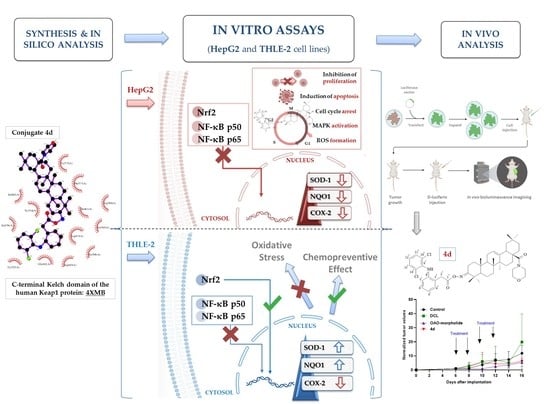
1. Introduction
Hepatocellular carcinoma (HCC) remains the most frequent primary liver cancer and is a leading cause of cancer-related deaths. Chronic inflammation resulting mainly from viral hepatitis or metabolic disorders is considered the major risk factor of HCC [
1]. Limited therapeutic options, particularly in the case of advanced forms of HCC, make necessary the development of new therapeutic and/or preventive strategies [
2]. The key roles in the induction of inflammation involve two signaling pathways: Nrf2-ARE and NF-κB. While activation of the latter leads to transcription of genes encoding pro-inflammatory proteins, including COX-2 and iNOS enzymes, Nrf2 is responsible for cell cytoprotection and is the major cellular defense against reactive oxygen (ROS) and electrophilic species [
3,
4]. Overexpression of COX-2 associated with increased cell growth and invasiveness is seen in human HCC [
5]. Therefore, targeting NF-κB activation is one of the possible therapeutic approaches. Usually, reduced activation and expression of NF-κB results in the opposite effect in the Nrf2 signaling pathway [
6]. However, it has also been demonstrated that Nrf2, due to genetic and epigenetic alterations, is overexpressed in cancer cells, including HCC and may lead to an enhanced invasiveness potential and chemo-and radio-resistance [
7].
Activation of both NF-κB and Nrf2 requires their translocation into the nucleus. Nrf2 activators enhance its binding to the Kelch-like ECH-associated protein-1 (Keap1), inducing a conformational modification that renders Keap1 incapable of promoting Nrf2 degradation and thus allowing its translocation [
8]. NF-κB is sequestered in the cytoplasm through an association with its inhibitor, the IκB protein. Its activation occurs after phosphorylation by IκB kinase beta (IKK), resulting in degradation of the inhibitory subunit, enabling released NF-κB heterodimers to migrate into the cell nucleus to bind to a specific sequence encoding target genes [
3].
Considering the inflammatory backgrounds of cancers such as HCC, nonsteroidal anti-inflammatory drugs (NSAIDs) are said to play a role in cancer treatment and chemoprevention. Most of these drugs act as COX-2 inhibitors, which besides their beneficial effects, i.e., decreasing the risk of certain types of cancer, are well-known for many unfavorable side effects [
9].
Our earlier studies showed that oleanolic acid oxime (OAO) derivatives are potent modulators of Nrf2 and NF-κB in HCC derived HepG2 cells [
10]. Moreover, conjugation with indomethacin (IND) improved that modulating impact, leading to a downregulation of these pathways. Significantly, treatment with those conjugates decreased activation of Nrf2 and NF-κB and expression of their active forms in HepG2 cells. In contrast, in normal hepatocytes, activation of Nrf2 increased, and that of NF-κB was diminished. Hence, conjugation of IND with OAO derivatives may preserve cancer cells against chemoresistance by inhibiting the Nrf2-ARE and NF-κB pathways, whilst at the same time exerting a chemopreventive impact in normal hepatocytes [
11].
Diclofenac (DCL) is another commonly used NSAID, which displays various effects on the immune system, the angiogenic cascade, chemo- and radio-sensitivity, and tumor metabolism. While it is a non-selective inhibitor of both isoforms of the cyclooxygenase enzyme (COX-1 and COX-2), DCL has a preferred binding to COX-2 [
12], which may explain its intermediate risk profile for gastrointestinal events in comparison to other NSAIDs [
13]. Its broad-spectrum of action makes DCL one of the most potent NSAIDs in the context of cancer treatment. So far, its effect on HCC has not been well documented. Still, the co-administration of DCL and sorafenib significantly increased Bax protein expression levels leading to an increased frequency of apoptosis in HepG2 cells [
14]. Moreover, a more recent study showed that DCL potentiated sorafenib-based treatment in hepatocellular cancer cells by enhancing oxidative stress [
15].
Therefore, this study aimed to design and evaluate the effect of the newly synthesized conjugates of previously investigated OAO derivatives with DCL (
Figure 1) on Nrf2 and NF-κB signaling in connection with cell cycle distribution, apoptosis, and proliferation in normal hepatocytes, and HCC cell line and tumor growth in vivo.
2. Results
2.2. Spectral Characteristics of the Oleanolic Acid Oximes and Their Conjugates with Diclofenac
In the IR spectra of conjugates 4a–4d, the presence of the NH group was confirmed by the presence of an absorption band located at about 3380 cm−1. The presence of aromatic systems within the DCL moiety was confirmed by the presence of an absorption band located at 3325–3335 cm−1. The signal observed at ν 1735–1740 cm−1 was assigned to the C=O group within the –COO function of the diclofenac moiety. The next characteristic absorption band, observed at ν 1720–1725 cm−1 was assigned to –N=C-3 of the acyloxyimino function.
The NMR analysis of conjugates 4a–4d confirmed the structures of these compounds. In the 13C NMR spectra from the above conjugates, the signals of a double chlorinated aromatic ring within the DCL moiety (C6H3Cl2-NH-C6H4-CH2-COON=C-3) were present at about δ 142 ppm (Cq, signal intensity: 2C), 138 ppm (Cq, intensity 1C), 131 ppm (CH, intensity 2C), and 124 ppm (CH, intensity 1C). The signals derived from the second aromatic ring within the DCL moiety (C6H3Cl2-NH-C6H4-CH2-COON=C-3) were observed at about 129 ppm (2xCH, both of 1C intensity), 128 ppm (Cq and CH, both of 1C intensity), 124 ppm (Cq, intensity: 1C), and 118 ppm (CH, also of intensity 1C). The acyl carbon (C6H3Cl2-NH-C6H4-CH2-COON=C-3) gave a signal present at about δ 176 ppm. The last carbons of the diclofenac system (C6H3Cl2-NH-C6H4-CH2-COON=C-3) formed signals present at about δ 60 ppm.
In the 1H NMR spectra from the conjugates 4a–4d, the protons attached to the double chlorinated aromatic (C6H3Cl2-NH-C6H4-CH2-COON=C-3) formed two singlets, both of intensity 1H, observed at δ 7.36–7.34 ppm. For the derivative 4c (benzyl ester), these signals were partially covered by signals from the aromatic ring of the benzyl moiety, so, in total, the signals from both aromatic systems (C6H3Cl2-NH-C6H4-CH2-COON=C-3 and -COOCH2C6H5) were observed in the form of a multiplet located at δ 7.41–7.31 ppm. The next signal derived from the lat proton attached to the double chlorinated aromatic ring was observed as a triplet of a doublet, at δ 7.13–7.14 ppm. Four protons of the second aromatic ring within the diclofenac moiety (C6H3Cl2-NH-C6H4-CH2-COON=C-3) formed a doublet of doublets (δ of about 7.3 ppm), two triplets (δ of about 6.96–7.01 ppm), and one doublet, present at a δ of about 6.6 ppm. The signal derived from the NH group was observed as a singlet, located at a δ of about 6.9 ppm. The protons of the –CH2– group within the DCL moiety (C6H3Cl2-NH-C6H4-CH2-COON=C-3) formed a singlet of intensity 2H, observed at a δ of 3.96–3.98 ppm. All signals confirming the presence of an oleanolate skeleton and characteristic functional groups were present in the 1H and 13C NMR spectra of the conjugates 4a–4d (with a diclofenac moiety).
