Metabolic Profiles of New Unsymmetrical Bisacridine Antitumor Agents in Electrochemical and Enzymatic Noncellular Systems and in Tumor Cells
Abstract
1. Introduction
2. Results
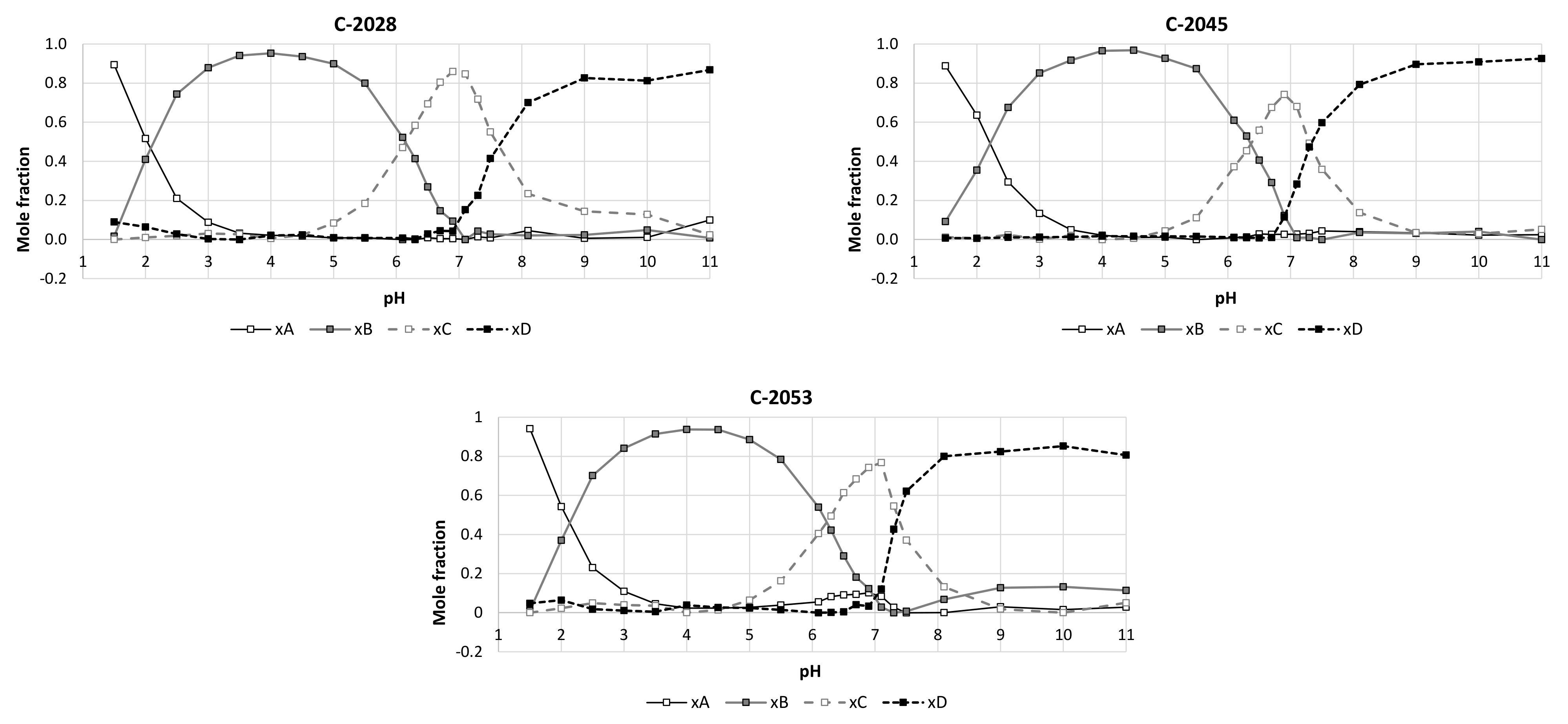
2.1. Physicochemical Properties
2.2. Transformations of the Compounds in Noncellular Systems
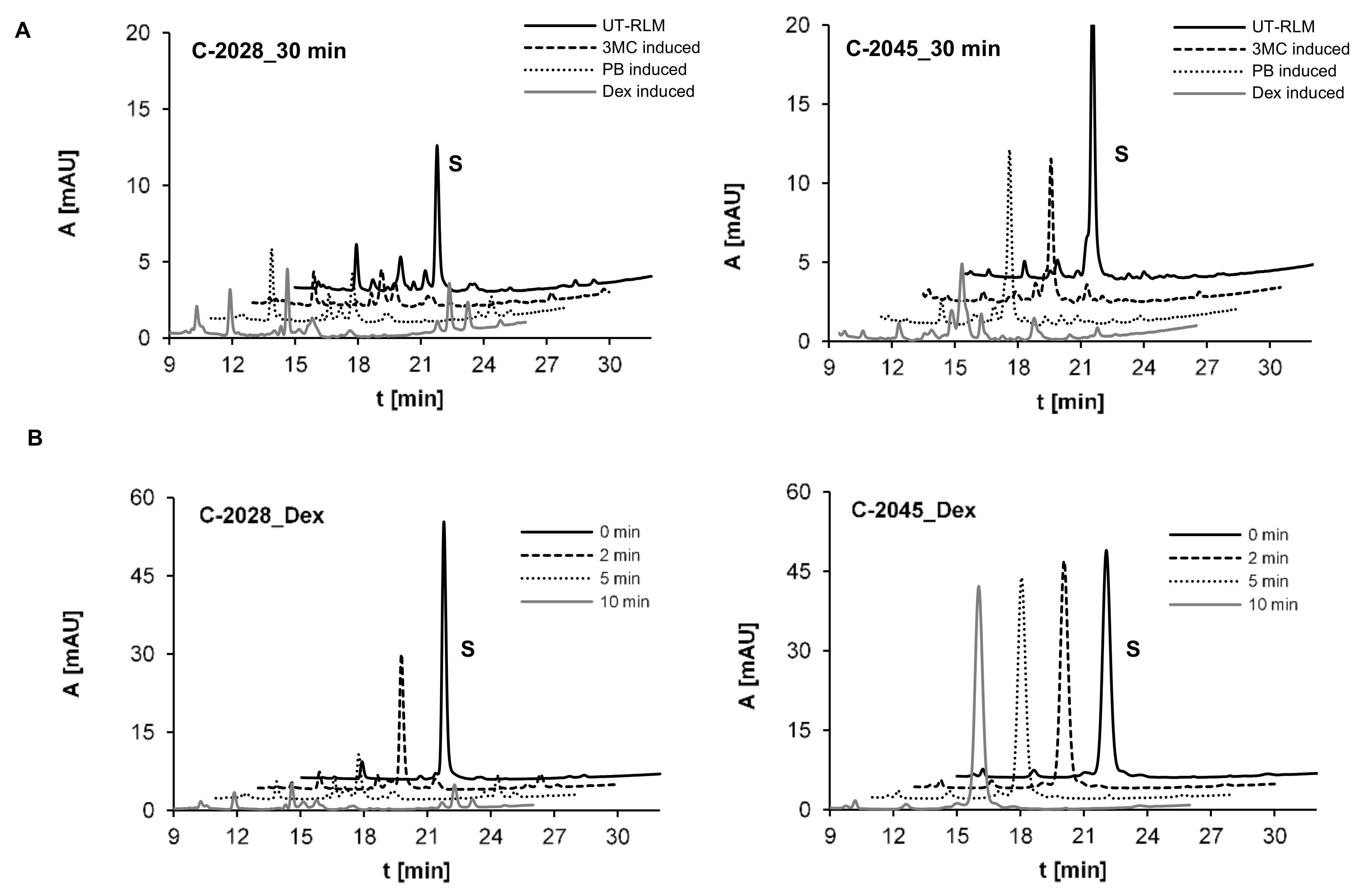
2.2.1. Preliminary Research on Metabolic Transformations of UAs with RLMs
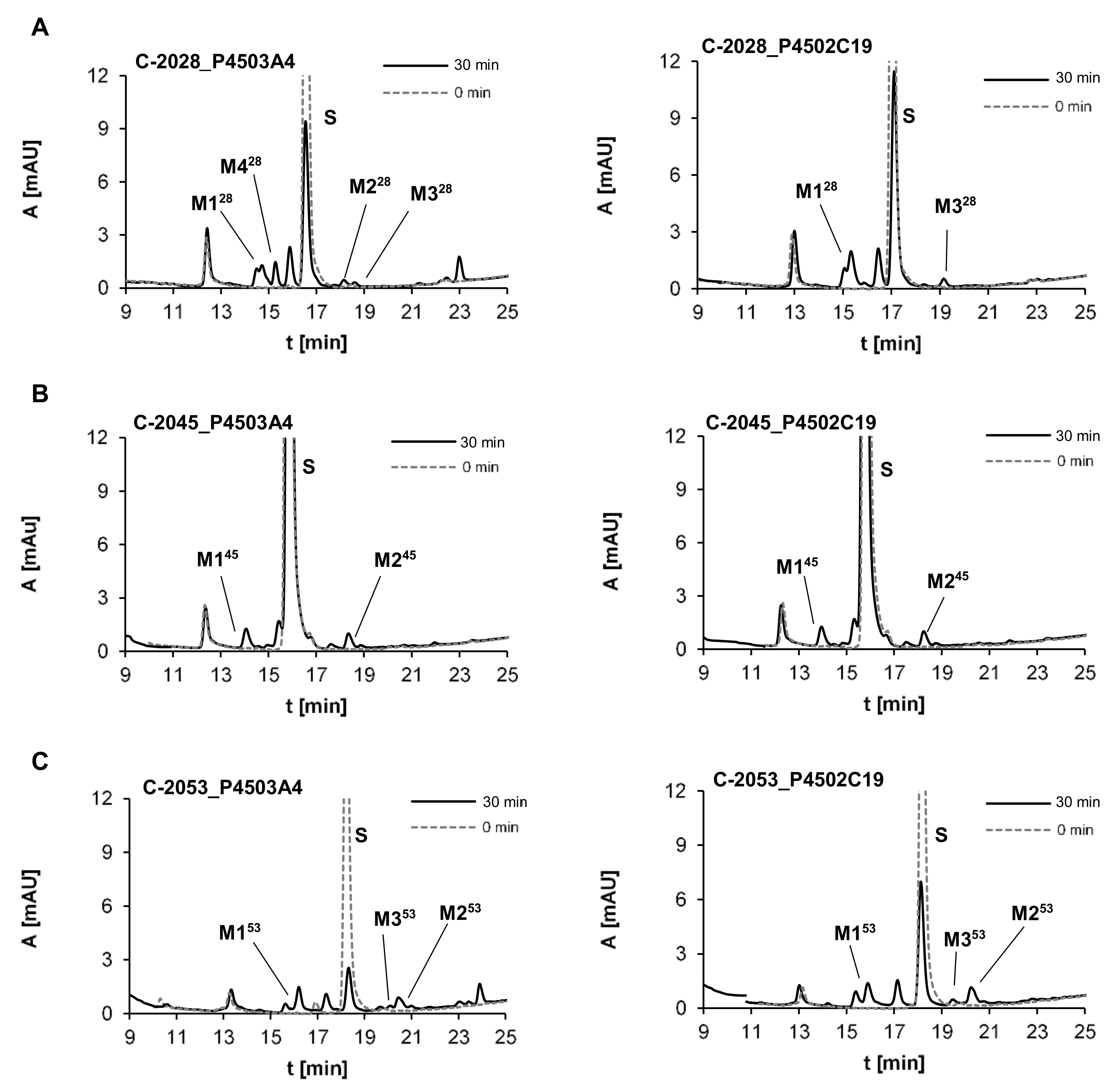
2.2.2. Metabolism of UAs with Human Recombinant P4503A4 and P4502C19 Isoenzymes
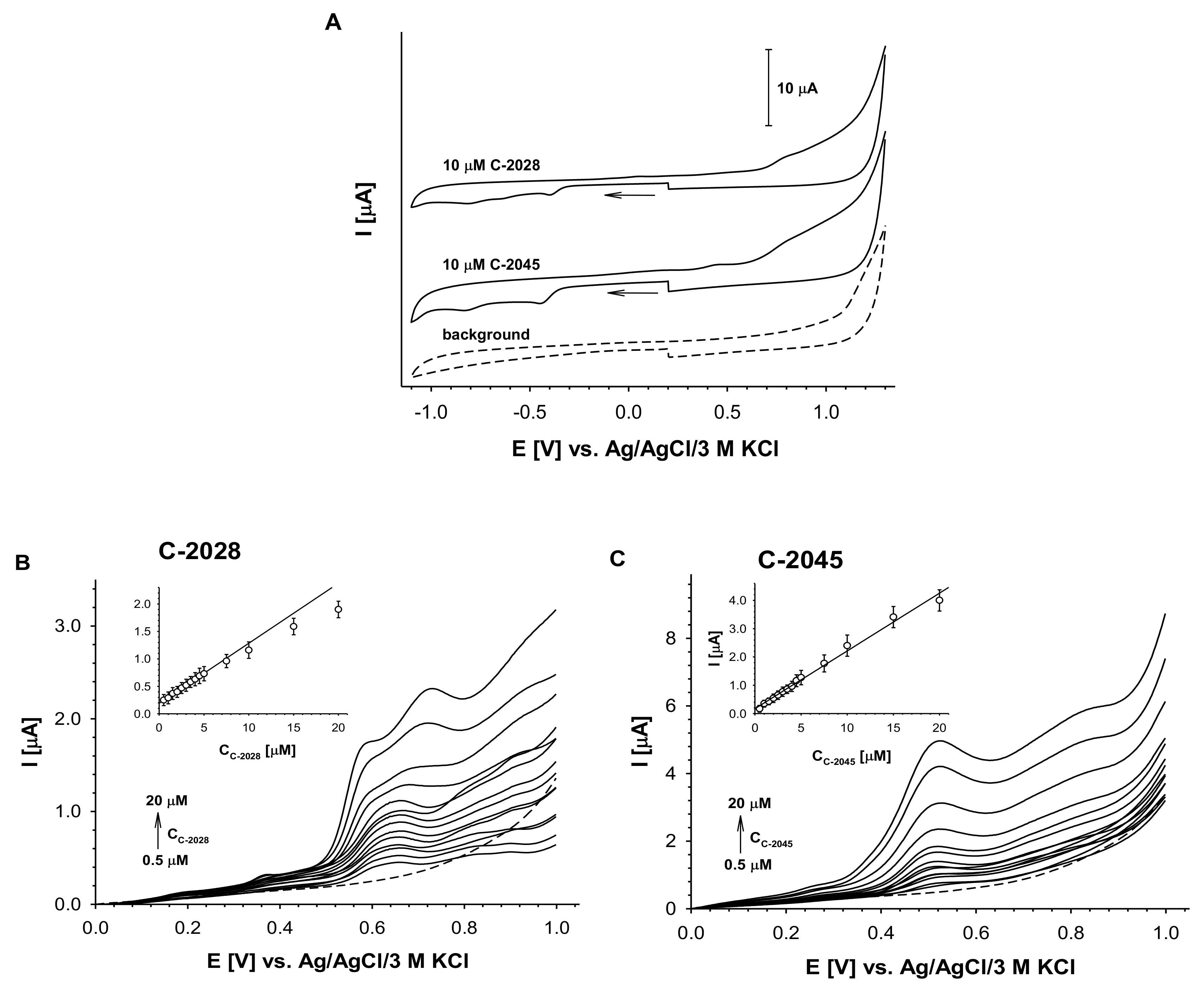
2.2.3. Results of Electrochemical Simulations of Drug Metabolism
2.2.4. Metabolism of UAs with Human Recombinant Flavin Monooxygenases, FMO
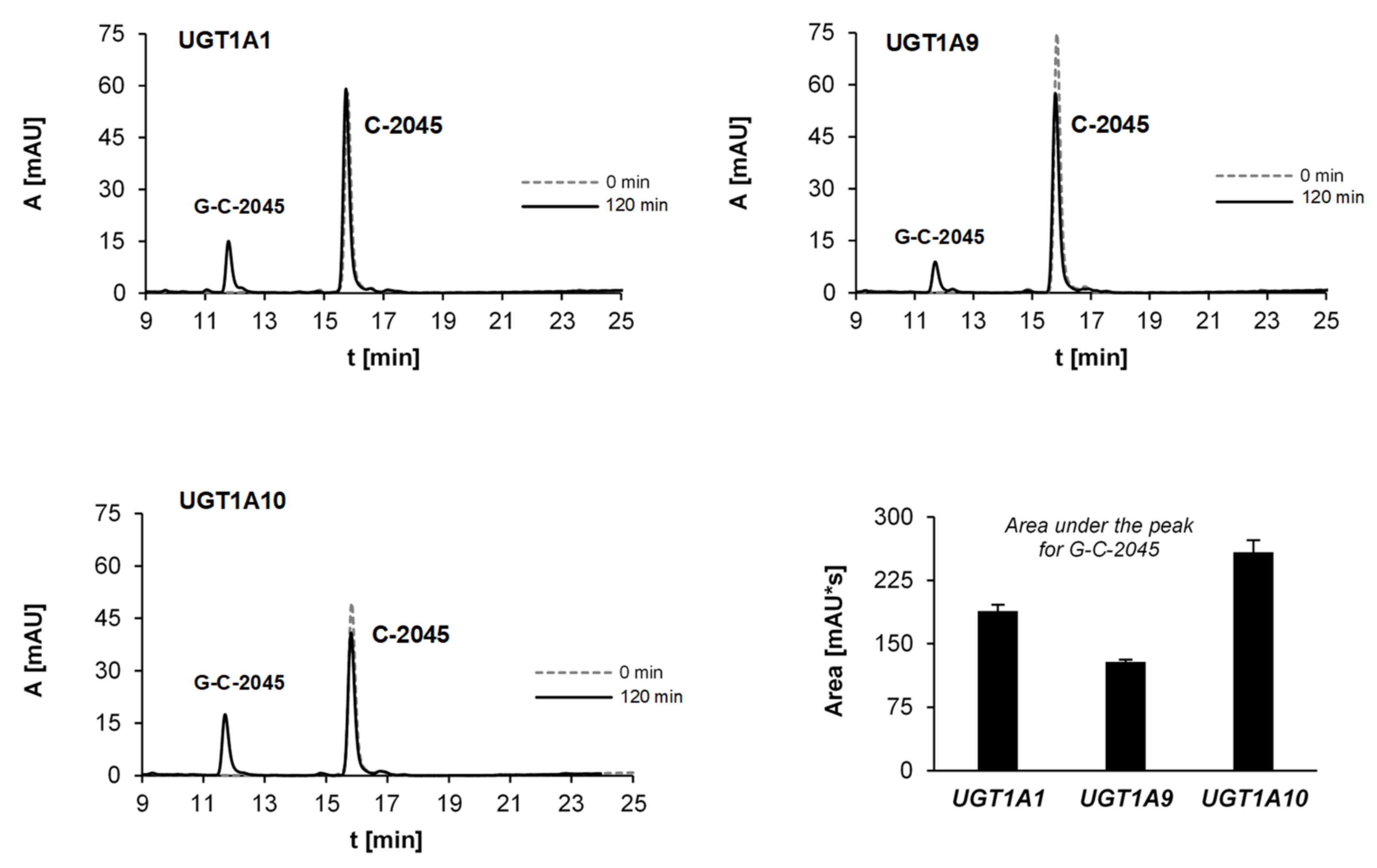
2.2.5. Glucuronidation of UAs with Human Recombinant UGT Isoenzymes
2.3. Metabolism of Studied UAs in Colon Tumor Cells
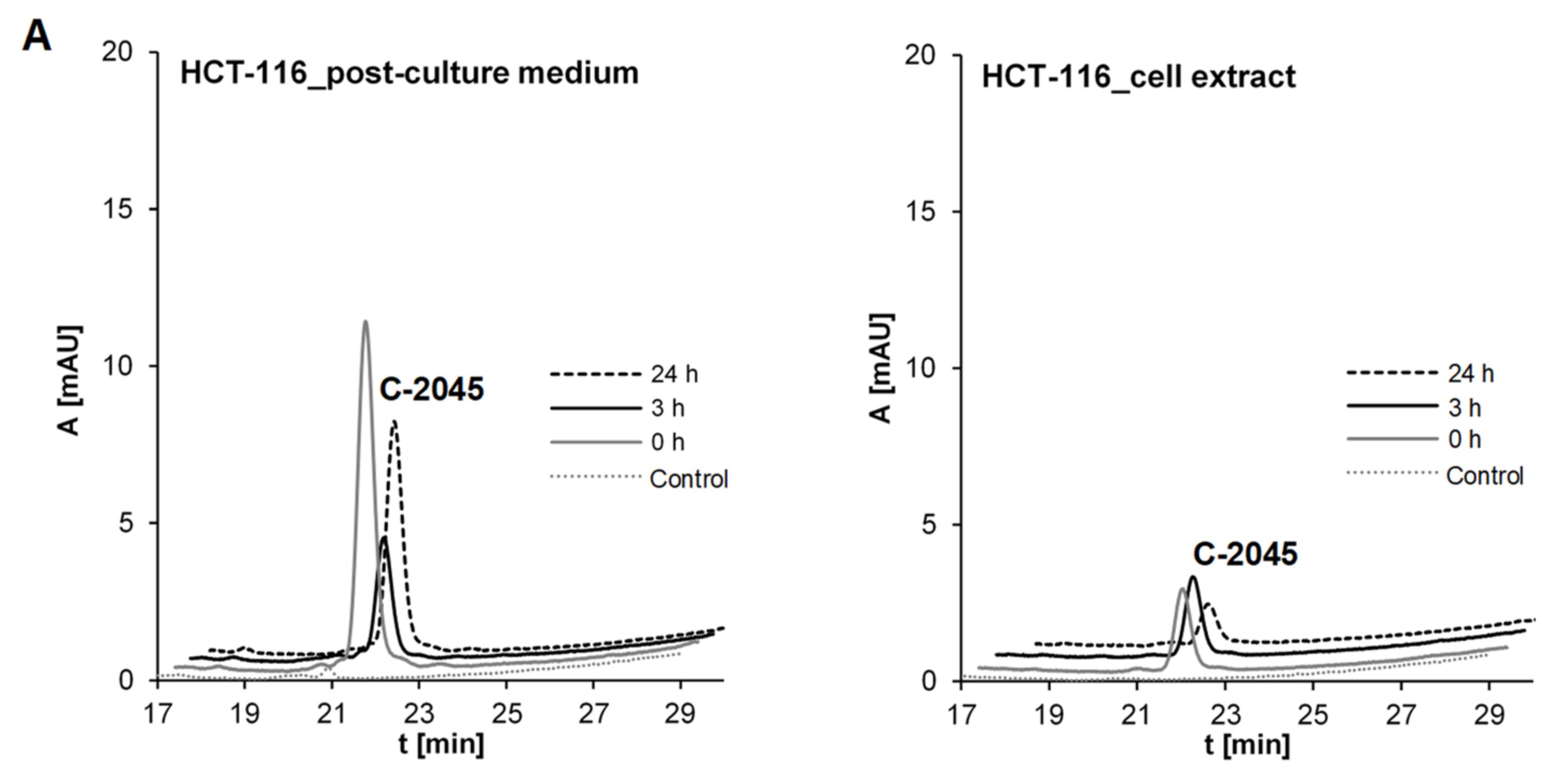
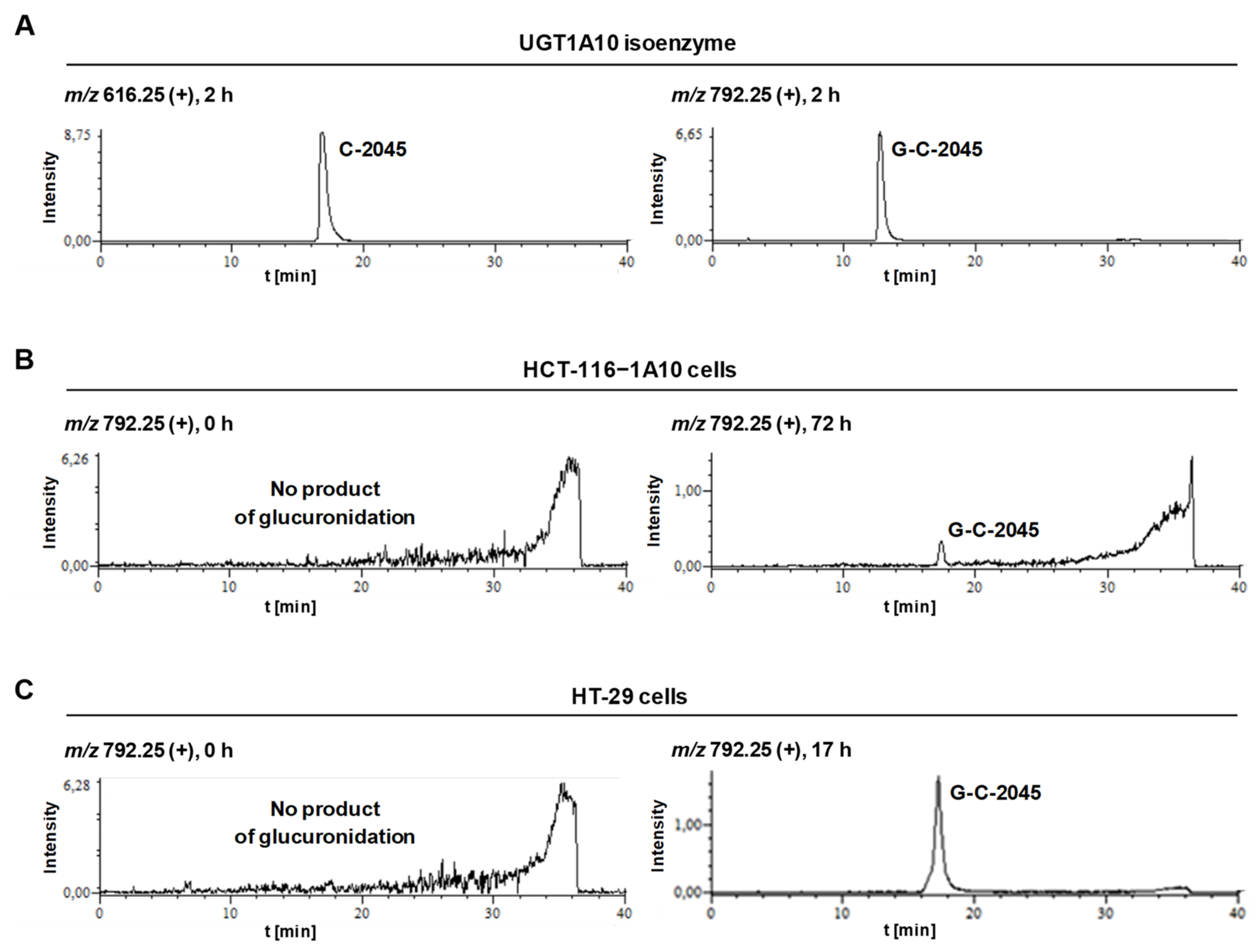
2.3.1. Transformations of C-2045 in Colon Cell Lines with Different UGT Expression
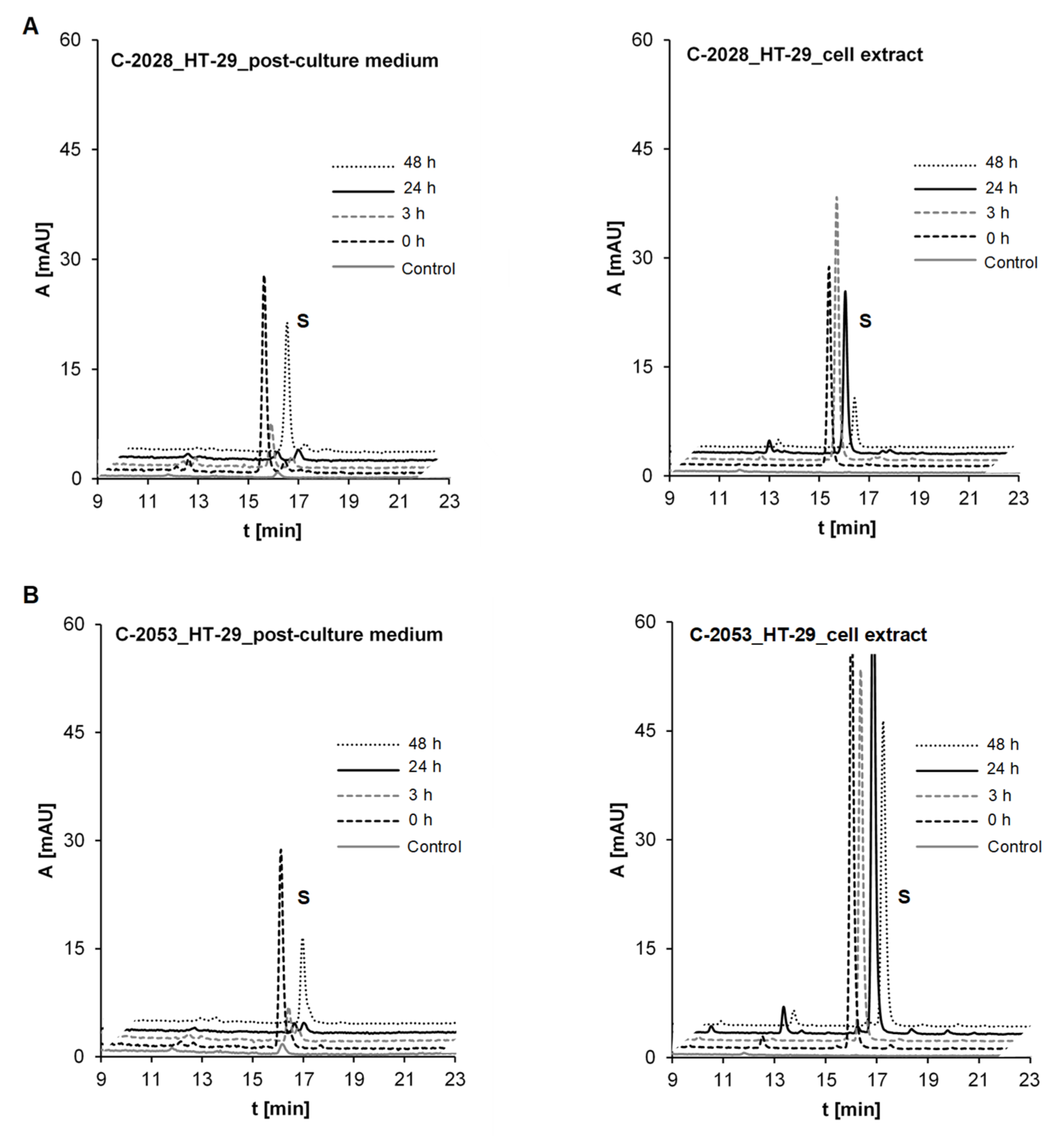
2.3.2. Metabolism of UAs in the HT-29 Tumor Cell Line of High UGT Expression
2.4. The Compound’s Impact on P450 and UGT Isoenzymes Activity
2.4.1. P4503A4 Inhibition by C-2045 and C-2053
2.4.2. UGT Isoenzymes Inhibition by Bisacridines Studied
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Enzymes
4.3. PKa Value Determinations
4.4. Voltammetric Analysis
4.5. Metabolism with Rat Liver Microsomes and Recombinant P450 Isoforms
4.6. Transformations with Recombinant FMO Isoforms
4.7. Transformations with Recombinant UGT Isoforms
4.8. Drug Metabolism in Colon Tumor Cell Lines HCT-116 and HT-29
4.9. Modulation of P4503A4 and UGT Activity In Vitro
4.10. HPLC-UV-vis Analysis—Products of Electrochemical and Metabolic Transformations
4.11. HPLC-ESI-MS, HPLC-ESI-MS/MS and HPLC-ESI-Q-TOF-MS Identification of the Transformation Products
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barraud, L.; Merle, P.; Soma, E.; Lefrançois, L.; Guerret, S.; Chevallier, M.; Dubernet, C.; Couvreur, P.; Trépo, C.; Vitvitski, L. Increase of doxorubicin sensitivity by doxorubicin-loading into nanoparticles for hepatocellular carcinoma cells in vitro and in vivo. J. Hepatol. 2005, 42, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.; Dullaart, A.; Bock, A.K.; Zweck, A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006, 24, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Cholody, W.M.; Martelli, S.; Konopa, J. 8-Substituted 5-[(aminoalkyl)amino]-6H-v-triazolo [4,5,1-de]acridin-6-ones as potential antineoplastic agents. Synthesis and biological activity. J. Med. Chem. 1990, 33, 2852–2856. [Google Scholar] [CrossRef] [PubMed]
- Cholody, W.M.; Martelli, S.; Paradziej-Lukowicz, J.; Konopa, J. 5-[(Aminoalkyl)amino]imidazo[4,5,1-de]acridin-6-ones as a novel class of antineoplastic agents. Synthesis and biological activity. J. Med. Chem. 1990, 33, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Cholody, W.M.; Martelli, S.; Konopa, J. Chromophore-modified antineoplastic imidazoacridinones. Synthesis and activity against murine leukemias. J. Med. Chem. 1992, 35, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Cholody, W.M.; Horowska, B.; Paradziej-Lukowicz, J.; Martelli, S.; Konopa, J. Structure-Activity Relationship for Antineoplastic Imidazoacridinones: Synthesis and Antileukemic Activity in Vivo. J. Med. Chem. 1996, 39, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Isambert, N.; Campone, M.; Bourbouloux, E.; Drouin, M.; Major, A.; Loadman, P.; Capizzi, R.; Grieshaber, C.; Fumoleau, P. Evaluation of the safety of C-1311 administered in a phase I dose-escalation trial as a weekly infusion for 3 consecutive weeks in patients with advanced solid tumors. J. Clin. Oncol. 2006, 24. [Google Scholar] [CrossRef]
