Fyn-Nrf2 signaling. Free Radic. Res. 2018, 52, 1020–1029. [Google Scholar] [CrossRef]
- Jensen, H.; Doughty, R.W.; Grant, D.; Myhre, O. The Effects of the Iodinated X-Ray Contrast Media Iodixanol, Iohexol, Iopromide, and Ioversol on the Rat Kidney Epithelial Cell Line NRK 52-E. Ren. Fail. 2011, 33, 426–433. [Google Scholar] [CrossRef] [Green Version]
- Glube, N.; Langguth, P. Caki-1 Cells as a Model System for the Interaction of Renally Secreted Drugs with OCT3. Nephron Physiol. 2008, 108, 18–28. [Google Scholar] [CrossRef]
- Glube, N.; Closs, E.; Langguth, P. OCTN2-Mediated Carnitine Uptake in a Newly Discovered Human Proximal Tubule Cell Line (Caki-1). Mol. Pharm. 2007, 4, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, S.E.; Chung, G.W.; Van Loon, E.; Bakar, N.S.; Dalzell, A.M.; Brown, C.D.A. The limitations of renal epithelial cell line HK-2 as a model of drug transporter expression and function in the proximal tubule. Pflügers Arch. Eur. J. Physiol. 2012, 464, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Simmons, N. Established human renal cell lines: Phenotypic characteristics define suitability for use in in vitro models for predictive toxicology. Toxicol. In Vitro 1993, 7, 129–136. [Google Scholar] [CrossRef]
- Toutain, H.; Courjault, F.; Vauclin-Jacques, N.; Morin, J.P. Adenylate Cyclase Responses and Biochemical Characterization of Primary Rabbit Proximal Tubular Cell Cultures and Llc-Pk1 Cells. Ren. Fail. 1990, 12, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Augustine, L.M.; Markelewicz, R.J., Jr.; Boekelheide, K.; Cherrington, N.J. Xenobiotic and endobiotic transporter mRNA expression in the blood-testis barrier. Drug Metab. Dispos. 2005, 33, 182–189. [Google Scholar] [CrossRef] [Green Version]
- Lash, L.H.; Putt, D.A.; Xu, F.; Matherly, L.H. Role of rat organic anion transporter 3 (Oat3) in the renal basolateral transport of glutathione. Chem. Interact. 2007, 170, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Prange, J.A.; Bieri, M.; Segerer, S.; Burger, C.; Kaech, A.; Moritz, W.; Devuyst, O. Human proximal tubule cells form functional microtissues. Pflug. Arch. 2016, 468, 739–750. [Google Scholar] [CrossRef]
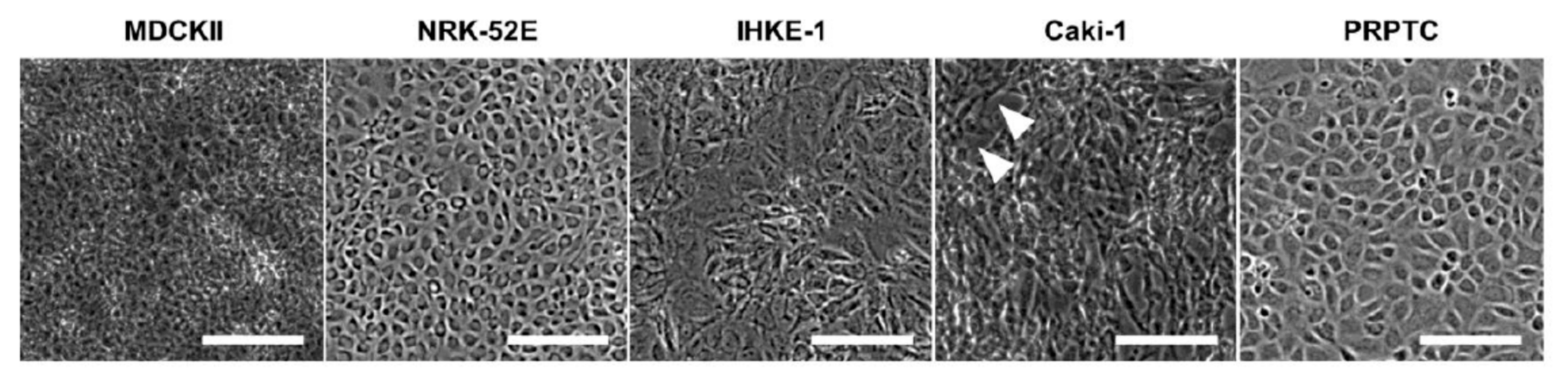


| Name | Species | Origin | Type | |
|---|---|---|---|---|
| MDCKII | Madin-Darby canine kidney II | dog | Kidney epithelium | immortalized |
| NRK-52E | Normal rat kidney-52E | rat | Kidney epithelium | immortalized |
| IHKE-1 | Immortalized human kidney epithelial-1 | human | Fetal kidney cortex | immortalized |