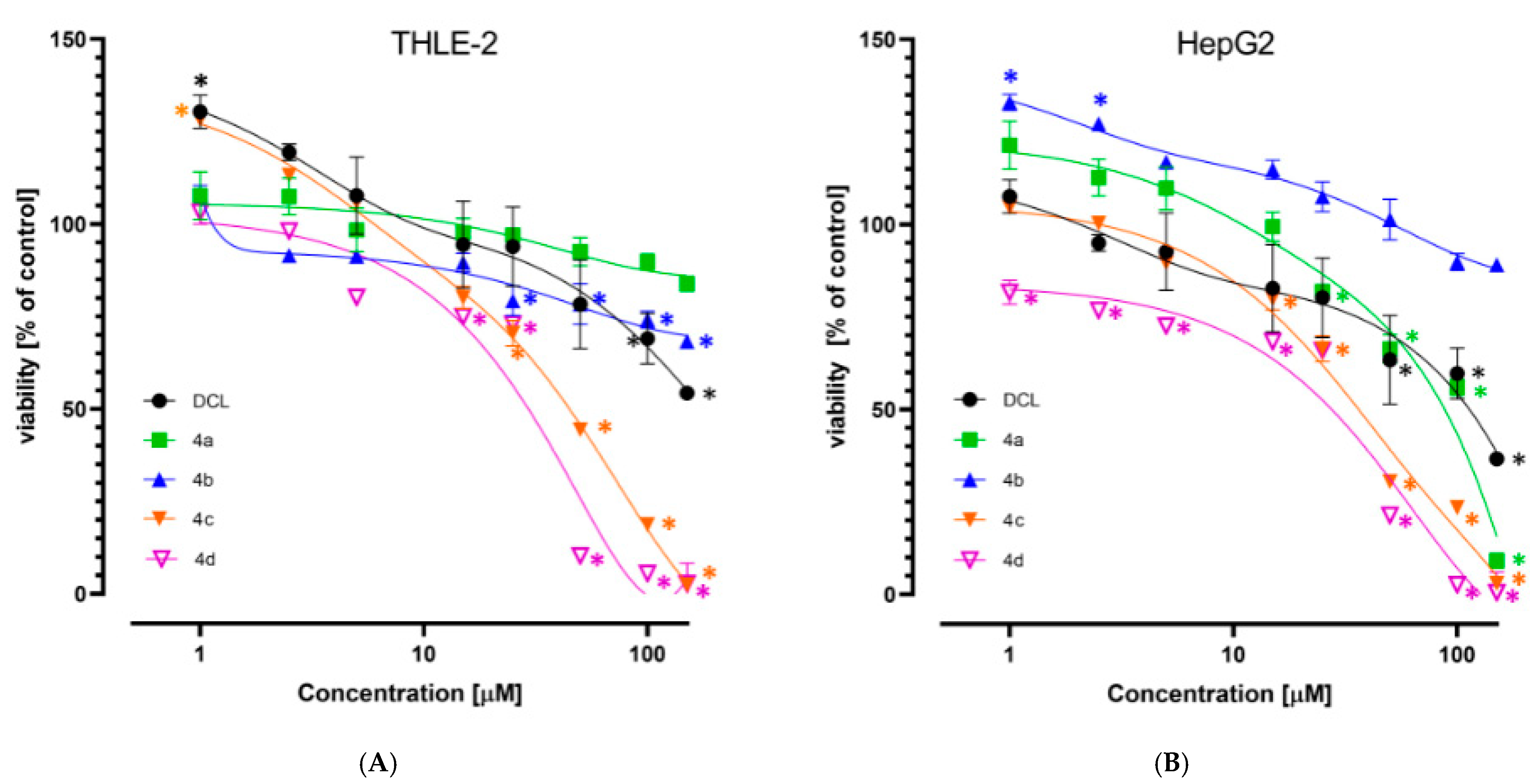
2.3. Conjugation with OAO Derivatives Increases the Cytotoxicity of Diclofenac
The viability of the immortalized hepatocytes line THLE-2, and HCC derived HepG2 cells was assessed after treatment with DCL or its conjugates at a concentration range of 1–150 µM. As shown in
Figure 2A,B, DCL and conjugates (
4a) and (
4b) at this concentration range assured ~70% viability of both cell types, exhibiting a slightly higher cytotoxicity toward the HepG2 cells. Compounds (
4c) and (
4d) were more cytotoxic, as indicated by the calculated IC
50 values (
Table 1). HepG2 cells were slightly more susceptible than THLE-2 to the cytotoxic activity of the tested compounds. Based on the MTT assay results, the 10 µM and 20 µM concentrations of DCL and its conjugated OAO derivatives were applied in further assays.
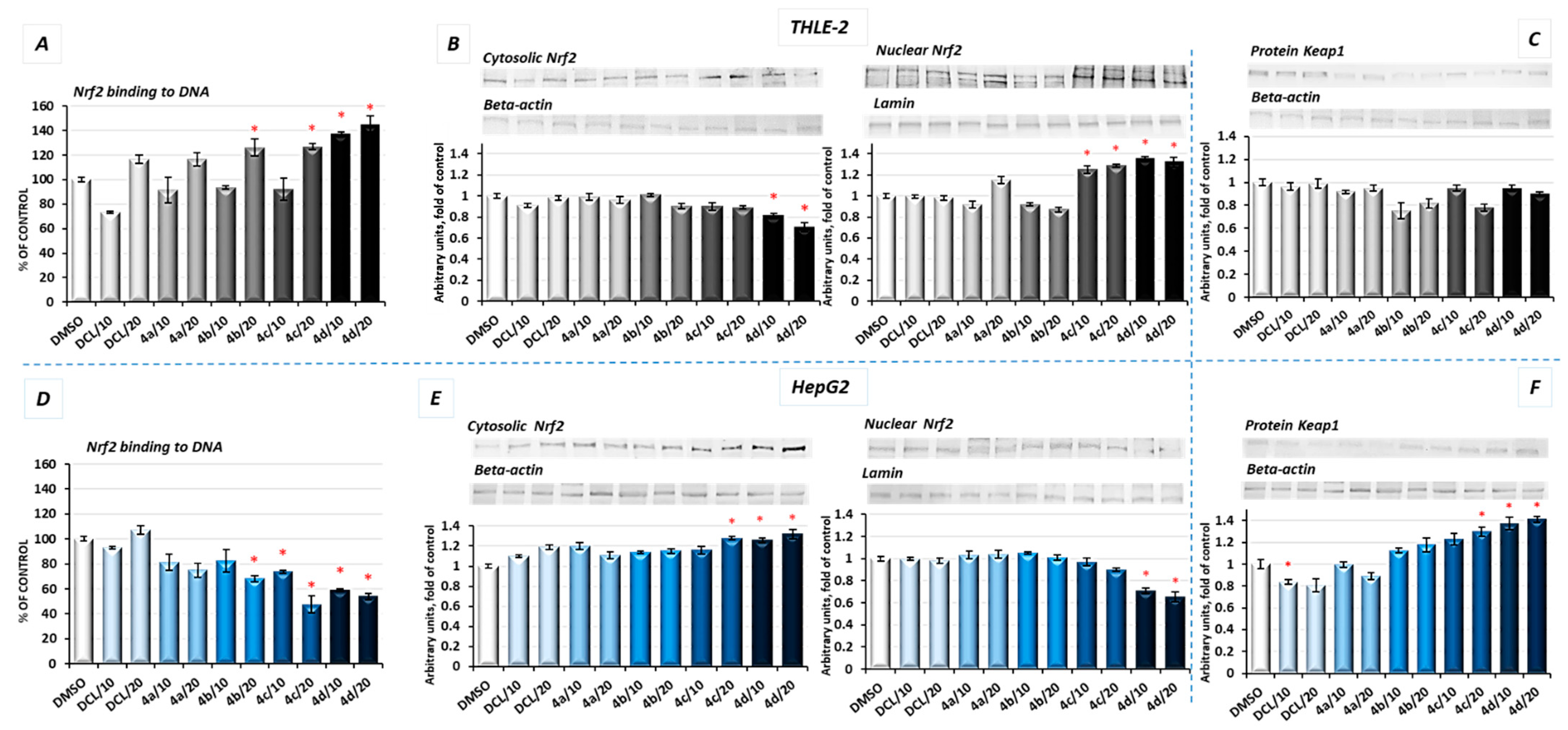
2.4. Conjugates of Diclofenac Differently Affects the Nrf2 Activation in THLE-2 and HepG2 Cells and Expression of Nrf2 Target Genes
The activation of Nrf2 was evaluated based on the amount of Nrf2 contained in the DNA binding complex to the ARE sequence and the translocation of its active form into the nucleus. The oligonucleotides containing the ARE consensus-binding site (5′-GTCACAGTGACTCAGCAGAATCTG-3′) for Nrf2 were immobilized on microplates as bait.