- Isambert, N.; Campone, M.; Bourbouloux, E.; Drouin, M.; Major, A.; Yin, W.; Loadman, P.; Capizzi, R.; Grieshaber, C.; Fumoleau, P. Evaluation of the safety of C-1311 (SYMADEX) administered in a phase 1 dose escalation trial as a weekly infusion for 3 consecutive weeks in patients with advanced solid tumours. Eur. J. Cancer 2010, 46, 729–734. [Google Scholar] [CrossRef]
- Thomas, A.L.; Anthoney, A.; Ahmed, S.; Drouin, M.; Major, A.; Capizzi, R.L.; Grieshaber, C.; Loadman, P. Evaluation of the safety of C-1311 administered in a phase 1 dose-escalation trial as a 1-hour infusion once every 3 weeks in patients with advanced solid tumors. J. Clin. Oncol. 2006, 24, 2576. [Google Scholar] [CrossRef]
- Thomas, A.L.; Anthoney, A.; Scott, E.; Ahmed, S.; Lundberg, A.S.; Major, A.; Capizzi, R.L.; Twelves, C.J. C-1311, a novel inhibitor of FLT3 and topoisomerase II: A phase 1 trial of a once every three week schedule in patients with advanced solid tumors. J. Clin. Oncol. 2008, 24, 2576. [Google Scholar] [CrossRef]
- Capizzi, R.L.; Roman, L.A.; Tjulandin, S.; Smirnova, I.; Manikhas, A.; Paterson, J.S.; Major, A.; Lundberg, A.S. Phase II trial of C1311, a novel inhibitor of topoisomerase II in advanced breast cancer. J. Clin. Oncol. 2008, 24, 1055. [Google Scholar] [CrossRef]
- WHO Chronicle. International Non-proprietary Names for Pharmaceutical Substances. Suppl. WHO Chron. 1976, 30, 1–18. [Google Scholar]
- Ashok, B.T.; Tadi, K.; Banerjee, D.; Konopa, J.; Iatropoulos, M.; Tiwari, R.K. Pre-clinical toxicology and pathology of 9-(2′-hydroxyethylamino)-4-methyl-1-nitroacridine (C-1748), a novel anti-cancer agent in male Beagle dogs. Life Sci. 2006, 79, 1334–1342. [Google Scholar] [CrossRef]
- Ashok, B.T.; Tadi, K.; Garikapaty, V.P.S.; Chen, Y.; Huang, Q.; Banerjee, D.; Konopa, J.; Tiwari, R.K. Preclinical toxicological examination of a putative prostate cancer-specific 4-methyl-1-nitroacridine derivative in rodents. Anticancer Drugs 2007, 18, 87–94. [Google Scholar] [CrossRef]
- Tadi, K.; Ashok, B.T.; Chen, Y.; Banerjee, Y.D.; Wysocka-Skrzela, B.; Konopa, J.; Darzynkiewicz, Z.; Tiwari, R.K. Pre-clinical evaluation of 1-nitroacridine derived chemotherapeutic agent that has preferential cytotoxic activity towards prostate cancer. Cancer Biol. Ther. 2007, 6, 1632–1637. [Google Scholar] [CrossRef]
- Składanowski, A.; Plisov, S.Y.; Konopa, J.; Larsen, A.K. Inhibition of DNA topoisomerase II by imidazoacridinones, new antineoplastic agents with strong activity against solid tumors. Mol. Pharmacol. 1996, 49, 772–780. [Google Scholar]
- Dziegielewski, J.; Konopa, J. Interstrand crosslinking of DNA induced in tumor cells by a new group of antitumor imidazoacridinones. Proc. Am. Assoc. Cancer Res. 1996, 37, 410. [Google Scholar]
- Pawlak, K.; Pawlak, J.; Konopa, J. Cytotoxic and antitumor activity of 1-nitroacridines as an aftereffect of their interstrand DNA cross-linking. Cancer Res. 1984, 44, 4289–4296. [Google Scholar]
