Anti-Metastatic and Anti-Angiogenic Effects of Curcumin Analog DK1 on Human Osteosarcoma Cells In Vitro
Abstract
1. Introduction
2. Results
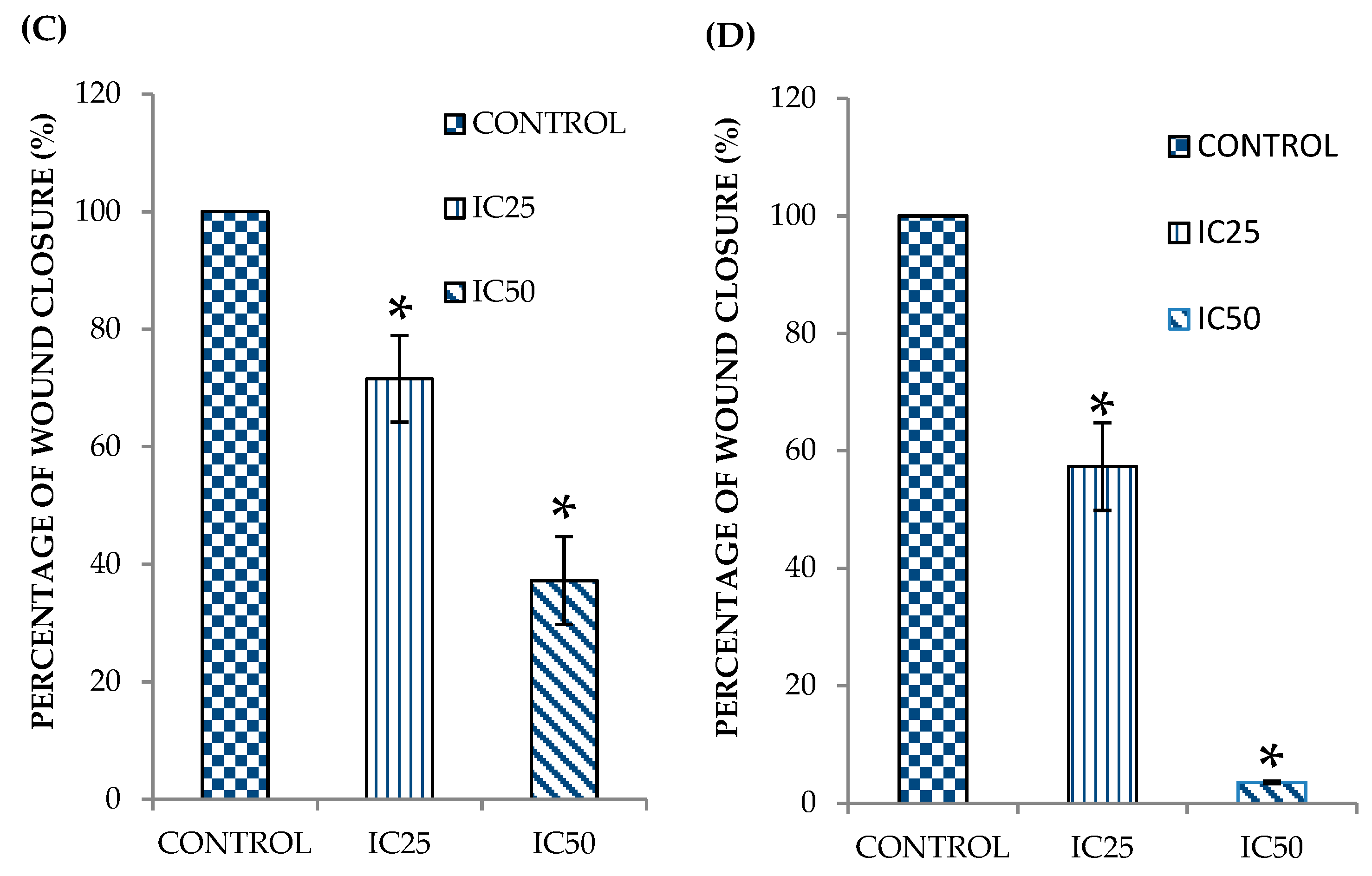
2.1. Curcumin Analog DK1 Inhibit Motility of Both OS Cell Lines
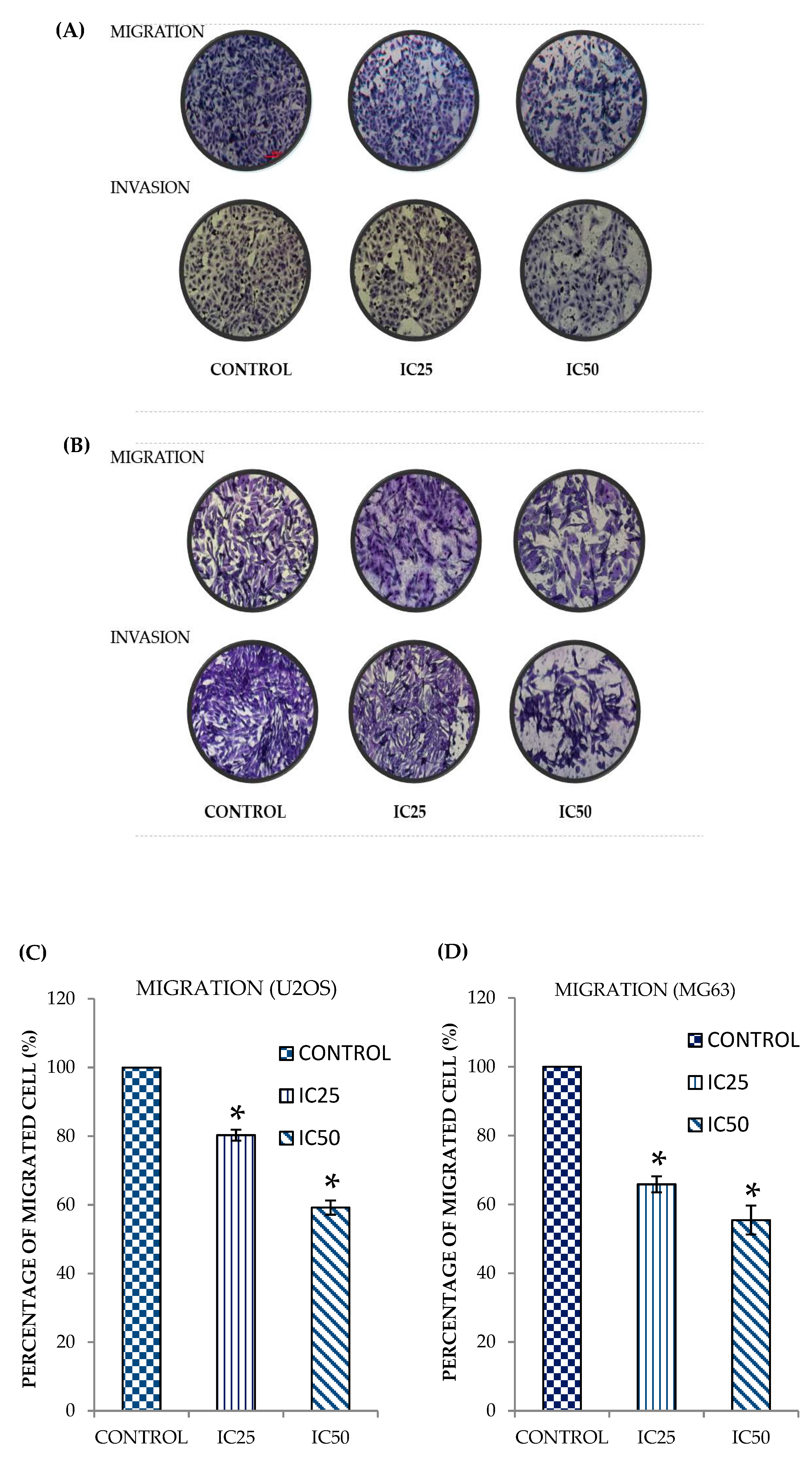
2.2. Curcumin Analog DK1 Inhibit Migration and Invasion Capability of Both OS Cell Lines
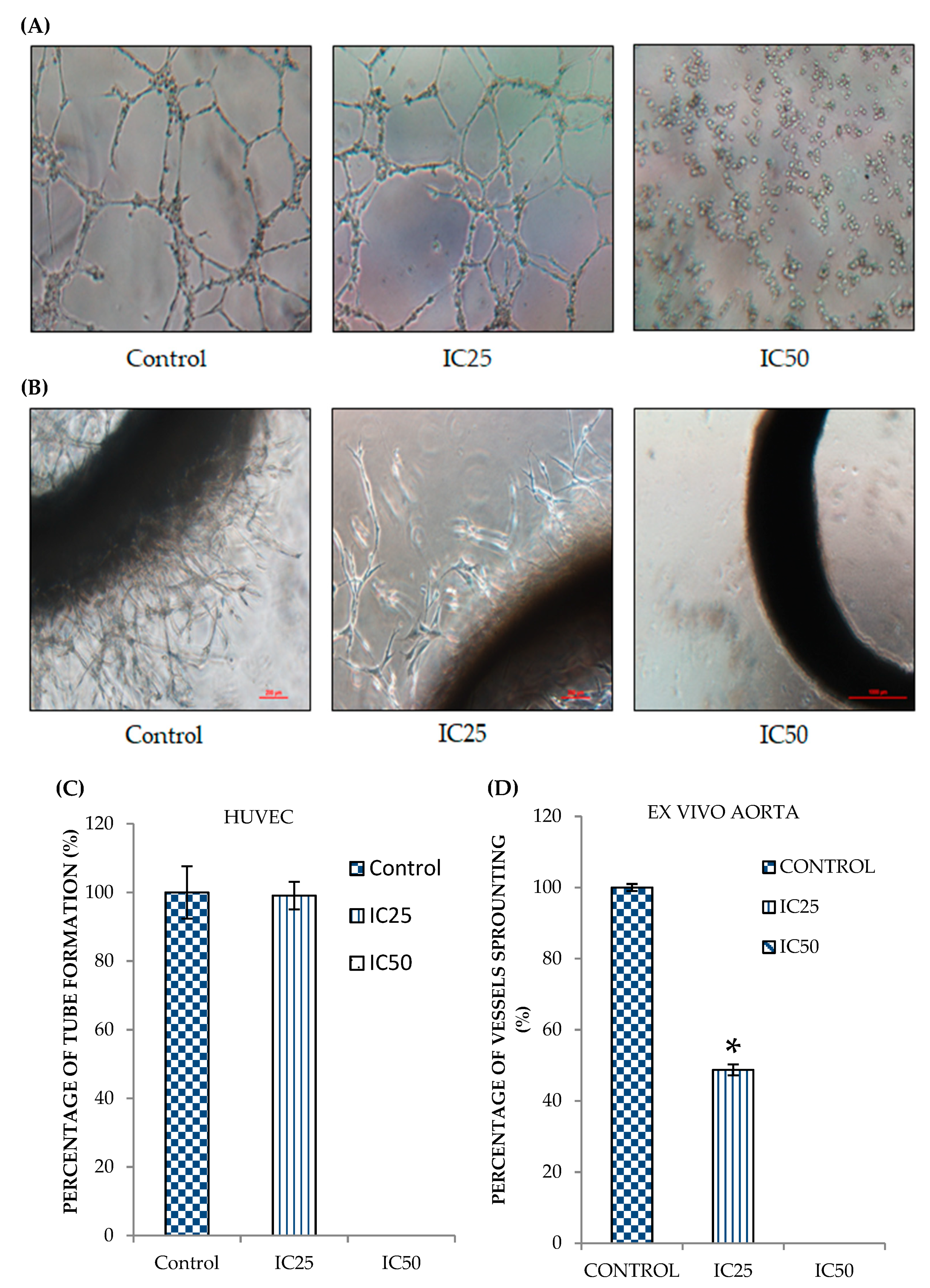
2.3. Curcumin Analog DK1 Possesses Anti-Angiogenesis effects
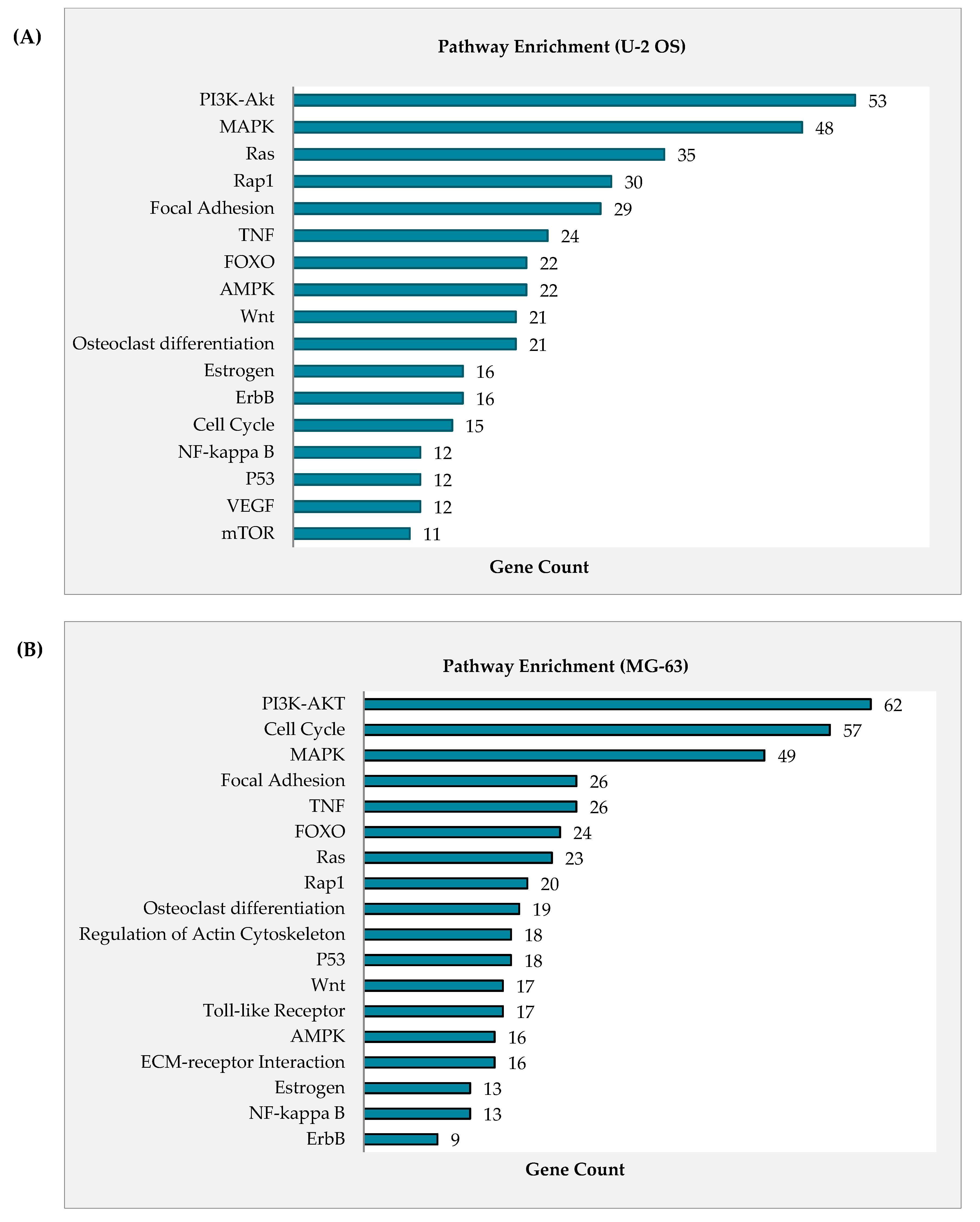
2.4. Curcumin Analog DK1 Induce Several Cancer-Related Pathways in Both OS Cell Lines
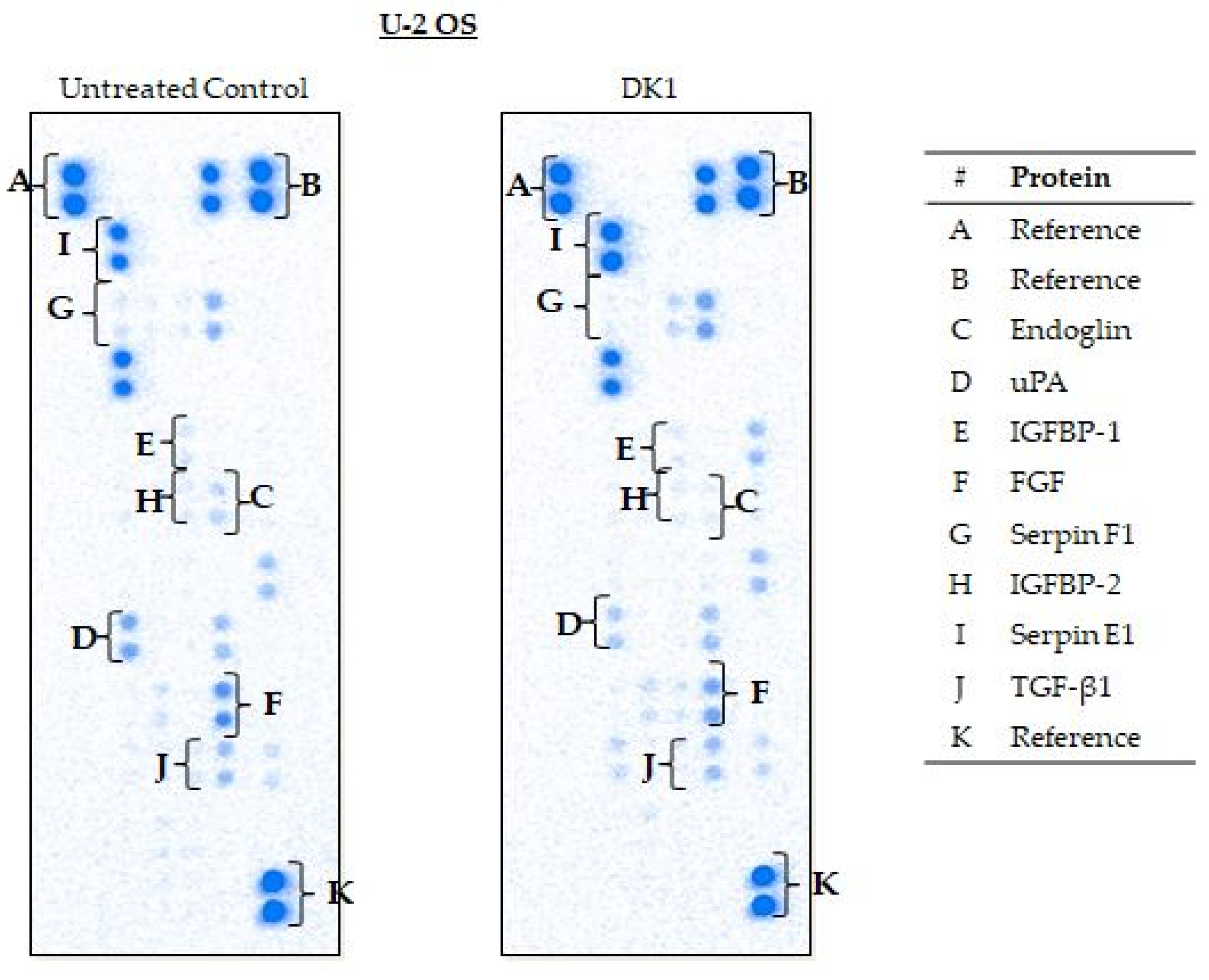
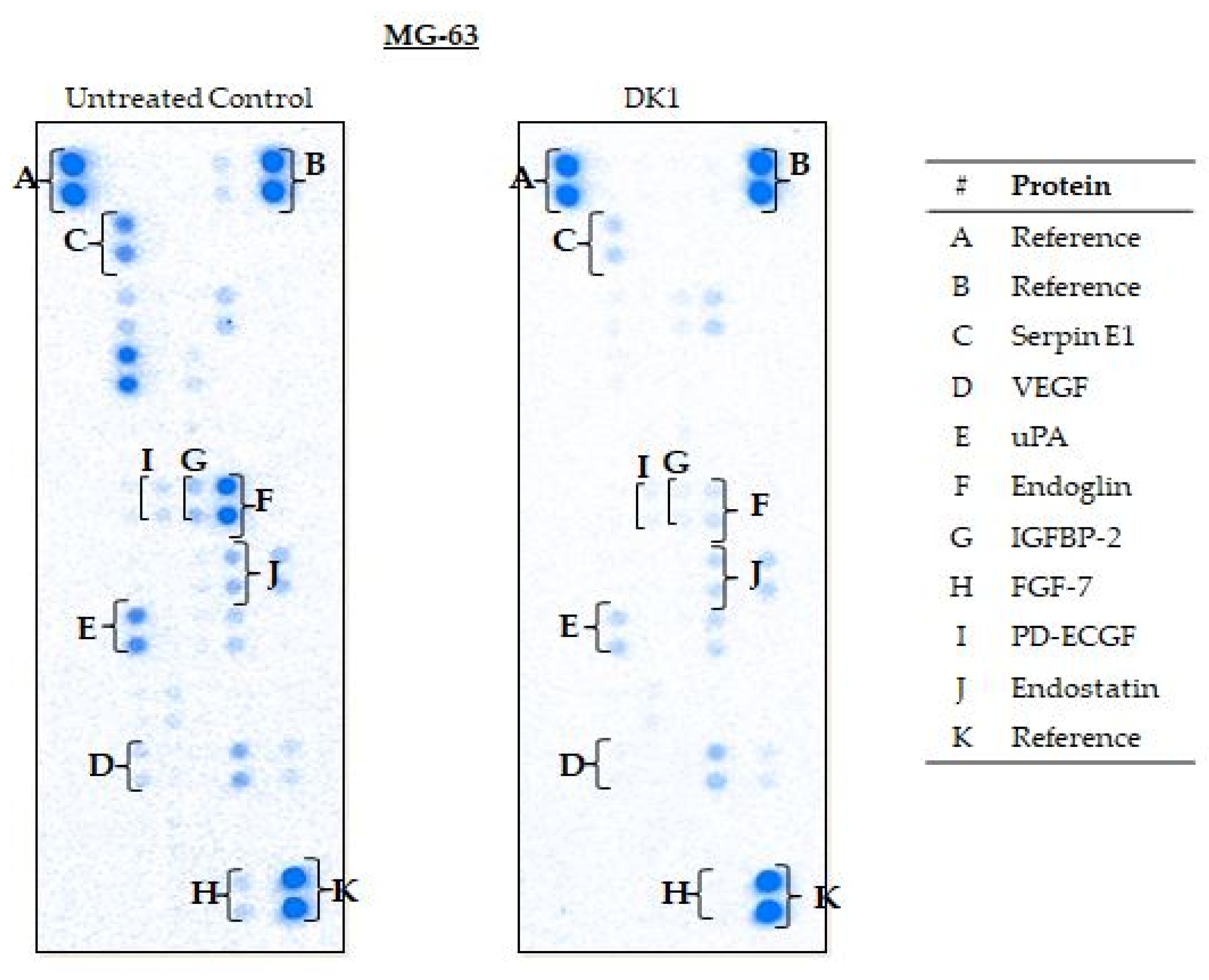
2.5. Curcumin Analog DK1 Regulates Several Genes and Protein Related to Metastasis and Angiogenesis
3. Discussion