As shown in
Figure 3A, in THLE-2 cells, all conjugates except (
4a) in a higher concentration (20 µM) increased the Nrf2 binding level. The most pronounced effect, an increase in the binding level by ~43%, was found in the case of DCL conjugated to the OAO morpholide derivative (
4d). In HepG2 cells, a significant decrease in the Nrf2 binding level was observed as a result of treatment with methyl ester (
4b), benzyl ester (
4c), and morpholide (
4d) derivatives by ~32%, ~52%, and ~46%, respectively, at the higher concentrations (
Figure 3D).
The increased activation of Nrf2 in the THLE-2 cells and decrease in HepG2 cells resulting from the treatment with DCL–OAO conjugates were further confirmed by Nrf2 translocation from the cytosol to the nucleus (
Figure 3B,E). Again, conjugates (
4c) and (
4d) were the most effective modulator of Nrf2 translocation reducing its level in the cytosol and increasing in the cell nucleus. The level of Keap1 protein decreased, but not significantly in THLE-2 cells (
Figure 3C). However, in cancer cells, a ~40% increase in the cytosolic Keap1 protein level as a result of treatment with the compound (
4d) was observed.
Activation of Nrf2 initiates the transcription of a battery of cytoprotective genes, including those encoding NAD(P)H: quinone oxidoreductase 1 (NQO1) and superoxide dismutase (SOD) [
8].
As a result of Nrf2 activation by the conjugate (
4d), significantly increased protein levels of SOD-1 and NQO1 in immortalized hepatic normal cells were observed (
Figure 4A,C).
In contrast, in the HepG2 cells, the same conjugates, i.e., (
4d) and (
4c), reduced the protein level of SOD-1 along with its gene transcript (
Figure 4B). The NQO1 expression was significantly diminished at the mRNA level after treatment with the (
4c) and (
4d) derivatives. The methyl ester derivative (
4b) also reduced the NQO1 protein level (
Figure 4D). DCL did not show any significant effect on Nrf2 activation neither in THLE-2 nor HepG2 cells.
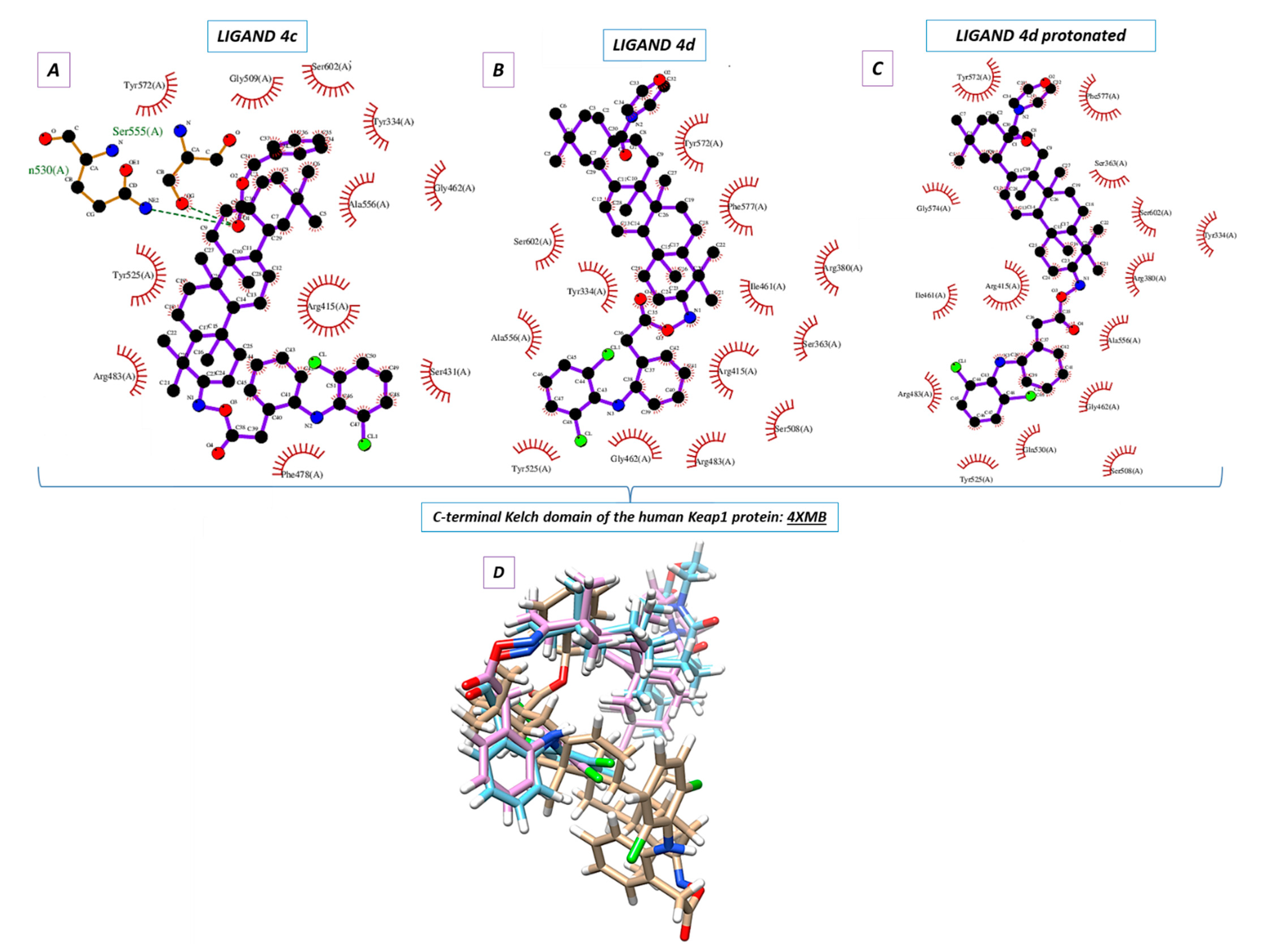
2.5. Molecular Docking
Molecular docking was used to elucidate whether the influence of the DCL–OAO derivative conjugates on Nrf2 activation was due to a specific interaction with the Keap-1 protein. The most active conjugates, (
4c) and (
4d), were the subject of the molecular docking. Additionally, the protonated version of the (
4d) ligand was analyzed (marked as 4d_H with the protonated nitrogen atom within the morpholine system). For this purpose, the small ligand-binding C-terminal Kelch domain of the human Keap1 (PDB entry: 4XMB) was selected based on the literature data [
19,
20].