- Paradziej-Łukowicz, J.; Skwarska, A.; Peszyńska-Sularz, G.; Brillowska-Dąbrowska, A.; Konopa, J. Anticancer imidazoacridinone C-1311 inhibits hypoxia-inducible factor-1 (HIF-1), vascular endothelial growth factor (VEGF) and angiogenesis. Cancer Biol. Ther. 2011, 12, 586–597. [Google Scholar] [CrossRef]
- Skwarska, A.; Augustin, E.; Befinger, M.; Wojtczyk, A.; Konicz, S.; Laskowska, K.; Polewska, J. Targeting of FLT3-ITD kinase contributes to high selectivity of imidazoacridinone C-1311 against FLT3-activated leukemia cells. Biochem. Pharmacol. 2015, 95, 238–252. [Google Scholar] [CrossRef]
- Zaffaroni, N.; De Marco, C.; Villa, R.; Riboldi, S.; Daidone, M.G.; Double, J.A. Cell growth inhibition, G2M cell cycle arrest and apoptosis induced by the imidazoacridinone C-1311 in human tumour cell lines. Eur. J. Cancer 2001, 37, 1953–1962. [Google Scholar] [CrossRef]
- Augustin, E.; Pawłowska, M.; Polewska, J.; Potega, A.; Mazerska, Z. Modulation of CYP3A4 activity and induction of apoptosis, necrosis and senescence by the anti-tumour imidazoacridinone C-1311 in human hepatoma cells. Cell Biol. Int. 2013, 37, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Polewska, J.; Skwarska, A.; Augustin, E.; Konopa, J. DNA-damaging imidazoacridinone C-1311 induces autophagy followed by irreversible growth arrest and senescence in human lung cancer cells. J. Pharmacol. Exp. Ther. 2013, 346, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Augustin, E.; Moś-Rompa, A.; Nowak-Ziatyk, D.; Konopa, J. Antitumor 1-nitroacridine derivative C-1748, induces apoptosis, necrosis or senescence in human colon carcinoma HCT8 and HT29 cells. Biochem. Pharmacol. 2010, 79, 1231–1241. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cholody, W.M.; Hernandez, L.; Hassner, L.; Scudiero, D.A.; Djurickovic, D.B.; Michejda, C.J. Bisimidazoacridones and Related Compounds: New Antineoplastic Agents with High Selectivity against Colon Tumors. J. Med. Chem 1995, 38, 3043–3052. [Google Scholar] [CrossRef]
- Denny, W.A.; Gamage, S.A.; Spicer, J.A.; Baguley, B.C.; Finlay, G.J. Bis(acridinecarboxamide) and bis(phenazinecarboxamide) as Antitumor Agents. U.S. Patent WO 9817650 A1, 5 September 2000. [Google Scholar]
- Spicer, J.A.; Gamage, S.A.; Atwell, G.J.; Finlay, G.J.; Baguley, B.C.; Denny, W.A. Dimeric Analogues of Non-Cationic Tricyclic Aromatic Carboxamides are a New Class of Cytotoxic Agents. Anticancer Drug Des. 1999, 14, 281–289. [Google Scholar]
- Gribble, G.W.; Mosher, M.D.; Jaycox, G.D.; Cory, M.; Fairley, T.A. Potential DNA bis-intercalating agents. Synthesis and antitumor activity of N,N-methylenedi-4,1-cyclohexanediyl-bis(9-acridinamine) isomers. Heterocycles 2014, 88, 535–546. [Google Scholar] [CrossRef]
- Konopa, J.K.; Horowska, B.; Paluszkiewicz, E.A.; Borowa-Mazgaj, B.; Agustin, E.A.; Skwarska, A.; Mazerska, Z. Asymmetric Bis-acridines with Antitumor Activity and Use Thereof. Gdansk University of Technology. European Patent EP 3070078, 4 October 2017. [Google Scholar]
- Konopa, J.K.; Horowska, B.; Paluszkiewicz, E.A.; Borowa-Mazgaj, B.; Agustin, E.A.; Skwarska, A.; Mazerska, Z. Asymmetric Bis-acridines with Antitumor Activity and Use Thereof. Gdansk University of Technology. U.S. Patent US10,202,349 B2, 12 February 2019. [Google Scholar]