| Caki-1 | Carcinoma kidney | human | Kidney carcinoma | immortalized |
| PRPTC | Primary rat proximal tubule cells | rat | Proximal tubule | primary |
| Protein | Cell Lines | Primary Cells | |||
|---|---|---|---|---|---|
| MDCKII | NRK-52E | IHKE-1 | Caki-1 | Cultured PRPTCs | |
| TJP1 | + | + | + | (+) | + |
| Occludin | + | + | + | (+) | + |
| Claudin-2 | + | − | (+) | (+) | − |
| Claudin-7 | + | − | − | − | + |
| Claudin-10 | − | − | − | − | − |
| Gene | Accession Number (RefSeq) | For/Rev | Sequence (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| Abcb1a | NM_133401.1 | for | CATCTTGGCGGACCTTACGGAGG | 183 |
| rev | ACCTGCATAGCGAAACATTGTGAGC | |||
| Abcc2 | NM_012833.1 | for | GCGCCCTGGGTGACTGACAAG | 166 |
| rev | CAGTCCTGCCCACTACGCCG | |||
| Abcc4 | NM_133411.1 | for | ACAGCTTACGGCTACGCGGC | 181 |
| rev | GGCCCGTGGTTGTCTTCCCC | |||
| Abcg2 | NM_181381.2 | for | AGGCGGAGGCAAGTCTTCGTTG | 103 |
| rev | ATTGGCAGGTTGAGGTGCCC | |||
| Alpl | NM_013059.1 | for | GTGGAAGGAGGCAGGATTGACCAC | 116 |
| rev | TTCTGGGAAGTCATGGTGCCCG | |||
| Anpep | NM_031012.1 | for | TCTGCCGCTGAGCAACACCC | 135 |
| rev | AGATAGCGCTTCATGGGGCCG | |||
| Cldn10 | NM_001106058.1 | for | TGTTCCATGACGGGCTGTTCCC | 96 |
| rev | GGCAGCCCCCAATTCATACTTTTGC | |||
| Cldn11 | NM_053457.2 | for | TTCTCCCCTGTATCCGAATG | 157 |
| rev | AAGCTCACGATGGTGATCTC | |||
| Cldn2 | NM_001106846.2 | for | AGAACAGCTCCGTTTTCTAGATGCC | 160 |
| rev | TGTTCGCTTGTCTTTTGGCTGCG | |||
| Cldn7 | NM_031702.1 | for | TGGCCATGTTTGTCGCCACGATG | 126 |
| rev | CCAAAGCAGCAAGACCTGCCACG | |||
| Gapdh | NM_017008.3 | for | ATCTTCTTGTGCAGTGCCAGCCTCG | 115 |
| rev | ACTTTGTCACAAGAGAAGGCAGCCC | |||
| Ggt1 | NM_053840.2 | for | TGAGCTGGCACACCAACGGC | 169 |
| rev | CCGGCAGAACACCTCGCACA | |||
| Hk1 | NM_012734.1 | for | ACACCTGCTTTTCCGCATCCCC | 124 |
| rev | AGCCGCATGGCGTACAGATAC | |||
| Ocln | NM_031329.2 | for | CGCCTCTGGTACCTGAAGTAGCCC | 159 |
| rev | GGTCTGGGTCTGTCCTCTTCGCC | |||
| Slc15a1 | NM_001079838.1 | for | TCTCTGCCTCTCTCTCACTTGCC | 166 |
| rev | TGTGACAGAGAAGACCACCTCGCC | |||
| Slc15a2 | NM_031672.1 | for | AAGAGGAACATGCAGAGGCACGG | 81 |
| rev | ACGTGGAAATCAAGCTCCCCGC | |||
| Slc22a1 | NM_012697.1 | for | GCTGATGGAAGTTTGGCAAGCCC | 158 |
| rev | TGGCCTTTGATTTCCTCCTCCCC | |||
| Slc22a12 | NM_001034943.1 | for | AGGAACCGAACGGGAACCAAGC | 107 |
| rev | GCCAAAGGCAAACCAGCACAGC | |||
| Slc22a2 | NM_031584.1 | for | ACTGGTTGTATTTGCTGTGGTTGGC | 120 |
| rev | CTTGGCCTCTGCATATTCTCGGC | |||
| Slc22a24 | NM_173302.1 | for | TACATGGGCCGCAGAACAACCC | 99 |
| rev | ACGCGGGATCTGCATTTCTTGAGTC | |||