We focused on the global chemical reactivity descriptors, providing insights into the chemical reactivity and stability of molecules [
21,
22]. Therefore, we considered the neutral compounds (
4c) and (
4d), and from this perspective, electronegativity (χ), chemical hardness (η), first ionization potential, and electronic potential were especially interesting. The equations relating to these parameters have been described in our previous paper [
23]. Computations at the B3LYP/6-311G++(d, p) level of theory proved that the mentioned descriptors were identical and equaled: χ = 3.40 eV, η = 2.40 eV, first ionization potential = 5.80 eV and electronic potential = −3.40 eV both for (
4c) and (
4d) derivatives. The results allowed us to assume which one of the analytes could be more potent in the chemical environment from the standpoint of their electronic nature. It was shown that (
4c) and (
4d) were able to interact similarly. Since the mutual ligand-amino acid contacts are crucial for determining biological activity, we concluded that particular amino acids forced these derivatives to occur in different orientations within the protein cavity during the docking protocol.
Nine poses were obtained for each of the DCL–OAO ligands from which the first poses had the lowest negative value for the binding affinity (
Table 2). Analyzing the previously optimized ligands (Gaussian 16 C.01 program [
24]) docked to the 4XMB.pdb protein, we noticed that the ligands (
4d) and
4d_H nearly overlapped (
Figure 5B,C). The compound (
4c) was oriented almost perpendicular to its morpholide analogs (
4d) and
4d_H. The resulting docking score (
Table 2) pointed to the morpholide derivative (
4d) and its protonated analog to be more potent and able to interact with the chemical environment within the cavity. Based on the literature data, the most interacting amino acid residues surrounding this cavity were selected [
19,
25,
26].
In the docking procedure, we considered the distance
d ≤ 4 Å between a proton and a heteroatom of the adjacent molecule (
Table 3,
Figure 5D) as an important factor that allows the hydrogen bonds to be formed. Thus, fitting the first poses of the ligands (
4c), (
4d), and
4d_H resulted in the formation of several bonds between the DCL–OAO derivative conjugates and amino acids within the cavity of the C-terminal Kelch domain of the Keap1 protein. Apart from the (
4c) conjugate, other derivatives accepted an H-bond from the
N-H guanidine nitrogens of Arg415 to the nitrogen atom of the morpholide (
4d) ligand and its protonated analog. Interaction of all analytes with Arg415 was additionally supported by contact between the chlorine atom of the DCL–OAO derivative conjugates and the N-H moiety of the arginine. Considering the distance
d ≤ 4 Å, we observed that shorter contacts of the carbonyl group within the (
4c) with Gln530 (2.269 Å) and Ser555 were not detected in the case of the morpholide analogs. On the contrary, the ligands (
4d) and
4d_H were able to interact with Arg415 (contact N
…H-N), Gly462 (contact C-H
…N), Gln530 (contact C-Cl
…H-N), or Ser555 (contact C-Cl
…H-O). Moreover, the morpholide compound’s protonation allowed forming its interaction with the hydroxyl functionality of Tyr572. Furthermore, we can assume that the Arg415 and Gln530 seem to be significant for the ligand-protein hydrogen bond formation process, and therefore for the biological response.
Based on the data from the docking protocol, ligands and residues involved in hydrogen bond formation were optimized (
Sections S4 and S4.1, Supplementary Materials, Tables S2–S4) to extend the in silico studies. In Summary, Arg415 and Gln530 were crucial for hydrogen bond formation in terms of the ligand-protein complex (4XMB.pdb) interactions of the analyzed ligands in the ligand cavity (
Sections S4 and S4.1, Supplementary Materials). The additional SAPT (symmetry-adapted perturbation theory) analysis of the ligand-amino acid complexes (
Sections S4 and S4.2, Supplementary Materials, Table S5) was performed, and the interaction energy was estimated (
Sections S4 and S4.3, Supplementary Materials, Table S6). As a result, it turned out that a more negative value of the total energy SAPT0 was obtained in the case of the derivative (
4d) (
Sections S4 and S4.2, Supplementary Materials), pointing it to be more potent for the interaction with the biological target. This conclusion was also supported by the heat of formation (HOF) computations (Mopac 2016 program and the Mozyme module [
27]). The data obtained regarding the enthalpy of interactions agreed with the binding affinity estimation during the docking procedure (
Sections S4 and S4.3, Supplementary Materials).
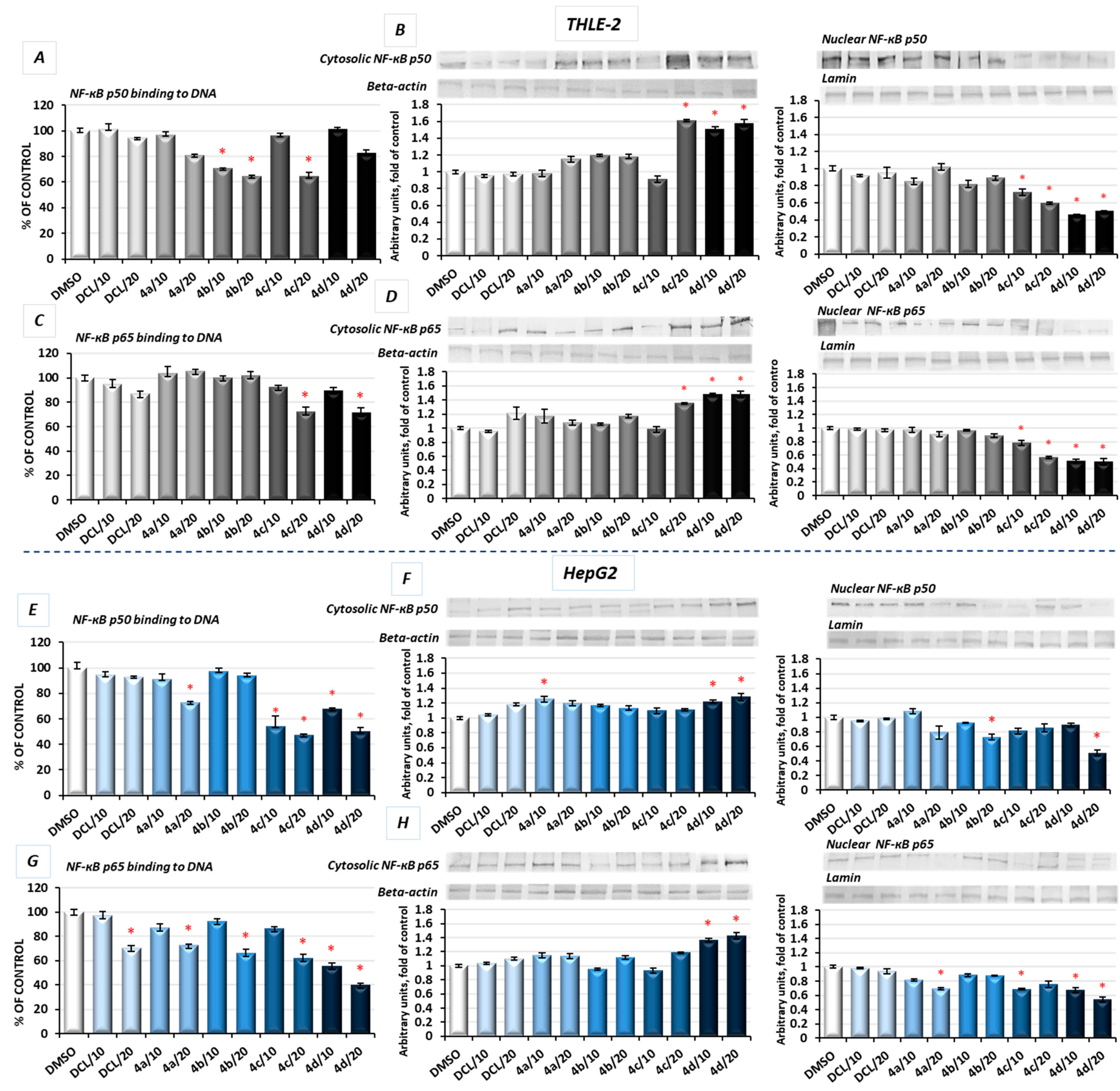
2.6. DCL–OAO Conjugates Diminish the Activation of NF-κB and COX-2 Expression in Normal THLE-2 Cells and Cancer HepG2 Cells
In unstimulated conditions, NF-κB is sequestered in the cytoplasm in association with its inhibitor, the IκB protein. Its activation takes place after phosphorylation of the IκB by IκB kinase beta (IKK), leading to a degradation of the inhibitory subunit, allowing free NF-κB heterodimers to migrate into the cell nucleus and to bind specific sequences of the target genes. The NF-κB generally refers to a p50-p65 heterodimer, depicting the major Rel/NF-κB complex in cells [
28]. Accordingly, the effect of the DCL conjugates on NF-κB activation was evaluated based on the binding of its p50 and p65 subunits to their immobilized consensus site and the translocation of the activated complex from the cytosol into the nucleus.