- Paluszkiewicz, E.; Horowska, B.; Borowa-Mazgaj, B.; Peszyńska-Sularz, G.; Paradziej-Łukowicz, J.; Augustin, E.; Konopa, J.; Mazerska, Z. Design, synthesis and high antitumor potential of new unsymmetrical bisacridine derivatives towards human solid tumors, specifically pancreatic cancers and their unique ability to stabilize DNA G-quadruplexes. Eur. J. Med. Chem. 2020, 204, 112599. [Google Scholar] [CrossRef]
- Mazerska, Z.; Mróz, A.; Pawłowska, M.; Augustin, E. The role of glucuronidation in drug resistance. Pharmacol. Ther. 2016, 159, 35–55. [Google Scholar] [CrossRef]
- Bejrowska, A.; Pawłowska, M.; Mróz, A.; Mazerska, Z. Modulation of UDP-glucuronidation by acridinone antitumor agents C-1305 and C-1311 in HepG2 and HT29 cell lines, despite slight impact in noncellular systems. Pharmacol. Rep. 2018, 70, 470–475. [Google Scholar] [CrossRef]
- Mróz, A.; Ryska, I.; Sominko, H.; Bejrowska, A.; Mazerska, Z. Drug-drug interaction potential of antitumor acridine agent C-1748: The substrate of UDP-glucuronosyltransferases 2B7, 2B17 and the inhibitor of 1A9 and 2B7. Pharmacol. Rep. 2018, 70, 972–980. [Google Scholar] [CrossRef]
- Preissner, S.; Simmaco, M.; Gentile, G.; Preissner, R. Personalized Cancer Therapy Considering Cytochrome P450 Variability. Adv. Pharmacol. 2015, 74, 113–130. [Google Scholar] [CrossRef]
- Rendic, S.; Di Carlo, F.J. Human cytochrome P450 enzyme: A status report summarizing their reactions, substrates, induction, and inhibitors. Drug. Metab. Rev. 1997, 29, 413–580. [Google Scholar] [CrossRef]
- McFadyen, M.C.E.; Melvin, W.T.; Murray, G.I. Cytohrome P450 enzymes: Novel options for cancer therapeutics. Mol. Cancer Ther. 2003, 3, 363–371. [Google Scholar]
- Sakurai, K.; Enomoto, K.; Matsuo, S.; Amano, S.; Shiono, M. CYP3A4 Expression to Peredict Treatment Response to Docetaxel for Metastasis and Recurrence of Primary Breast Cancer. Surg. Today 2011, 41, 674–679. [Google Scholar] [CrossRef]
- Scott, S.A.; Sangkuhl, K.; Shuldiner, A.R.; Hulot, J.; Thorn, C.F.; Altman, R.B.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenet. Genom. 2012, 22, 159–165. [Google Scholar] [CrossRef]
- Flaten, H.K.; Kim, H.S.; Campbell, J.; Hamilton, L.; Monte, A. CYP2C19 drug-drug and drug-gene interactions in ED patients. Am. J. Emerg. Med. 2016, 34, 245–249. [Google Scholar] [CrossRef]
- Sequeira, D.; Strobel, H.W. High-performance liquid chromatographic method for the analysis of imipramine metabolism in vitro by liver and brain microsomes. J. Chromatogr. B. Biomed. Appl. 1995, 673, 251–258. [Google Scholar] [CrossRef]
- Mazerska, Z.; Zamponi, S.; Marassi, R.; Sowiński, P.; Konopa, J. The products of electro- and photochemical oxidation of 2-hydroxyacridinone, the reference compound of antitumor imidazoacridinone drivatives. J. Electroanal. Chem. 2002, 521, 144–154. [Google Scholar] [CrossRef]
- Mazerska, Z.; Sowiński, P.; Konopa, J. Molecular mechanism of the enzymatic oxidation investigated for imidazoacridinone entitumor drug, C-1311. Biochem. Pharmacol. 2003, 66, 1727–1736. [Google Scholar] [CrossRef]
- Potęga, A.; Garwolińska, D.; Nowicka, A.M.; Fau, M.; Kot-Wasik, A.; Mazerska, Z. Phase I and phase II metabolism simulation of antitumor-active 2-hydroxyacridinone with electrochemistry coupled on-line with mass spectrometry. Xenobiotica 2019, 49, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Potęga, A.; Paczkowski, S.Z.; Paluszkiewicz, E.; Mazerska, Z. Electrochemical simulation of metabolic reduction and conjugation reactions of unsymmetrical bisacridine antitumor agents, C-2028 and C-2053. J. Pharm. Biomed. Anal. 2021, 179, 113970. [Google Scholar] [CrossRef] [PubMed]