4. Materials and Methods
4.1. Preparation of Curcumin Analogue DK1
4.2. Cell Treatment
4.3. In Vitro Scratch Assay
4.4. In Vitro Transwell Migration and Invasion Assay
4.5. In Vitro HUVEC Tube Formation Assay
4.6. Ex Vivo Rat Aortic Ring Assay
4.7. Microarray-Based Gene Expression Analysis
4.8. Real-time Quantitative PCR Analysis
4.9. Angiogenesis-Related Proteome Profiling
4.10. Statictical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Genes | Accession Number | Forward Primers | Reverse Primers | Amplicon Size (bp) | Primer Efficiency (%) |
|---|---|---|---|---|---|
| NFKBIA | NM_020529 | GTTACCTACCAGGGCTATTCTC | CTGTGAACTCCGTGAACTCT | 163 | 98 |
| NFKBIE | NM_004556 | GAAGCACTCACTTACATCTC | GCTGTCTGGTAAAGGTTATT | 143 | 105 |
| TP73 | NM_005427 | AGCAGCCCATCAAGGAGGAGTT | TCCTGAGGCAGTTTTGGACACA | 132 | 95 |
| CDKN1A | NM_078467 | GGGGTTATCTCTGTGTTAGGG | AGCCTCTACTGCCACCATCT | 179 | 101 |
| PTEN | NM_000314 | CACTTGTGGCAACAGATAAG | GTCCAGAGTCCAGCATAAA | 138 | 110 |
| MMP3 | NM_002422 | AAGATGCTGTTGATTCTGCTGT | ATTGGTCCCTGTTGTATCCTTT | 211 | 110 |
| COL1A1 | NM_000088 | TCTGCGACAACGGCAAGGTG | GACGCCGGTGGTTTCTTGGT | 146 | 103 |
| FGF1 | NM_000800 | TGAGAAGAAGACACCAAGTGGA | TTGTGGCGCTTTCAAGACTA | 110 | 95 |
| CDK1 | NM_001170406 | CAGACTAGAAAGTGAAGAGGAAGG | ACTGACCAGGAGGGATAGAATC | 191 | 97 |
| Cycs | NM_018947 | ACCTTCCATCTTGGCTAGTTGTG | ATCGCTTGAGCCTGGGAAATAG | 129 | 100 |
| Casp 3 | NM_004346 | AGAACTGGACTGTGGCATTGAG | GCTTGTCGGCATACTGTTTCAG | 191 | 103 |
| Casp 9 | NM_001229 | CTTCGTTTCTGCGAACTAACAGG | GCACCACTGGGGTAAGGTTT | 75 | 106 |
| SERPINE1 | NM_000602 | ACTTTTCAGAGGTGGAGAGA | CCAGGATGTCGTAGTAATGG | 300 | 107 |