As shown in
Figure 6A, in THLE-2 cells, treatment with the DCL–OAO methyl ester (
4b) and benzyl ester (
4c) derivative conjugates significantly diminished (by 29–36%) the content of the NF-ĸB p50 subunit in the DNA-binding complex. However, translocation of this NF-ĸB p50 subunit into the nucleus was the most affected by the DCL–OAO morpholide derivative (
4d) (
Figure 6B). Similarly, binding of the p65 subunit in concert with an increased level of its protein in the cytosol and a decrease in the nucleus was observed as a result of treatment with the DCL–OAO morpholide derivative (
4d), and to a lesser extent the DCL–OAO benzyl derivative conjugate (
4c) (
Figure 6C,D).
DCL did not change the level of binding to DNA or translocation of both the NF-κB subunits in the THLE-2 cells.
Basically, a similar pattern was observed in the HepG2 cells (
Figure 6E–H). The compounds (
4d) and (
4c) diminished the most significantly with binding of both NF-κB subunits to DNA. However, this effect was more pronounced in the case of the p65 subunit (up to a ~60% reduction as a result of treatment with (
4d)) (
Figure 6G). These observations were confirmed by the diminished translocation of both subunits into the nucleus (
Figure 6F,H).
DCL at a concentration of 20 µM reduced the content of p65 in the DNA-binding complex but did not affect its translocation.
COX-2 is one of the NF-κB target genes and is often overexpressed in HCC [
5]. Therefore, as a result of diminished activation of NF-κB in both tested cell lines by the compound (
4d), a reduced level of COX-2 protein was observed. In HepG2 cells, the mRNA level of the
COX-2 was significantly decreased after treatment with all DCL–OAO derivative conjugates, similarly to its protein level, and especially after treatment with the (
4d) conjugate (up to ~55% reduction) (
Figure 7B).
These data indicate that the DCL–OAO conjugates, especially morpholide, are more efficient inhibitors of NF-ĸB in normal immortalized hepatocytes and human hepatocellular cancer cells than DCL, which, at the same concentration, showed an insignificant effect or none.
2.7. Conjugates of Diclofenac and OAO Derivatives Diminish the Transcription of Nrf2, NF-κB p50, and p65 in HepG2 Cells
The evaluation of the transcript levels of the Nrf2 and NF-ĸB subunits in HepG2 cells was performed to determine if the DCL–OAO conjugates may affect their expression. As shown in
Figure 7, only the conjugates (
4c) and (
4d) reduced the Nrf2 and NF-ĸB p50 mRNA levels, while (
4b) and (
4d) significantly decreased the p65 mRNA level. DCL alone tended to increase Nrf2 transcript levels and decrease both NF-ĸB subunits, but the observed differences in comparison with untreated control cells were not statistically significant. These results indicate that DCL–OAO conjugates, particularly with the benzyl and morpholide groups at the C-17 position, not only diminished the activation of key signaling pathways involved in HCC development but also affected the expression of the transcription factors participating in the final stages of these pathways.
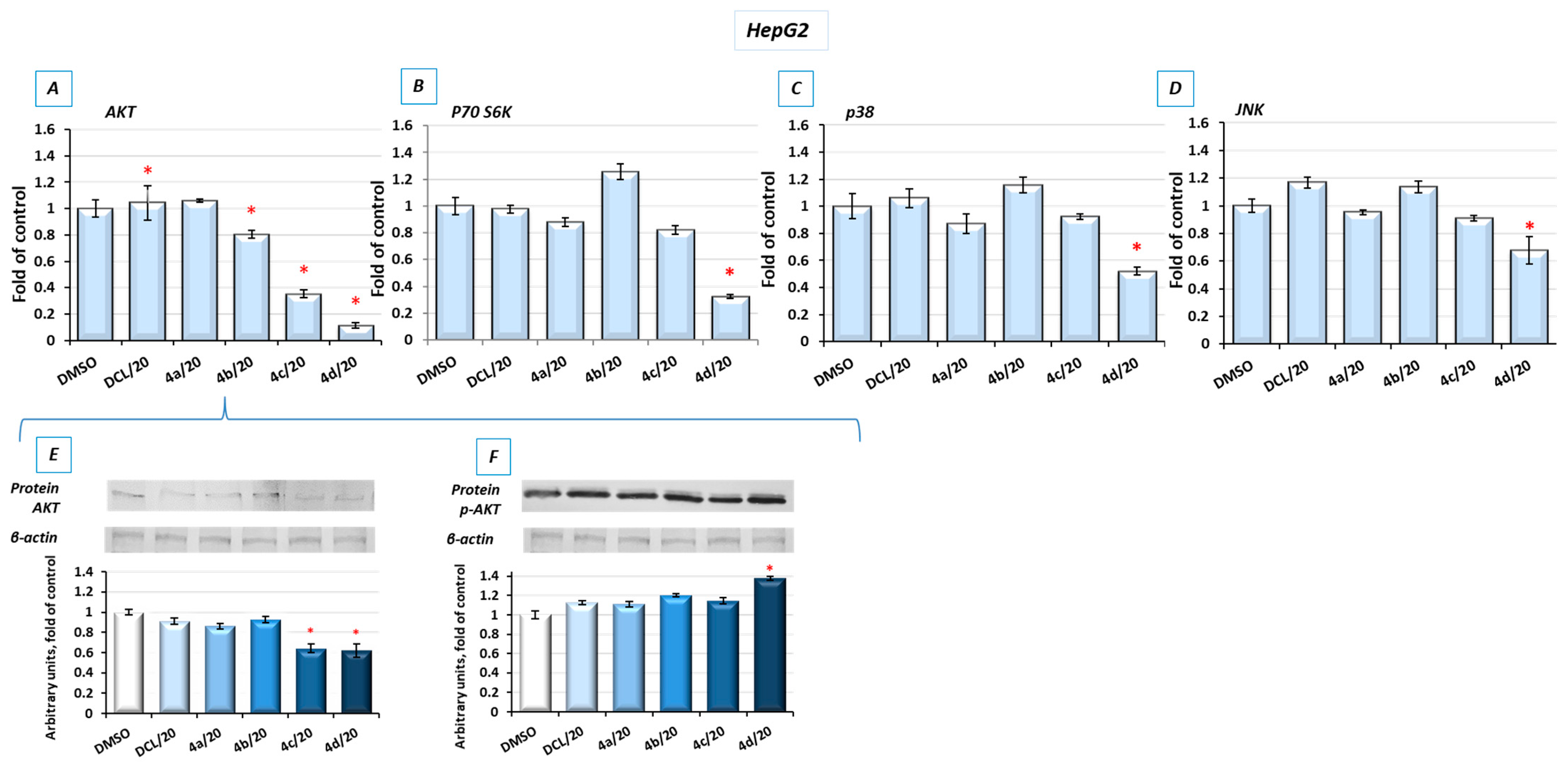
2.8. Bead-Based Multiplex Immunoassay Revealed Possible Downregulation of Protein Regulating Several Signaling Pathways
To look into possible interferences of the tested conjugates with the other signaling pathways involved in cell survival, the bead-based multiplex immunoassay was applied. As shown in
Figure 8, conjugates (
4d) and, to a lesser extent, (
4c) reduced the level of kinases from the MAPK family, p38 and JNK, AKT, and P70S6K involved in multiple cellular processes, including apoptosis, cell proliferation, and protein synthesis [
29], respectively. For verification, the results of the bead-based multiplex immunoassay, and a Western blot analysis of AKT and p-AKT were performed (
Figure 8E,F). The reduction in AKT levels was also noticeable, although the observed changes were not as significant as with the use of the bead-based immunoassay. The level of phosphorylated AKT (p-AKT) in this assay was slightly increased.