- Gorlewska, K.; Mazerska, Z.; Sowiński, P.; Konopa, J. Products of metabolic activation of the antitumor drug ledakrin (nitracrine) in vitro. Chem. Res. Toxicol. 2001, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, A.; Niemira, M.; Jagiełło, K.; Potęga, A.; Swist, M.; Henderson, C.; Skwarska, A.; Augustin, E.; Konopa, J.; Mazerska, Z. Diminished toxicity of C-1748, 4-methyl-9-hydroxyethylamino-1-nitroacridine, compared with its demethyl analog, C-857, corresponds to its resistance to metabolism in HepG2 cells. Biochem. Pharmacol. 2012, 84, 30–42. [Google Scholar] [CrossRef]
- Wiśniewska, A.; Chrapkowska, A.; Kot-Wasik, A.; Konopa, J.; Mazerska, Z. Metabolic transformations of antitumor imidazoacridinone, C-1311, with microsomal fractions of rat and human liver enzymes. Acta Bioch. Polonica 2007, 54, 831–838. [Google Scholar] [CrossRef]
- Potęga, A.; Dąbrowska, E.; Niemira, M.; Kot-Wasik, A.; Ronseaux, S.; Henderson, C.J.; Wolf, C.R.; Mazerska, Z. The imidazoacridinone antitumor Drug, C-1311, is Metabolized by Flavin Monooxygenases not by Cytochrome P450s. Drug Metab. Dispos. 2011, 39, 1423–1432. [Google Scholar] [CrossRef]
- Cashman, J.R.; Zhang, J. Interindividual differences of human flavin-containing monooxygenase 3: Genetic polymorphism and functional variation. Drug Metab. Dispos. 2002, 30, 1043–1052. [Google Scholar] [CrossRef]
- Philips, I.A.; Shephard, E.A. Flavin containing monooxygenase 3 (FMO3): Genetic variants and their consequences for drug metabolism and disease. Xenobiotica 2020, 50, 19–33. [Google Scholar] [CrossRef]
- Cataluci, G.; Querio, G.; Sadeghi, S.J.; Gilardi, G.; Levi, R. Enzymatically produced trimethylamine N-oxide: Conserving it or eliminating it. Catalysts 2019, 9, 1028. [Google Scholar] [CrossRef]
- Thodberg, S.; Neilson, E.H.J. The “green” FMOs: Diversity, functionality and application of plant flavoproteins. Catalysts 2020, 10, 329. [Google Scholar] [CrossRef]
- Williams, J.A.; Hyland, R.; Jones, B.C.; Smith, D.A.; Hurst, S.; Goosen, T.C.; Peterkin, V.; Koup, J.R.; Ball, S.E. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab. Dispos. 2004, 32, 1201–1208. [Google Scholar] [CrossRef]
- Fujiwara, R.; Sumida, K.; Kutsuno, Y. Species differences in drug glucuronidation: Humanized UDP-glucuronosyltransferase 1 mice and their application for predicting drug glucuronidation and drug-induced toxicity in humans. Drug Metab. Pharmacokinet. 2015, 30, 82–88. [Google Scholar] [CrossRef]
- Yang, N.; Sun, R.; Liao, X.; Aa, J.; Wang, G. UDP-glucuronosyltransferases (UGTs) and their related metabolic cross-talk with internal homeostasis: A systematic review of UGT isoforms for precision medicine. Pharmacol. Res. 2017, 30, 169–183. [Google Scholar] [CrossRef]
- Ohno, S.; Nakajin, S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 2009, 37, 32–40. [Google Scholar] [CrossRef]
- Court, M.H.; Zhang, X.; Ding, X.; Yee, K.K.; Hesse, L.M.; Finel, M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica 2012, 42, 266–277. [Google Scholar] [CrossRef]