| IKBKE | NM_014002 | GTCAAACGACCTCATCACA | CCAAGCATAGAAAGAACAGG | 142 | 106 |
| AKT2 | NM_001626 | ACCTCTGGGTGTTTGGAGTT | CCCTGGTCTGAAATGAGTGT | 231 | 107 |
| BCL2 | NM_000657 | TTGACAGAGGATCATGCTGTACTT | ATCTTTATTTCATGAGGCACGTT | 103 | 110 |
| VEGFA | NM_001025370 | AGGGCAGAATCATCACGAAGT | AGGGTCTCGATTGGATGGCA | 75 | 105 |
| IKBKB | NM_001556 | CTACTTTGGTGGTTGTCCTC | ATACCCTTTCTGTCCCTCCT | 176 | 107 |
| GUSB | NM_000181.4 | CACCAGGGACCATCCAATACC | GCAGTCCAGCGTAGTTGAAAAA | 99 | 103 |
| ACTB | NM_001101.3 | AGAGCTACGAGCTGCCTGAC | AGCACTGTGTTGGCGTACAG | 184 | 110 |
Appendix B
References
- American Cancer Society. Ypes of Cancer that Develop in Children. 2019. Available online: https://www.cancer.org/cancer/cancer-in-children/types-of-childhood-cancers.html (accessed on 1 November 2020).