2.9. Conjugates of Diclofenac and OAO Derivatives Change the Cell Cycle Distribution, Induce Apoptosis and Affect Cell Proliferation in HepG2 Cells
Figure 9A shows the effect of the DCL–OAO derivatives on the cell cycle distribution in the HepG2 cells. Conjugates of DCL with OAO derivatives substituted with benzyl (
4c) and morpholide (
4d) groups increased the number of cells significantly in the G2/M and subG1 phases. They also decreased the number of cells in the G0/G1 phases (
Figure 9A). However, compared to the reference compound, topotecan, which at low concentrations induces cell cycle arrest in S and G2/M phases, the effect of the DCL–OAO derivative conjugates was much weaker. DCL, similarly to compounds (
4a) and (
4b), did not affect the cell cycle distribution.
Treatment with the DCL–OAO derivatives (
4a) and (
4b) increased the percentage of early and late apoptotic cells to an extent comparable to DCL. However, in the case of the (
4c) and (
4d) conjugates, the proapoptotic effect exceeded even that observed after treatment with topotecan. The highest percentage of total apoptotic cells after treatment with (
4c) and (
4d) was marked by ~67% and ~77%, respectively, compared to the results from the DMSO-treated cells (
Figure 9B).
The pro-apoptotic effect of these compounds was further confirmed by an increased level of the Bax and Caspase-3 proteins.
The effect of the DCL conjugates on cell proliferation was evaluated based on the Ki67 protein expression, allowing the assessment of the cell’s proliferation rate. The most pronounced anti-proliferative effect among the tested derivatives was observed, similarly as in the previous assays, as a result of treatment with the compounds (
4c) and (
4d). In this regard, the highest percentage of resting cells (43.5%) (exhibiting the lowest Ki67 expression) was found after treatment with the (
4d) derivative at its higher concentration. The percentage of resting cells treated with these compounds exceeded the amount observed in the starved cells lacking FBS in DMEM (
Figure 9C).
Reduced activation of both transcription factors, NF-κB, which may enhance anti-apoptotic gene expression, and Nrf2 controlling genes whose products are involved in cell proliferation and differentiation may be responsible for the cell cycle arrest and induction of apoptosis by the DCL–OAO derivative conjugates.
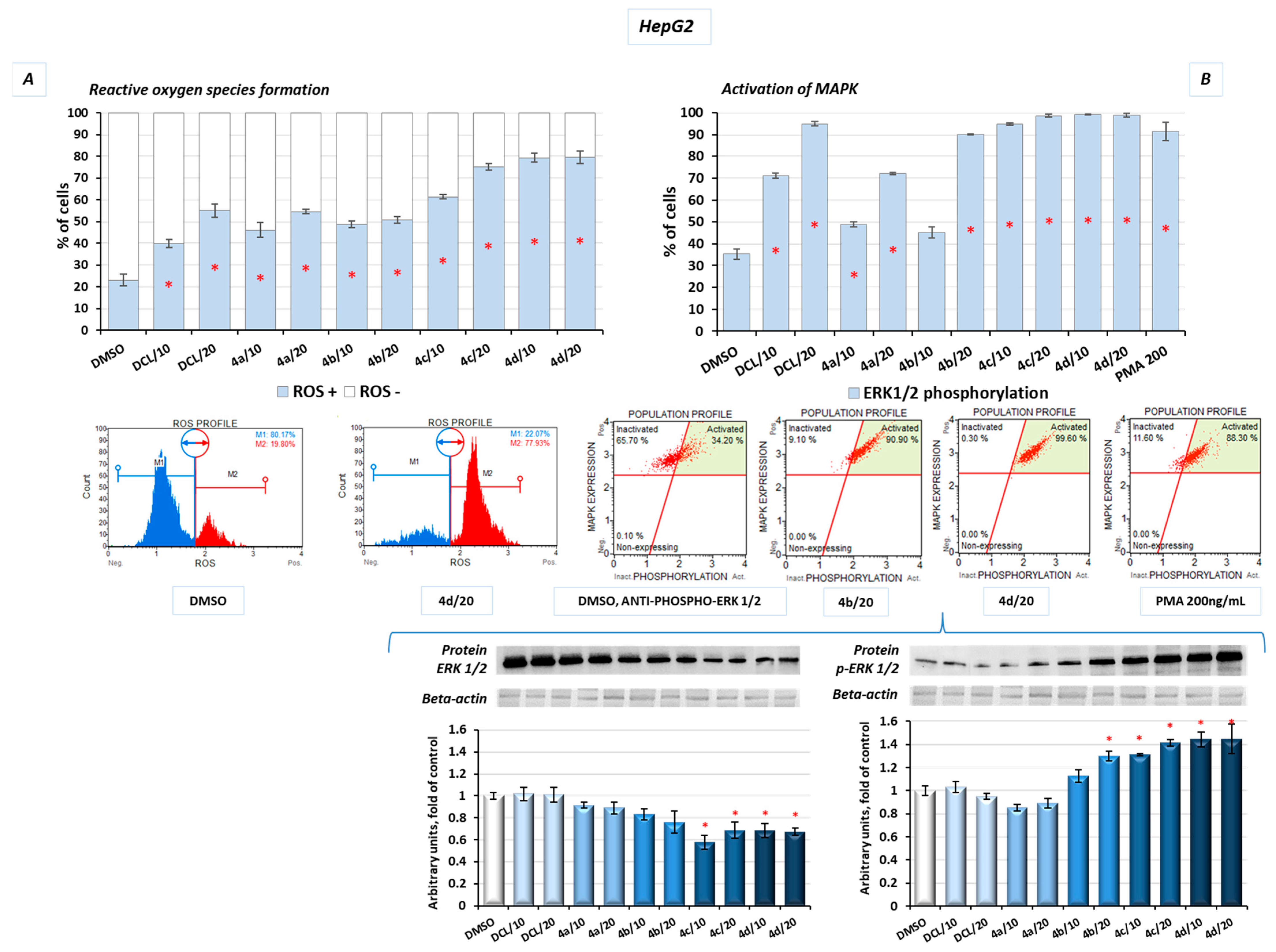
2.10. Conjugates of Diclofenac and OAO Derivatives Affect the Phosphorylation of ERK and Formation of ROS in HepG2 Cells
Additionally, in the HepG2 cells, after treatment with the DCL–OAO derivative conjugates, the levels of reactive oxygen species (ROS) were assessed. Induction of oxidative stress was dose-dependent, with the most significant increase in ROS levels resulting from treatment with the derivatives (
4c) and (
4d) at a concentration of 20 µM, by ~57% and ~60%, respectively, compared to the DMSO-treated cells (
Figure 10A).