- Patana, A.; Kurkela, M.; Finel, M.; Goldman, A. Mutation analysis in UGT1A9 suggests a relationship between substrate and catalytic residues in UDP-glucuronosyltransferases. Protein Eng. Des. Sel. 2008, 21, 537–543. [Google Scholar] [CrossRef]
- Korprasertthaworn, P.; Rowland, A.; Lewis, B.C.; Mackenzie, P.I.; Yoovathaworn, K.; Miners, J.O.; Goldman, A. Effects of amino acid substitutions at positions 33 and 37 of UDP-glucuronosyltransferase 1A9 (UGT1A9) activity and substrate selectivity. Biochem. Pharmacol. 2012, 84, 1511–1521. [Google Scholar] [CrossRef]
- Mojarabi, B.; Mackenzie, P.I. Characterization of Two UDP Glucuronosyltransferases That Are Predominantly Expressed in Human Colon. Biochem. Biophys. Res. Commun. 1998, 247, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Pilch, J.; Matysiak-Brynda, E.; Kowalczyk, A.; Bujak, P.; Mazerska, Z.; Nowicka, A.; Augustin, E. New unsymmetrical bisacridine derivatives noncovalently attached to quaternary quantum dots improve cancer therapy by enhancing cytotoxicity toward cancer cells and protecting normal cells. ACS Appl. Mater. Interfaces 2020, 12, 17276–17289. [Google Scholar] [CrossRef]
- Pawłowska, M.; Chu, R.; Fedejko-Kap, B.; Augustin, E.; Mazerska, Z.; Radominska-Pandya, A.; Chambers, T.C. Metabolic transformation of antitumor acridinone C-1305 but not C 1311 via selective cellular expression of UGT1A10 increases cytotoxic response: Implications for clinical use. Drug Metab. Dispos. 2013, 41, 414–421. [Google Scholar] [CrossRef]
- Cosa, G.; Scaiano, J.C. Reactivity of adrenaline toward alkoxyl radicals and karbonyl triplet states. Org. Biomol. Chem. 2008, 6, 4609–4614. [Google Scholar] [CrossRef] [PubMed]
- Smythies, J.; Galzigna, L. The oxidative metabolism of catecholamines in the brain: A review. Biochim. Biophys. Acta 1998, 1382, 159–162. [Google Scholar] [CrossRef]
- Pawłowska, M.; Augustin, E.; Mazerska, Z. CYP3A4 overexpression enhances apoptosis induced by anticancer agent imidazoacridinone C-1311, but does not change the metabolism of C-1311 in CHO cells. Acta Pharmacol. Sin. 2014, 35, 98–112. [Google Scholar] [CrossRef]
- Cunningham, D.; Maroun, J.; Vanhoefer, U.; Van Cutsem, E. Optimizing the use of irinotecan in colorectal cancer. Oncologist 2001, 6, 17–23. [Google Scholar] [CrossRef]
- Bouché, O.; Raoul, J.L.; Bonnetain, F.; Giovannini, M.; Etienne, P.L.; Lledo, G.; Arsène, D.; Paitel, J.F.; Guérin-Meyer, V.; Mitry, E.; et al. Randomized multicenter phase II trial of a biweekly regimen of fluorouracil and leucovorin (LV5FU2), LV5FU2 plus cisplatin, or LV5FU2 plus irinotecan in patients with previously untreated metastatic gastric cancer: A Federation Francophone de Cancerologie Digestive Group Study-FFCD 9803. J. Clin. Oncol. 2004, 22, 4319–4328. [Google Scholar] [CrossRef]
- Wang, L.Z.; Ramirez, J.; Yeo, W.; Chan, M.Y.; Thuya, W.L.; Lau, J.Y.; Wan, S.C.; Wong, A.L.; Zee, Y.K.; Lim, R.; et al. Glucuronidation by UGT1A1 is the dominant pathway of the metabolic disposition of belinostat in liver cancer patients. PLoS ONE 2013, 8, e54522. [Google Scholar] [CrossRef]
- Wen, Z.; Tallman, M.N.; Ali, S.Y.; Smith, P.C. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for etoposide glucuronidation in human liver and intestinal microsomes: Structural characterization of phenolic and alcoholic glucuronides of etoposide and estimation of enzyme kinetics. Drug Metab. Dispos. 2007, 35, 371–380. [Google Scholar] [CrossRef]