| Slc22a4 | NM_022270.1 | for | AGGCGGGAAGCATGAGGGACTATG | 195 |
| rev | ACGCGCTGCTCAAGTTCACG | |||
| Slc22a5 | NM_019269.1 | for | TCCTGTGGCTGACCATATCAGTGGG | 132 |
| rev | ACAGCCAGGCCAACACATAGGC | |||
| Slc22a6 | NM_017224.2 | for | CCACAGTGATTCGGCAGACAGGC | 148 |
| rev | GCACTGGCGACCACAGGGAC | |||
| Slc22a7 | NM_053537.2 | for | GGCTGTTGCTGCCCAAAGTTGC | 135 |
| rev | CCTCCTGGGTACTCTTCCTCTCCAC | |||
| Slc22a8 | NM_031332.1 | for | TTCCCAATGACACCCAGAGGGC | 101 |
| rev | GCTGCACACCAAGTCCCACTCTATC | |||
| Slc47a1 | NM_001014118.2 | for | CAGCTTCCTGAGTGGTATCCTTGGC | 139 |
| rev | TGCACCCAACGCATTTCCCACC | |||
| Slco1a1 | NM_017111.1 | for | GATGTGTGATTATGGGCCTGGGG | 157 |
| rev | TGCGTTGGCTTTAAGGTCTGTGTTC | |||
| Slco1a3 | NM_030837.1 | for | CACTACCTCATTTCCTCATGGGCCG | 139 |
| rev | ACACACTCTGCTGGGTCTTGCG | |||
| Slco4c1 | NM_001002024.1 | for | CCACGGGATTTGCCACGTTTTTACC | 89 |
| rev | AACAGCCCCTCCAAGTGTTGCC | |||
| Tjp1 | NM_001106266.1 | for | TTCAGTTCGCTCCCATGACAGGC | 132 |
| rev | TCCTCTCTGCTCCGGAGACTGCC | |||
| Vil1 | NM_001108224.2 | for | TGCCCAATCGGACCTCAGGC | 161 |
| rev | GCGCCACACCTGCACTTCCC |
| Gene | Accession Number (RefSeq) | for/rev | Sequence (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|---|
| ABCB1 | NM_000927.4 | for | CTCTTTGCCACAGGAAGCCTGAGC | 175 |
| rev | CTTCAAGATCCATTCCGACCTCGC | |||
| ABCC2 | NM_000392.3 | for | AACGCACATGAAGAGAGAGCTGC | 151 |
| rev | TTCAACATCTTCCAGGACAAGGGC | |||
| ABCC4 | NM_001105515.1 | for | ACTTCAGAGCTGGTGCTCACTGG | 111 |
| rev | TGTTTGCCCAGTATGAAAGCCACC | |||
| ABCG2 | NM_001257386.1 | for | CGATATGGATTTACGGCTTTGCAGC | 191 |
| rev | ACAATCATACAAGCCAAGGCCACG | |||
| ALPL | NM_000478.4 | for | CAGGGATAAAGCAGGTCTTGGG | 102 |
| rev | CTTTCTCTTTCTCTGGCACTAAGG | |||
| ANPEP | NM_001150.2 | for | ACAGCCAGTATGAGATGGACAGC | 104 |
| rev | CACCACCTTTCTGACATTGCCC | |||
| CLDN10 | NM_182848.3 | for | TATTCATACTGTCAGGGCTGTGC | 135 |
| rev | CTGCCCATCCAATAAACAGAGCG | |||
| CLDN11 | NM_005602.5 | for | CTGACTGTTCTTCCCTGCATCC | 101 |
| rev | CAGAGAGCCAGCAGAATGAGC | |||
| CLDN2 | NM_001171092.1 | for | TGGGAAGGCTAGGGAGTCAAAGGG | 197 |
| rev | TAGAAGACCTGAATGGCCTGTGAGC | |||
| CLDN7 | NM_001185022.1 | for | GGACAGTGGGTCGCCGAGAGC | 182 |
| rev | AAGGGTTTGGGCCAGGTGAATGC | |||
| GAPDH | NM_001256799 | unknown a | 95 | |
| NM_002046 | unknown a | 95 | ||
| GGT1 | NM_001032364.2 | for | CCCAGAAGTGAGAGCAGTTGG | 95 |
| rev | CTGCACCCGCTATGATGTCC | |||
| HK1 | NM_000188.2 | for | CAGATCGAGAGTGACCGATTAGC | 170 |
| rev | ATCTTATCCACAACCGCAGCC | |||
| OCLN | NM_001205254.1 | for | AGTTGCGGCGAGCGGATTGG | 113 |