The mitogen-activated protein kinases (MAPK) family, including extracellular signal-regulated kinases (ERK), is known to be directly activated by the ROS and regulates the growth and survival of cancer cells. MAPK pathways are frequently deregulated in human hepatocarcinogenesis. Increased ERK phosphorylation relating to ROS production was observed as a result of treatment with the conjugates tested in this study. The results confirmed that prolonged ERK activation requires the presence of the ROS, and almost the whole population of the HepG2 cells presented MAPK signaling activation (increased phosphorylation of ERK 1/2) in flow cytometric analysis after treatment with the DCL–OAO derivatives with benzyl ester and morpholide at both tested concentrations (
Figure 10B). A similar increase was obtained in the protein levels, where we observed a ~30% and ~40% increase of p-ERK after treatment with the conjugates (
4c) and (
4d), respectively (
Figure 10B).
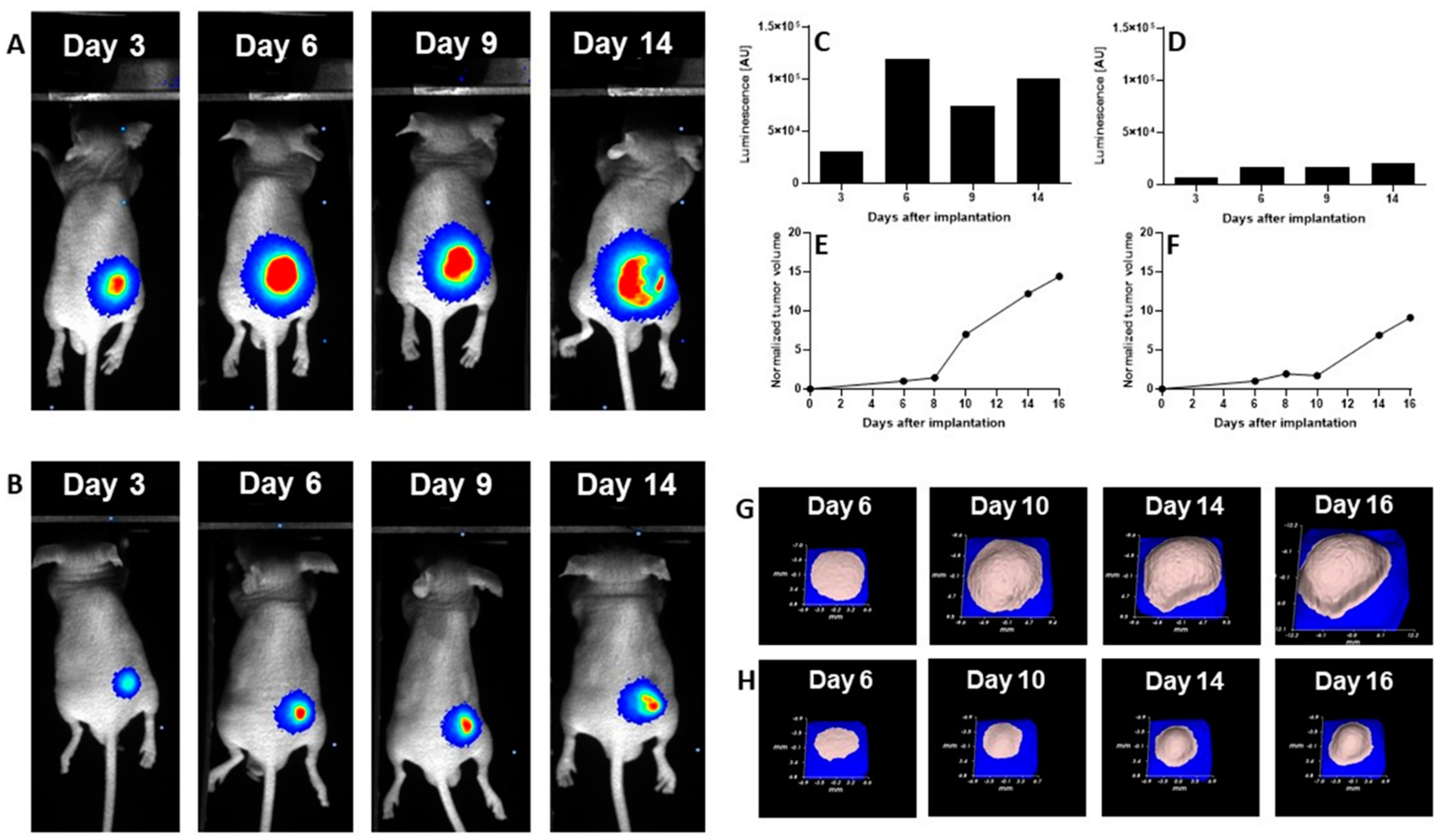
2.11. Antitumor Efficacy of DCL–OAO Derivatives in Human Hepatic Tumor Xenograft Growth
We evaluated the anti-cancer efficacy of the DCL–OAO derivative conjugates in nude mice bearing HepG2 tumor xenografts. As shown in
Figure 11, DCL did not significantly affect the tumor growth compared to the untreated mice. At sacrifice, tumor volumes were significantly reduced by the OAO morpholide derivative and to a lesser extent by its conjugation to DCL. No significant difference in mice weights at sacrifice was observed between the different treatment groups.
It should be noted that
Figure 11 shows the number of animals at the end of the experiment. At the beginning, each group included eight mice. Unfortunately, the treatment with two tested compounds (OAO morpholide and
4d) was toxic for the animals. The tendency to develop signs of necrosis in certain xenografts impeded the measurement of the luminescence signal. However, the obtained data confirmed the results of the biometric measurements (
Figure 12).
3. Discussion
Diclofenac, a widely used NSAID, has shown several biological activities which might be useful in cancer therapy or prophylaxis [
13]. Moreover, recent studies showed that in combination with other drugs such as sorafenib, diclofenac displayed greater antiproliferative efficacy in HCC cells and anti-tumor activity in vivo in comparison to DCL or sorafenib alone [
15]. Our study focused on the evaluation of synthetic hybrids of DCL with OAO derivatives in the context of modifications to signaling pathways critical for cell survival. The results showed that the conjugation of DCL, particularly with OAO substituted with morpholide in C-17 (
4d), significantly increased toxicity toward both cancer HepG2 and immortalized non-tumor THLE-2 cells. However, in the THLE-2 cells, this effect was slightly less pronounced.