- Worldwide Cancer Statistic. Cancer Research UK. 2018. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/worldwide-cancer (accessed on 1 November 2020).
- Mansor, A.; Yap, T.Y. A Teen Thought he was Going to Lose His Leg Because of this Common Bone Cancer, The Star Online. 24 May 2017. Available online: https://www.thestar.com.my/lifestyle/family/2017/05/24/osteosarcoma-bone-cancer-children (accessed on 1 November 2020).
- Osteosarcoma. Bone Cancer Research Trust UK. 2016. Available online: http://www.bcrt.org.uk/information/information-by-type/osteosarcoma/ (accessed on 1 November 2020).
- Messerschmitt, P.J.; Garcia, R.M.; Abdul Karim, F.W.; Greenfield, E.M.; Getty, P.J. Osteosarcoma. J. Am. Acad. Orthop. Surg. 2009, 17, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Taran, R.; Taran, S.; Malipatil, N. Pediatric osteosarcoma: An updated review. Indian J. Med Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.S.; Khanna, C. A Review of the Association between Osteosarcoma Metastasis and Protein Translation. J. Comp. Pathol. 2012, 146, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Mikulić, D.; Ilić, I.; Cepulić, M.; Orlić, D.; Giljević, J.S.; Fattorini, I.; Seiwerth, S. TUMOR ANGIOGENESIS AND OUTCOME IN OSTEOSARCOMA. Pediatr. Hematol. Oncol. 2004, 21, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma treatment—Where do we stand? A state of the art review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yuan, M.; Yuan, H.; Huang, X.; Sui, X.; Cui, X.; Tang, F.; Peng, J.; Chen, J.; Lu, S.; et al. Co-encapsulation of magnetic Fe3O4 nanoparticles and doxorubicin into biodegradable PLGA nanocarriers for intratumoral drug delivery. Int. J. Nanomed. 2012, 7, 1697–1708. [Google Scholar]
- Janeway, K.A.; Grier, H.E. Sequelae of osteosarcoma medical therapy: A review of rare acute toxicities and late effects. Lancet Oncol. 2010, 11, 670–678. [Google Scholar] [CrossRef]
- Harake, D.; Franco, V.I.; Henkel, J.M.; Miller, T.L.; Lipshultz, S.E. Cardiotoxicity in childhood cancer survivors: Strategies for prevention and management. Future Cardiol. 2012, 8, 1–37. [Google Scholar] [CrossRef]
- Adams, B.K.; Ferstl, E.M.; Davis, M.C.; Herold, M.; Kurtkaya, S.; Camalier, R.F.; Hollingshead, M.G.; Kaur, G.; Sausville, E.A.; Rickles, F.R.; et al. Synthesis and biological evaluation of novel curcumin analogs as anti-cancer and anti-angiogenesis agents. Bioorganic Med. Chem. 2004, 12, 3871–3883. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Shao, L.; Wang, Y.; Zhao, C.; Chu, Y.; Xiao, J.; Zhao, Y.; Li, X.; Yang, S. Exploration and synthesis of curcumin analogues with improved structural stability both in vitro and in vivo as cytotoxic agents. Bioorganic Med. Chem. 2009, 17, 2623–2631. [Google Scholar] [CrossRef] [PubMed]
- Jantarat, C. Bioavailability enhancement techniques of herbal medicine: A case example of curcumin. Int. J. Pharm. Pharm. Sciences 2013, 5, 493–500. [Google Scholar]
- Nagahama, K.; Utsumi, T.; Kumano, T.; Maekawa, S.; Oyama, N.; Kawakami, J. Discovery of a new function of curcumin which enhances its anticancer therapeutic potency. Sci. Rep. 2016, 6, 30962. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.P.; Hubbard, R.B., IV; Ehlers, T.J.; Arbiser, J.L.; Goldsmith, D.J.; Bowen, J.P. Synthesis and biological evaluation of aromatic enones related to curcumin. Bioorganic Med. Chem. 2005, 13, 4007–4013. [Google Scholar] [CrossRef] [PubMed]
- Faião-Flores, F.; Suarez, J.A.Q.; Maria-Engler, S.S.; Soto-Cerrato, V.; Pérez-Tomás, R.; Maria, D.A. The curcumin analog DM-1 induces apoptotic cell death in melanoma. Tumor Biol. 2013, 34, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.M.; Yeap, S.K.; Abu, N.; Lim, K.L.; Ky, H.; Zaim, A.; Ho, W.Y.; Tan, S.W.; Alan-Ong, H.K.; Zareen, S.; et al. Synthetic curcumin derivative DK1 possessed G2 / M arrest and induced apoptosis through accumulation of intracellular ROS in MCF-7 breast cancer cells. Cancer Cell Int. 2017, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zamrus, S.N.H.; Akhtar, M.N.; Yeap, S.K.; Quah, C.K.; Loh, W.-S.; Alitheen, N.B.; Zareen, S.; Tajuddin, S.N.; Hussin, Y.; Shah, S.A.A. Design, synthesis and cytotoxic effects of curcuminoids on HeLa, K562, MCF-7 and MDA-MB-231 cancer cell lines. Chem. Central J. 2018, 12, 31. [Google Scholar] [CrossRef]
- Aziz, M.N.M.; Hussin, Y.; Rahim, N.F.C.; Nordin, N.; Mohamad, N.E.; Yeap, S.K.; Yong, C.Y.; Masarudin, M.J.; Cheah, Y.K.; Abu, N.; et al. Curcumin Analog DK1 Induces Apoptosis in Human Osteosarcoma Cells In Vitro through Mitochondria-Dependent Signaling Pathway. Molecules 2018, 23, 75. [Google Scholar] [CrossRef]