| rev | AGGTGGACTTTCAAGAGGCCTGG | |||
| SLC15A1 | NM_005073.3 | for | ATGAAGTCGGTGCTTCAGGCAGG | 111 |
| rev | AATGTACTCGGCCCACTGTTTGC | |||
| SLC15A2 | NM_001145998.1 | for | AGCCATCTCCGACAATCTGTGGC | 186 |
| rev | AACAGAGGCTGCTGAAGGCATGG | |||
| SLC22A1 | NM_003057.2 | for | CCACATTCGTCAGGAACCTCGG | 170 |
| rev | TGGAAGAAGTAGCGTCACTCCCG | |||
| SLC22A12 | NM_144585.2 | for | ATCTCCACGTTGTGCTGGTTCGC | 129 |
| rev | CATCTTGGCTGGGATGTCCACG | |||
| SLC22A2 | NM_003058.3 | for | TAGCAGACAGGTTTGGCCGTAAGC | 144 |
| rev | AGCCTGCTTTGCTGACCAGTCC | |||
| SLC22A4 | NM_003059.2 | for | TCCATTGGTCTGGTCATGCTGGG | 188 |
| rev | AGCATTCTGTTGTAAGCACCGAGG | |||
| SLC22A5 | NM_003060.3 | for | GCCAATGGGATTGTTGTGCCTTCC | 165 |
| rev | GCCCACTGATATGGTCATCCACAGC | |||
| SLC22A6 | NM_004790.4 | for | CATCTACGGTGCTGTTCCTGTGGC | 158 |
| rev | GCGTCTGTTTCCCTTTCCTGGG | |||
| SLC22A7 | NM_006672.3 | for | TAGCTGTCACCCTGCCTTGTGC | 181 |
| rev | ACTTTGCTCACAGCCTCCTGGC | |||
| SLC22A8 | NM_001184732.1 | for | AGAAAGGCTCAGCTTGGAGGAGC | 128 |
| rev | GGGAAAGACAGAAGGTCATGCGGC | |||
| SLC22A9 | NM_080866.2 | for | CCGAGTGGGCAACACACAGATTCC | 183 |
| rev | CAGACTCTAGCAGCCAACTTGAGG | |||
| SLC47A1 | NM_018242.2 | for | AGCTTCCTCAGTGGCATCCTCG | 138 |
| rev | AGCACCCAGAGCGTTTCCTACC | |||
| SLC47A2 | NM_001099646.1 | for | CGGTGGCCTTTGTCAATGTCTGC | 190 |
| rev | ATGTGCTGGGTGTTGAGGAAGAGC | |||
| SLCO4C1 | NM_180991.4 | for | ACTGTCGCTGTCCGAGTTTGAGG | 135 |
| rev | ACTACAATACCTTGCGTGACGGC | |||
| TJP1 | NM_003257.3 | for | TGCCATTACACGGTCCTCTGAGC | 155 |
| rev | CTGCTTTCTGTTGAGAGGCTGGC | |||
| VIL1 | NM_007127.2 | for | CATTGTGGTGAAGCAGGGACACG | 154 |
| rev | TGTGACCTCAGCAGTGATCTGGC | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lechner, C.; Mönning, U.; Reichel, A.; Fricker, G. Potential and Limits of Kidney Cells for Evaluation of Renal Excretion. Pharmaceuticals 2021, 14, 908. https://doi.org/10.3390/ph14090908
Lechner C, Mönning U, Reichel A, Fricker G. Potential and Limits of Kidney Cells for Evaluation of Renal Excretion. Pharmaceuticals. 2021; 14(9):908. https://doi.org/10.3390/ph14090908
Chicago/Turabian StyleLechner, Christian, Ursula Mönning, Andreas Reichel, and Gert Fricker. 2021. "Potential and Limits of Kidney Cells for Evaluation of Renal Excretion" Pharmaceuticals 14, no. 9: 908. https://doi.org/10.3390/ph14090908
APA StyleLechner, C., Mönning, U., Reichel, A., & Fricker, G. (2021). Potential and Limits of Kidney Cells for Evaluation of Renal Excretion. Pharmaceuticals, 14(9), 908. https://doi.org/10.3390/ph14090908