Indeed, as demonstrated in our earlier study, OAO derivatives, particularly OAO- morpholide, showed cytotoxic effects in HepG2 cells and to a lesser extent in immortalized normal hepatocyte THLE-2 cells [
10].
It is worth noting the marked differences between the ASP, IND, and DCL conjugates on the parameters evaluated in the hepatic cells. Therefore, for DCL, similarly, as for the other NSAIDs studied so far, conjugation with the OAO derivatives is certainly an advantage (e.g., decreased IC50 values) and might improve the DCL therapeutic potential.
It should be pointed out that at the lowest concentrations an increased cell proliferation was observed indicating a hormetic effect [
30]. Such an effect might be related to a dysfunction in redox homeostasis and an initial compensation reaction in the presence of the lowest concentration of compounds, particularly those with antioxidant potential [
31].
One of the reasons leading to cell death may be an excessive generation of ROS. Indeed, in HepG2 cells, the ROS levels were significantly increased as a result of treatment with all tested compounds, but the highest percentage of ROS-positive cells (>80%) was found in the case of the most cytotoxic—(4c) and (4d)—conjugates.
The results of an earlier analysis of the transcriptome in primary human hepatocytes revealed that a majority of intrinsic hepatotoxic drugs activated the Nrf2 transcriptional program. Furthermore, strong Nrf2 activation corresponded with the downregulation of genes under the direct control of NF-κB [
32]. This mechanism may explain the cytotoxicity of the DCL–OAO derivative hybrids (
4c) and (
4d) toward the THLE-2 cells, in which increased activation of Nrf2 and subsequently increased expression of its target genes was observed with concomitant reduced activation of NF-κB. This cell line represents the immortalized human liver epithelial (non-tumor) cells characterized by an overall higher metabolic activity in comparison to the HepG2 cells. Besides, although they have normal hepatic epithelial cell morphology, their features make them a good model of liver tumor promotion and chemoprevention [
33]. The activation of Nrf2 in these cells as a result of treatment with potential chemopreventive agents such as betanin was found in our earlier studies [
34].
HepG2 cells are derived from hepatocellular carcinomas and represent the later stages of hepatocarcinogenesis. In these cells, the conjugates (
4c) and (
4d) decreased the Nrf2 activation in contrast to the OAO-derivatives alone observed in our earlier studies [
35]. Several mechanisms may be involved in Nrf2 activation and inhibition.
Probst et al. [
36] demonstrated that pharmacological Keap1 inhibition by the synthetic triterpenoid RTA 408, is distinct from those resulting from the loss of functional Keap1 in tumor cells and does not increase cellular proliferation or survival.
The results of the molecular docking performed in this study demonstrated that the compound (
4d) is able to bind to Keap1 and this suggests that in normal cells, Nrf2 activation may occur through the stabilization of Keap1. In cancer cells, increased ERK phosphorylation relating to ROS production was observed as a result of treatment with the DCL–OAO derivative conjugates, indicating the contribution of this mechanism to their cytotoxicity. On the other hand, the relationship between ERK and Nrf2 activation was described [
37]. While in the case of the OAO derivatives alone, both mechanisms, i.e., the down-regulation of Keap1 and activation of ERK, might contribute to Nrf2 activation and cytotoxicity, their conjugation with DCL or indomethacin [
11] results in the opposite effect. Therefore, the mechanism of Nrf2 inhibition by the DCL–OAO derivative hybrids in HCC cells requires further study. Nrf2 activation leads to the transcription of several genes, including expression of the
SOD-1 and
NQO1 genes, whose products are important for protection against ROS. SOD-1 catalyzes the dismutation of the superoxide-free radical anion into molecular oxygen and hydrogen peroxide, the substrate of catalase, and/or glutathione peroxidase [
38]. Similarly,
NQO1, besides its catalytic action in reducing quinones, may also directly scavenge superoxide [
39]. Therefore, reduced expression of both
SOD-1 and
NQO1 in HepG2 cells may contribute to the increased ROS levels in these cells and ultimately cell death.
Sustained Nrf2 activity in cancer cells may promote cancer growth and increase chemoresistance [
7]. Thus, the reduced activation of Nrf2 along with an increased generation of ROS in cancer cells is highly desirable, and the compounds (
4c) and (
4d) seem to act in this way. It is worth noting that DCL did not show any significant effects on Nrf2 activation in neither the THLE-2 nor HepG2 cells.
The activation of NF-κB measured in terms of binding of its active subunits p65 and p50 into DNA, and their translocation from the cytosol into the nucleus was reduced as an effect from treatment with the DCL–OAO morpholide derivative (
4d) and to a lesser extent the conjugate with the OAO benzyl ester (
4c). The p65 subunit is basically responsible for the transcription initiation, and its binding to DNA was more affected. The p50 subunit serves only as a helper in NF-κB DNA binding [
40,
41].
The disproportionate increase in the active p65 subunits and ultimately of effector molecules is fundamental to the pathogenesis of many diseases, including cancer. Henceforth, the NF-κB p65 signaling pathway is recognized as an important target for novel drug design, discovery, and development [
38]. Notably, DCL-conjugates decreased the level of
COX-2 protein in both lines but most significantly in the HepG2 cells. Since an overexpression of
COX-2 is related to increased cell growth and is observed in human HCC, the most potent conjugate of DCL (
4d) might be considered one of the candidates for new therapeutics. Again, DCL alone did not affect the NF-κB activation in THLE-2 and only slightly in the HepG2 cells, indicating that conjugation to the OAO derivatives may enhance its anti-cancer activity.
To explain the mechanism of cell death induced by these compounds, their effect on cell cycle distribution and apoptosis was assessed. While cell cycle distribution was not affected, apoptosis was induced by all conjugates, but the proapoptotic effect of compounds (4c) and (4d) exceeded induction of that of topotecan used as a reference compound. Reduced activation of both transcription factors, NF-κB, which may enhance proapoptotic gene expression, and Nrf2 controlling genes whose products are involved in cell proliferation and differentiation, along with reduced activation of kinases from the MAPK family, may also be responsible for the induction of apoptosis by the DCL–OAO derivative conjugates. Inhibition of these signaling pathways by the compounds (4c) and (4d) also resulted in reduced proliferation of the HepG2 cells.
Importantly, the anti-cancer activity of the DCL–OAO derivatives was confirmed in vivo. While DCL had little effect on tumor volume, its conjugate with the OAO morpholide decreased, although to a lesser extent than this derivative alone. The relatively high mortality level observed after treatment with these compounds was observed in mice bearing HepG2 tumor xenografts.
One reason which might explain this effect not occurring in normal mice is their metabolism to more toxic derivatives. In support of this concept in several tumors, a temporary decrease in the luminescence signal during the experiment was observed while the tumor volume was continuously increasing. This effect may be related to the disordered distribution of luciferin in the tumor due to blood vessel disruption or necrotic core formation. Further studies are required to explain the mechanism of antitumor activity of the DCL and OAO–morpholide conjugates in vivo. The data obtained in this experiment will be useful in designing a new protocol for HepG2 cell implantation as well as for compound delivery and dosing regimen, which should fully demonstrate their therapeutic potential.
In summary, this study showed that the conjugation of DCL with OAO derivatives, particularly OAO–morpholide, enhanced DCL anti-cancer activity in HCC. Reduced activation of Nrf2 and NF-κB along with other signaling molecules leading to apoptosis and limitations to cell proliferation may be responsible for these effects. In normal immortalized cells, through activation of Nrf2, the same conjugate may exert a chemopreventive effect. Although the results of the tumor xenografts experiment are promising, further studies are needed to confirm in vitro observations.