- Hussin, Y.; Aziz, M.N.M.; Rahim, N.F.C.; Yeap, S.K.; Mohamad, N.E.; Masarudin, M.J.; Nordin, N.; Rahman, N.M.A.-N.A.; Yong, C.Y.; Akhtar, M.N.; et al. DK1 Induces Apoptosis via Mitochondria-Dependent Signaling Pathway in Human Colon Carcinoma Cell Lines In Vitro. Int. J. Mol. Sci. 2018, 19, 1151. [Google Scholar] [CrossRef]
- Decaestecker, C.; Debeir, O.; Ham, P.V.a.n.; Kiss, R. Can Anti-Migratory Drugs Be Screened In Vitro? A Review of 2D and 3DAssays for the Quantitative Analysis of Cell Migration. Med. Res. Rev. 2006, 27, 149–176. [Google Scholar]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial Steps of Metastasis: Cell Invasion and Endothelial Transmigration. Mutat. Res.-Rev. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef]
- Geiger, T.R.; Peeper, D.S. Metastasis mechanisms. BBA-Rev. Cancer 2009, 1796, 293–308. [Google Scholar] [CrossRef]
- Abu, N.; Akhtar, M.N.; Yeap, S.K.; Lim, K.L.; Ho, W.Y.; Zulfadli, A.J.; Omar, A.R.; Sulaiman, M.R.; Abdullah, M.P.; Alitheen, N.B. Flavokawain A Induces Apoptosis in MCF-7 and MDA-MB231 and Inhibits the Metastatic Process In Vitro. PLoS ONE 2014, 9, e105244. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Chen, C.G.; Iozzo, R.V. A simplified aortic ring assay: A useful ex vivo method to assess biochemical and functional parameters of angiogenesis. Matrix Biol. Plus 2020, 6–7, 100025. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nat. Cell Biol. 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Mohseny, A.B.; Machado, I.; Cai, Y.; Schaefer, K.-L.; Serra, M.; Hogendoorn, P.C.W.; Llombart-Bosch, A.; Cleton-Jansen, A.-M. Functional characterization of osteosarcoma cell lines provides representative models to study the human disease. Lab. Investig. 2011, 91, 1195–1205. [Google Scholar] [CrossRef]
- Heng, H.H.Q.; Bremer, S.W.; Stevens, J.B.; Ye, K.J.; Liu, G.U.O.; Ye, C.J. Genetic and Epigenetic Heterogeneity in Cancer: A Genome-Centric Perspective. J. Cell. Physiol. 2009, 220, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Abu, N.; Hon, K.W.; Jeyaraman, S.; Yahaya, A.; Abdullah, N.M.; Mustangin, M.; A Sulaiman, S.; Jamal, R.; Ab-Mutalib, N.-S. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics 2019, 11, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Hewlings, S.J.; Douglas, S.K. Curcumin: A Review of Its’ Effects on Human Health. Foods 2017, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Review Article Therapeutic Roles of Curcumin: Lessons Learned from Clinical Trials. AAPS J. 2013, 15, 195–218. [Google Scholar] [CrossRef]
- Lozada-García, M.C.; Enríquez, R.G.; Ramírez-Apán, T.O.; Nieto-Camacho, A.; Palacios-Espinosa, J.F.; Custodio-Galván, Z.; Soria-Arteche, O.; Pérez-Villanueva, J. Synthesis of curcuminoids and evaluation of their cytotoxic and antioxidant properties. Molecules 2017, 22, 633. [Google Scholar] [CrossRef] [PubMed]
- Lauvrak, S.U.; Munthe, E.; Kresse, S.H.; Stratford, E.W.; Namløs, H.M.; Meza-Zepeda, L.A.; Myklebost, O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer 2013, 109, 2228–2236. [Google Scholar] [CrossRef]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef]
- Gupta, G.P.; Massagué, J. Review Cancer Metastasis: Building a Framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, M.; Mousa, S.A. The Role of Angiogenesis in Cancer Treatment. Biomedicines 2017, 5, 34. [Google Scholar] [CrossRef]
- Zuazo-Gaztelu, I.; Casanovas, O. Unraveling the Role of Angiogenesis in Cancer Ecosystems. Front. Oncol. 2018, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, A.; Pius, A.; Gopi, S. Biological activities of curcuminoids, other biomolecules from turmeric and their derivatives—A review. J. Tradit. Complement. Med. 2017, 7, 205–233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, X.-H.; Yan, Y.-G.; Wang, C.; Wang, W.-J. PI3K/Akt signaling in osteosarcoma. Clin. Chim. Acta 2015, 444, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Mireuta, M.; Darnel, A.; Pollak, M. IGFBP-2 expression in MCF-7 cells is regulated by the PI3K/AKT/ mTOR pathway through Sp1-induced increase in transcription. Growth Factors 2010, 28, 243–255. [Google Scholar] [CrossRef]
- Usatyuk, P.V.; Fu, P.; Mohan, V.; Epshtein, Y.; Jacobson, J.R.; Gomez-Cambronero, J.; Wary, K.K.; Bindokas, V.; Dudek, S.M.; Salgia, R.; et al. Role of c-Met/Phosphatidylinositol 3-Kinase (PI3k)/Akt Signaling in Hepatocyte Growth Factor (HGF)-mediated Lamellipodia Formation, Reactive Oxygen Species (ROS) Generation, and Motility of Lung Endothelial Cells. J. Biol. Chem. 2014, 289, 13476–13491. [Google Scholar] [CrossRef] [PubMed]