- Kamiyama, N.; Takagi, S.; Yamamoto, C.; Kudo, T.; Nakagawa, T.; Takahashi, M.; Nakanishi, K.; Takahashi, H.; Todo, S.; Iseki, K. Expression of ABC Transporters in Human Hepatocyte Carcinoma Cells with Cross-resistance to Epirubicin and Mitoxantrone. Anticancer Res. 2006, 26, 885–888. [Google Scholar]
- Paul, D.; Standifer, K.M.; Inturrisi, C.E.; Pasternak, G.W. Pharmacological characterization of morphine-6 beta-glucuronide, a very potent morphine metabolite. Pharmacol. Exp. Ther. 1989, 251, 477–483. [Google Scholar]
- Stone, A.N.; Mackenzie, P.I.; Galetin, A.; Houston, J.B.; Miners, J.O. Isoform selectivity and kinetics of morphine 3-and 6-glucuronidation by human UDP-glucuronosyltransferases: Evidence for atypical glucuronidation kinetics by UGT2B7. Drug Metab. Dispos. 2003, 31, 1086–1089. [Google Scholar] [CrossRef]
- Ogura, K.; Ishikawa, Y.; Kaku, T.; Nishiyama, T.; Ohnuma, T.; Muro, K.; Hiratsuka, A. Quaternary ammonium-linked glucuronidation of trans-4-hydroxytamoxifen, an active metabolite of tamoxifen, by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem. Pharmacol. 2006, 71, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Chen, G.; Dellinger, R.W.; Duncan, K.; Fang, J.L.; Lazarus, P. Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res. 2006, 8, R50. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Sharma, A.K.; Dellinger, R.W.; Blevis-Primeau, A.S.; Balliet, R.M.; Chen, G.; Boyiri, T.; Amin, S.; Lazarus, P. Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab. Dispos. 2007, 35, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Starlard-Davenport, A.; Lyn-Cook, B.; Beland, F.A.; Pogribny, I.P. The role of UDP-glucuronosyltransferases and drug transporters in breast cancer drug resistance. Exp. Oncol. 2010, 32, 172–180. [Google Scholar] [PubMed]
- Ortiz de Montellano, P.R. Cytochrome P450: Structure, Mechanism and Bio-Chemistry; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 33–69. [Google Scholar]
- Pawłowska, M.; Kwaśniewska, A.; Mazerska, Z.; Augustin, E. Enhanced activity of P4503A4 and UGT1A10 induced by acridinone derivatives C-1305 and C-1311 in MCF-7 and HCT116 cancer cells: Consequences for the drug’cytotoxicity, metabolism and cellular response. Int. J. Mol. Sci. 2020, 21, 3954. [Google Scholar] [CrossRef]
- Rios Martines, C.H.; Dardonville, C. Rapid Determination of Ionization Constants (pKa) by UV Spectroscopy Using 96-Well Microtiter Plates. ACS Med. Chem. Lett. 2013, 4, 142–145. [Google Scholar] [CrossRef]
- Rover, L., Jr.; Fernandes, J.C.B.; Neto, G.; Kubota, L.T.; Katekawa, E.; Serrano, S.H.P. Study of NADH Stability Using Ultraviolet–Visible Spectrophotometric Analysis and Factorial Design. Anal. Biochem. 1998, 260, 50–55. [Google Scholar] [CrossRef]
- Silva, F.V.L.; Resende, S.; Araujo, A.N.; Prior, J.A.V. Determination of pKa(s) of nilutamide through UV-visible spectroscopy. Microchem. J. 2018, 138, 303–308. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 13, 374. [Google Scholar] [CrossRef]
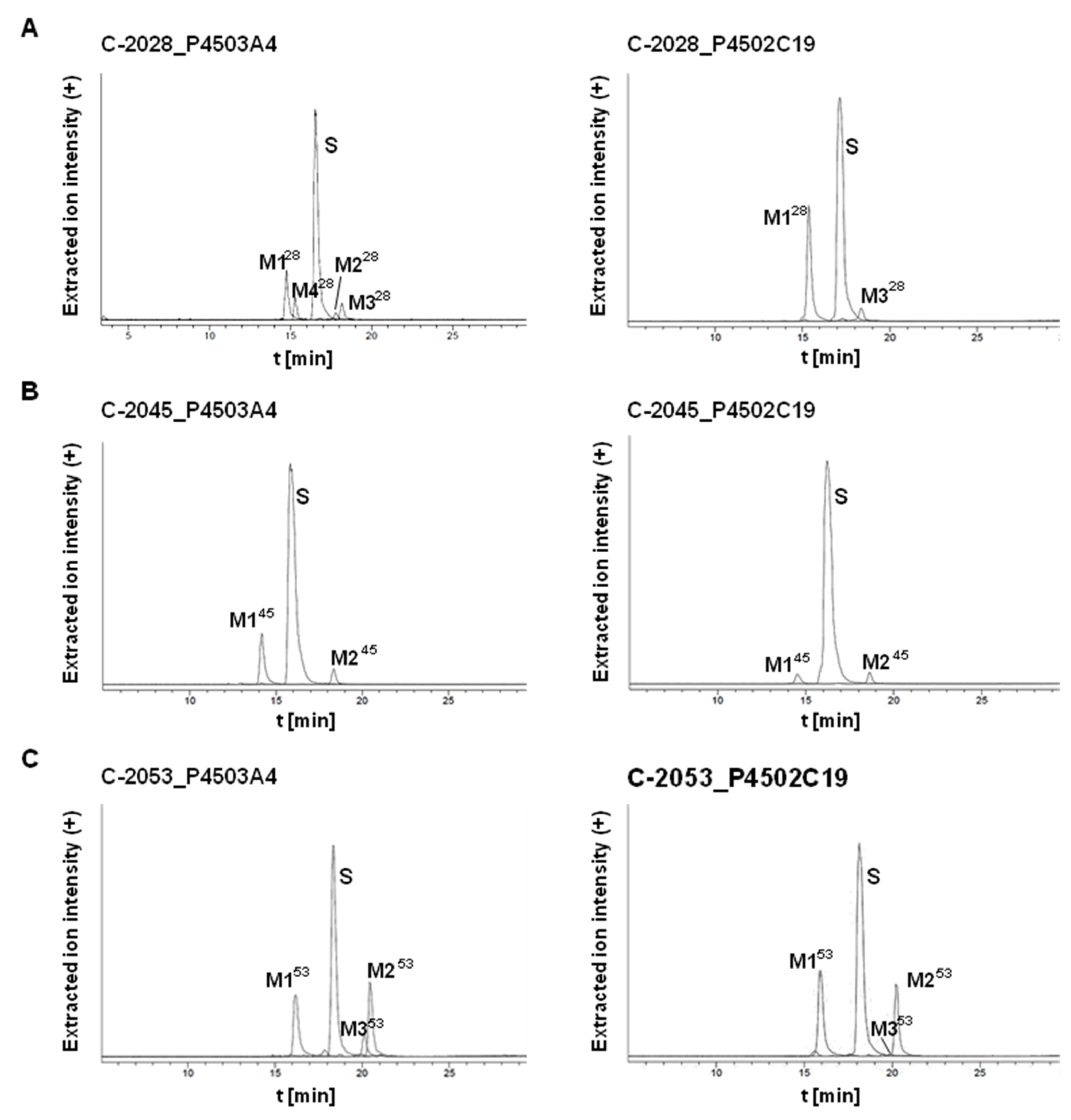
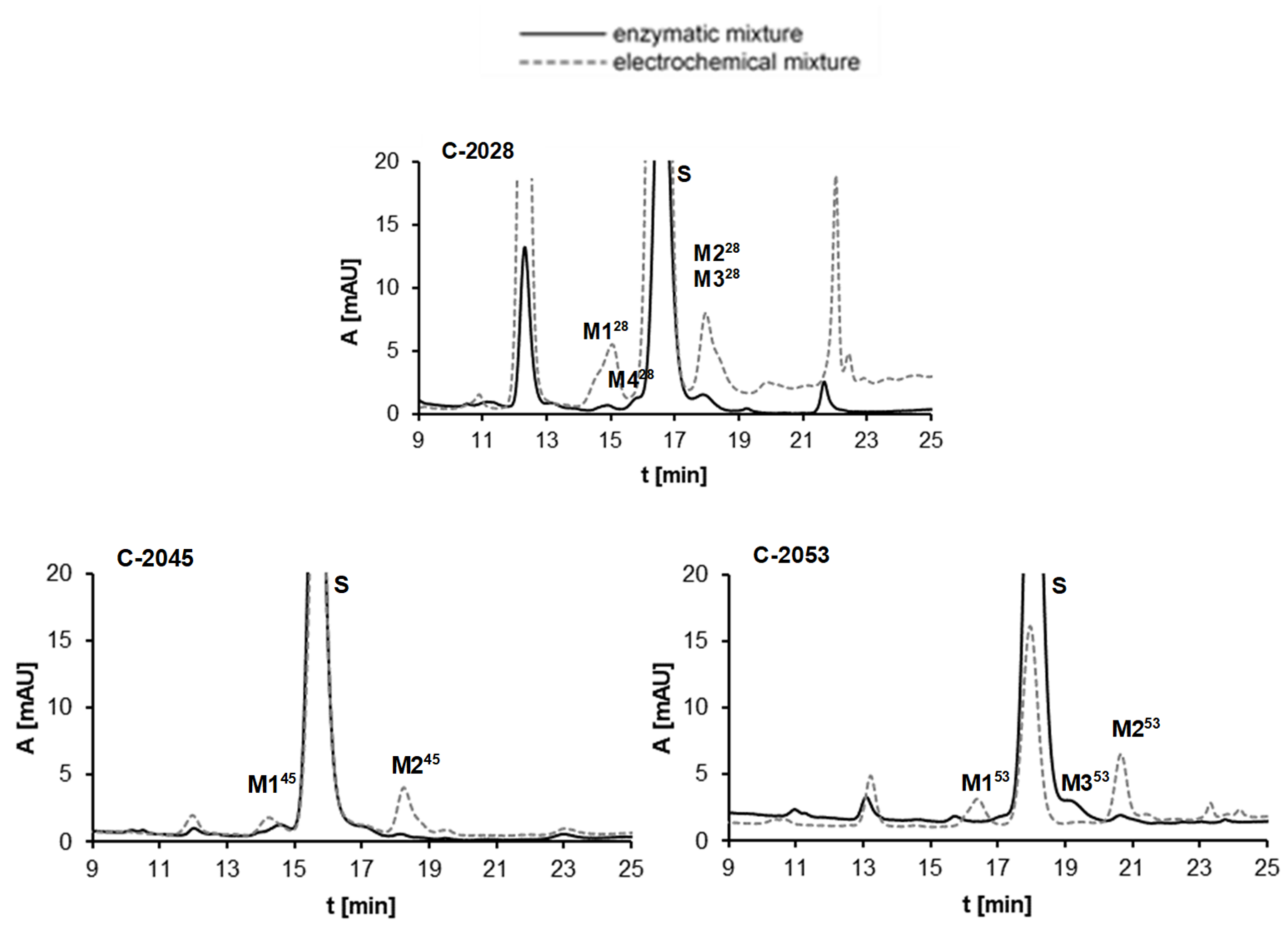
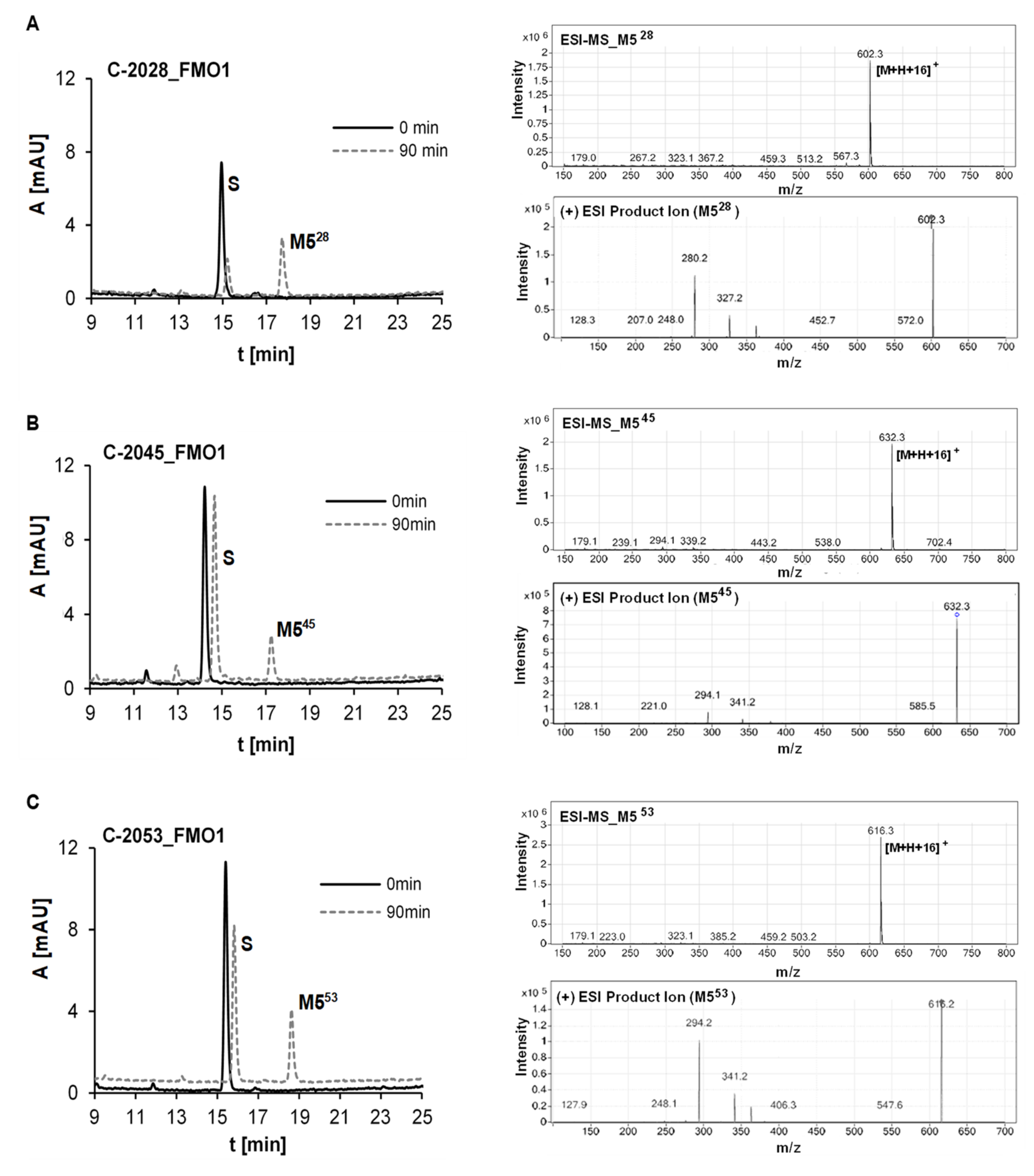
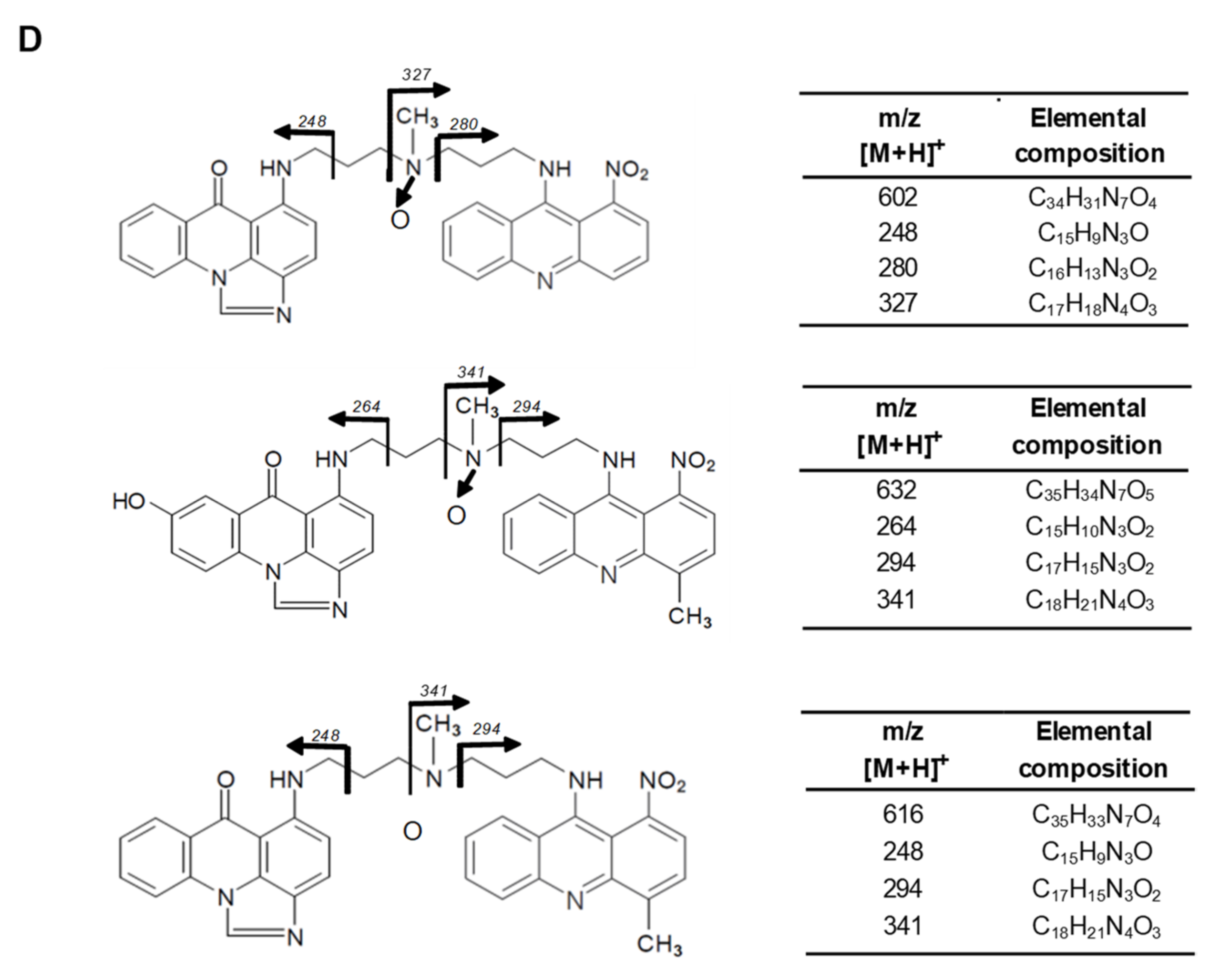


| Symbol | Structure | HPLC Retention Time [min] | UV-Vis Spectrum | ESI-MS m/z |
|---|---|---|---|---|