- Wheler, J.J.; Atkins, J.T.; Janku, F.; Moulder, S.L.; Philip, J.; Yelensky, R.; Valero, V.; Miller, V.; Kurzrock, R.; Meric-Bernstam, F. Presence of both alterations in FGFR/FGF and PI3K/AKT/ mTOR confer improved outcomes for patients with metastatic breast cancer treated with PI3K/AKT/ mTOR inhibitors. Oncoscience 2016, 3, 164–172. [Google Scholar] [CrossRef]
- Mirzaei, H.; Masoudifar, A.; Sahebkar, A.; Zare, N.; Nahand, S.J.; Bahman, R.; Mehrabian, E.; Mohammadi, M.; Mirzaei, H.R.; Jaafari, R.M. MicroRNA: A novel target of curcumin in cancer therapy. Cell. Physiol. 2018, 233, 3004–3015. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Rane, G.; Kanchi, M.M.; Arfuso, F.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; Tan, B.K.H.; Kumar, A.P.; Sethi, G. The Multifaceted Role of Curcumin in Cancer Prevention and Treatment. Molecules 2015, 20, 2728–2769. [Google Scholar] [CrossRef]
- Singh, S.; Khar, A. Biological Effects of Curcumin and Its Role in Cancer Chemoprevention and Therapy. Anti-Cancer Agents Med. Chem. 2006, 6, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Dolcet, X.; Llobet, D.; Pallares, J.; Matias-guiu, X. NF-kB in development and progression of human cancer. Virchows Arch. 2005, 446, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Akeda, K.; Matsubara, T.; Kusuzaki, K.; Matsumine, A.; Masuda, K.; Gemba, T.; Uchida, A.; Sudo, A. Transfection of NF- kB decoy oligodeoxynucleotide suppresses pulmonary metastasis by murine osteosarcoma. Cancer Gene Ther. 2011, 18, 250–259. [Google Scholar] [CrossRef]
- Reddy, K.B.; Nabha, S.M.; Atanaskova, N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003, 22, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Chandhanayingyong, C.; Kim, Y.; Staples, J.R.; Hahn, C.; Lee, F.Y. MAPK/ERK Signaling in Osteosarcomas, Ewing Sarcomas and Chondrosarcomas: Therapeutic Implications and Future Directions. Sarcoma 2012, 2012, 404810. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Jiao, Y.; Postlethwaite, A.; Stuart, J.M.; Wang, Y.; Sun, D.; Gu, W. Dual-specificity phosphatases 2: Surprising positive effect at the molecular level and a potential biomarker of diseases. Genes Immun. 2013, 14, 1–6. [Google Scholar] [CrossRef]
- Low, H.B.; Zhang, Y. Regulatory Roles of MAPK Phosphatases in Cancer. Immune Netw. 2016, 16, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zheng, H.; Du, X.; Yang, J. Characterization of FGFR signaling pathway as therapeutic targets for sarcoma patients. Cancer Biol. Med. 2016, 13, 260–268. [Google Scholar] [CrossRef]
- Pavón, M.A.; Arroyo-solera, I.; Céspedes, M.V.; Casanova, I.; León, X.; Mangues, R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget 2016, 7, 57351–57366. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.S.; Gordon, M.S.; Robert, F.; Matei, D.E. Endoglin for Targeted Cancer Treatment. Curr. Oncol. Rep. 2014, 16, 1–9. [Google Scholar] [CrossRef]
- Guo, C.; Lu, H.; Gao, W.; Wang, L.; Lu, K.; Wu, S.; Pataer, A.; Huang, M.; El-Zein, R.; Lin, T.; et al. Insulin-Like Growth Factor Binding Protein-2 Level Is Increased in Blood of Lung Cancer Patients and Associated with Poor Survival. PLoS ONE 2013, 8, e74973. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-C. Boyden Chamber Assay. Cell Migration 2005, 294, 015–022. [Google Scholar] [CrossRef]




| Gene Bank | Gene Symbol | Gene Name | Fold Change > 2 |
|---|---|---|---|
| Pro-metastasis | |||
| NM_016653 | ZAK | Homo sapiens mitogen-activated protein kinase kinase kinase 20 (MAP3K20) | -3.53 |
| NM_002982 | CCL2 | Homo sapiens C-C motif chemokine ligand 2 | −4.28 |
| NM_002422 | MMP3 | Homo sapiens matrix metallopeptidase 3 | −4.44 |
| NM_000800 | FGF1 | Homo sapiens fibroblast growth factor 1 | −5.23 |
| NM_002006 | FGF2 | Homo sapiens fibroblast growth factor 2 | −3.86 |
| NM_000088 | COL1A1 | Homo sapiens collagen type I alpha 1 chain | −3.08 |
| NM_001626 | AKT2 | Homo sapiens AKT serine/threonine kinase 2 | −2.30 |
| NM_004655 | AXIN2 | Homo sapiens axin 2 | −2.02 |
| NM_006739 | MCM5 | Homo sapiens minichromosome maintenance complex component 5 | −2.40 |
| NM_001105209 | LAMA4 | Homo sapiens laminin subunit alpha 4 | −8.16 |
| NM_033131 | WNT3A | Homo sapiens Wnt family member 3A | −4.23 |
| NM_001303429 | PIK3R3 | Homo sapiens phosphoinositide-3-kinase regulatory subunit 3 | −6.54 |
| Metastasis Inhibition | |||
| NM_020529 | NFKBIA | Homo sapiens NFKB inhibitor alpha | 2.50 |
| NM_004556 | NFKBIE | Homo sapiens NFKB inhibitor epsilon | 2.24 |
| NM_005427 | TP73 | Homo sapiens tumor protein p73 | 3.41 |
| NM_078467 | CDKN1A | Homo sapiens cyclin dependent kinase inhibitor 1A | 5.03 |
| NM_000314 | PTEN | Homo sapiens phosphatase and tensin homolog | 2.14 |
| NM_005194 | CEBPB | Homo sapiens CCAAT enhancer binding protein beta | 2.65 |
| NM_014417 | BBC3 | Homo sapiens BCL2 binding component 3 | 3.08 |