| C-2028 | 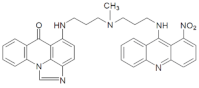 | 16.84 | 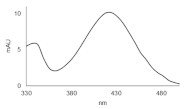 | 586.25 [M + H]+ |
| C-2045 | 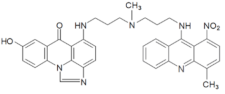 | 15.42 | 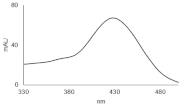 | 616.25 [M + H]+ |
| C-2053 | 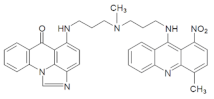 | 18.66 | 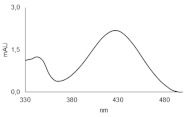 | 600.25 [M + H]+ |
| Compound | pKa 1 | Standard Deviation | pKa 2 | Standard Deviation | pKa 3 | Standard Deviation |
|---|---|---|---|---|---|---|
| C-2028 | 2.22 | ±0.01 | 6.01 | ±0.005 | 7.41 | ±0.02 |
| C-2045 | 2.33 | ±0.007 | 6.37 | ±0.005 | 7.50 | ±0.02 |
| C-2053 | 2.19 | ±0.01 | 6.19 | ±0.003 | 7.49 | ±0.01 |
| Metabolite | HPLC Retention Time [min] | ESI-MS m/z | ESI-Q-TOF-MS m/z | Proposed Structure |
|---|---|---|---|---|
| M128 | 15.06 | 556.25 [M + H-30]+ | 556.2799 [M+H-30]+ | 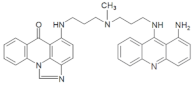 |
| M228 | 18.10 | 568.25 [M+H-18]+ | 568.2458 [M+H-18]+ | 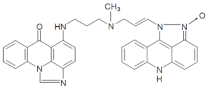 |
| M328 | 18.42 | 572.25 [M+H-14]+ | 572.2403 [M+H-14]+ |  |
| M428 | 15.64 | 307.25 | 307.1545 | 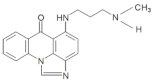 |
| M145 | 14.10 | 586.25 [M+H-30]+ | 586.2928 [M+H-30]+ | 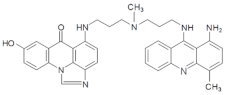 |
| M245 | 18.40 | 598.25 [M+H-18]+ | 598.2554 [M+H-18]+ | 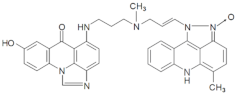 |
| M153 | 16.23 | 570.25 [M+H-30]+ | 570.2975 [M+H-30]+ | 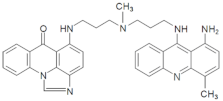 |
| M253 | 20.50 | 582.25 [M+H-18]+ | 582.2612 [M+H-18]+ | 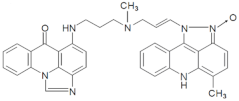 |
| M353 | 20.16 | 586.25 [M+H-14]+ | 586.2563 [M+H-14]+ |  |
| UGT 1A1 | UGT 1A6 | UGT 1A9 | UGT 1A10 | UGT 2B4 | UGT 2B7 | UGT 2B15 | UGT 2B17 | |
|---|---|---|---|---|---|---|---|---|
| C-2028 | − | − | − | − | − | − | − | − |
| C-2045 | + | +/− | + | + | − | − | − | − |
| C-2053 | − | − | − | − | − | − | − | − |
| Metabolite | HPLC Retention Time [min] | UV-vis Spectrum | ESI-MS m/z | Proposed Structure |
|---|---|---|---|---|
| G-C-2045 | 11.47 |  | 792.25 [M+H+176]+ |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mieszkowska, A.; Nowicka, A.M.; Kowalczyk, A.; Potęga, A.; Pawłowska, M.; Kosno, M.; Augustin, E.; Mazerska, Z. Metabolic Profiles of New Unsymmetrical Bisacridine Antitumor Agents in Electrochemical and Enzymatic Noncellular Systems and in Tumor Cells. Pharmaceuticals 2021, 14, 317. https://doi.org/10.3390/ph14040317
Mieszkowska A, Nowicka AM, Kowalczyk A, Potęga A, Pawłowska M, Kosno M, Augustin E, Mazerska Z. Metabolic Profiles of New Unsymmetrical Bisacridine Antitumor Agents in Electrochemical and Enzymatic Noncellular Systems and in Tumor Cells. Pharmaceuticals. 2021; 14(4):317. https://doi.org/10.3390/ph14040317
Chicago/Turabian StyleMieszkowska, Anna, Anna M. Nowicka, Agata Kowalczyk, Agnieszka Potęga, Monika Pawłowska, Michał Kosno, Ewa Augustin, and Zofia Mazerska. 2021. "Metabolic Profiles of New Unsymmetrical Bisacridine Antitumor Agents in Electrochemical and Enzymatic Noncellular Systems and in Tumor Cells" Pharmaceuticals 14, no. 4: 317. https://doi.org/10.3390/ph14040317
APA StyleMieszkowska, A., Nowicka, A. M., Kowalczyk, A., Potęga, A., Pawłowska, M., Kosno, M., Augustin, E., & Mazerska, Z. (2021). Metabolic Profiles of New Unsymmetrical Bisacridine Antitumor Agents in Electrochemical and Enzymatic Noncellular Systems and in Tumor Cells. Pharmaceuticals, 14(4), 317. https://doi.org/10.3390/ph14040317