| NM_004073 | PLK3 | Homo sapiens polo like kinase 3 | 7.29 |
| NM_170662 | CBLB | Homo sapiens Cbl proto-oncogene B | 2.23 |
| NM_001455 | FOXO3 | Homo sapiens forkhead box O3 | 2.77 |
| NM_001924 | GADD45A | Homo sapiens growth arrest and DNA damage inducible alpha | 3.92 |
| NM_004292 | RIN1 | Homo sapiens Ras and Rab interactor 1 | 2.77 |
| NM_004418 | DUSP2 | Homo sapiens dual specificity phosphatase 2 | 4.76 |
| NM_007207 | DUSP10 | Homo sapiens dual specificity phosphatase 10 | 4.95 |
| Gene Bank | Gene Symbol | Gene Name | Fold Change > 2 |
|---|---|---|---|
| Pro-metastasis | |||
| NM_000602 | SERPINE1 | Homo sapiens serpin family E member 1 | −8.96 |
| NM_001025370 | VEGFA | Homo sapiens vascular endothelial growth factor A | −2.67 |
| NM_003377 | VEGFB | Homo sapiens vascular endothelial growth factor B | −2.56 |
| NM_001626 | AKT2 | Homo sapiens AKT serine/threonine kinase 2 | −3.90 |
| NM_000657 | BCL2 | Homo sapiens BCL2 apoptosis regulator | −3.54 |
| NM_001798 | CDK2 | Homo sapiens cyclin dependent kinase 2 | −4.68 |
| NM_001556 | IKBKB | Homo sapiens inhibitor of nuclear factor kappa B kinase subunit beta | −2.23 |
| NM_014002 | IKBKE | Homo sapiens inhibitor of nuclear factor kappa B kinase subunit epsilon | −3.77 |
| NM_000597 | IGFBP2 | Homo sapiens insulin like growth factor binding protein 2 | −2.33 |
| NM_002009 | FGF7 | Homo sapiens fibroblast growth factor 7 | −8.49 |
| NM_001105209 | LAMA4 | Homo sapiens laminin subunit alpha 4 | −5.96 |
| NM_001010931 | HGF | Homo sapiens hepatocyte growth factor | −10.78 |
| Metastasis Inhibition | |||
| NM_002015 | FOXO1 | Homo sapiens forkhead box O1 | 4.03 |
| NM_018947 | CYCS | Homo sapiens cytochrome c | 2.82 |
| NM_001229 | CASP9 | Homo sapiens caspase 9 | 2.14 |
| NM_004346 | CASP3 | Homo sapiens caspase 3 | 2.16 |
| NM_001924 | GADD45A | Homo sapiens growth arrest and DNA damage inducible alpha | 8.90 |
| NM_015999 | ADIPOR1 | Homo sapiens adiponectin receptor 1 | 2.54 |
| NM_170662 | CBLB | Homo sapiens Cbl proto-oncogene B | 5.41 |
| NM_006705 | GADD45G | Homo sapiens growth arrest and DNA damage inducible gamma | 8.40 |
| NM_005359 | SMAD4 | Homo sapiens SMAD family member 4 | 2.46 |
| NM_000314 | PTEN | Homo sapiens phosphatase and tensin homolog | 2.34 |
| NM_004073 | PLK3 | Homo sapiens polo like kinase 3 | 5.36 |
| NM_001040619 | ATF3 | Homo sapiens activating transcription factor 3 | 13.86 |
| NM_004418 | DUSP2 | Homo sapiens dual specificity phosphatase 2 | 2.27 |
| NM_007207 | DUSP10 | Homo sapiens dual specificity phosphatase 10 | 5.27 |
| Cell Lines | Proteins | Relative Intensity (Fold Change) | Regulation |
|---|---|---|---|
| U-2 OS | Endoglin | −7.6 * ± 0.04 | Down |
| uPA | −2.4 * ± 0.04 | Down | |
| IGFBP-2 | −1.7 * ± 0.07 | Down | |
| FGF | −1.4 * ± 0.01 | Down | |
| Serpin F1 | −1.4 * ± 0.05 | Down | |
| IGFBP-1 | −1.5 * ± 0.06 | Down | |
| Serpin E1 | 1.7 * ± 0.13 | Up | |
| TGF-ꞵ1 | −1.4 * ± 0.04 | Down | |
| MG-63 | Serpin E1 | −3.6 * ± 0.02 | Down |
| VEGF | −2.0 * ± 0.01 | Down | |
| uPA | −3.3 * ± 0.04 | Down | |
| Endoglin | −12.5 * ± 0.01 | Down | |
| IGFBP-2 | −6.2 * ± 0.13 | Down | |
| FGF-7 | −3.2 * ± 0.12 | Down | |
| PD-ECGF | −4.7 * ± 0.01 | Down | |
| Endostatin | −1.9 * ± 0.07 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aziz, M.N.M.; Rahim, N.F.C.; Hussin, Y.; Yeap, S.K.; Masarudin, M.J.; Mohamad, N.E.; Akhtar, M.N.; Osman, M.A.; Cheah, Y.K.; Alitheen, N.B. Anti-Metastatic and Anti-Angiogenic Effects of Curcumin Analog DK1 on Human Osteosarcoma Cells In Vitro. Pharmaceuticals 2021, 14, 532. https://doi.org/10.3390/ph14060532
Aziz MNM, Rahim NFC, Hussin Y, Yeap SK, Masarudin MJ, Mohamad NE, Akhtar MN, Osman MA, Cheah YK, Alitheen NB. Anti-Metastatic and Anti-Angiogenic Effects of Curcumin Analog DK1 on Human Osteosarcoma Cells In Vitro. Pharmaceuticals. 2021; 14(6):532. https://doi.org/10.3390/ph14060532
Chicago/Turabian StyleAziz, Muhammad Nazirul Mubin, Nurul Fattin Che Rahim, Yazmin Hussin, Swee Keong Yeap, Mas Jaffri Masarudin, Nurul Elyani Mohamad, Muhammad Nadeem Akhtar, Mohd Azuraidi Osman, Yoke Kqueen Cheah, and Noorjahan Banu Alitheen. 2021. "Anti-Metastatic and Anti-Angiogenic Effects of Curcumin Analog DK1 on Human Osteosarcoma Cells In Vitro" Pharmaceuticals 14, no. 6: 532. https://doi.org/10.3390/ph14060532
APA StyleAziz, M. N. M., Rahim, N. F. C., Hussin, Y., Yeap, S. K., Masarudin, M. J., Mohamad, N. E., Akhtar, M. N., Osman, M. A., Cheah, Y. K., & Alitheen, N. B. (2021). Anti-Metastatic and Anti-Angiogenic Effects of Curcumin Analog DK1 on Human Osteosarcoma Cells In Vitro. Pharmaceuticals, 14(6), 532. https://doi.org/10.3390/ph14060532









