miR-16-5p Promotes Erythroid Maturation of Erythroleukemia Cells by Regulating Ribosome Biogenesis
Abstract
1. Introduction
2. Results
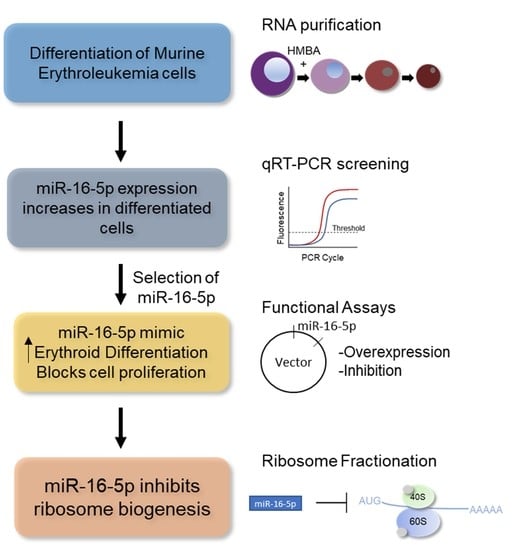
2.1. A Comprehensive miRNA Gene Expression Screening Uncovers miR-16-5p as a Potential Regulator of the MEL Cell Differentiation Program
2.2. miR-16-5p Overexpression Suppresses the Proliferation Potential of Various Cancer Cell Types
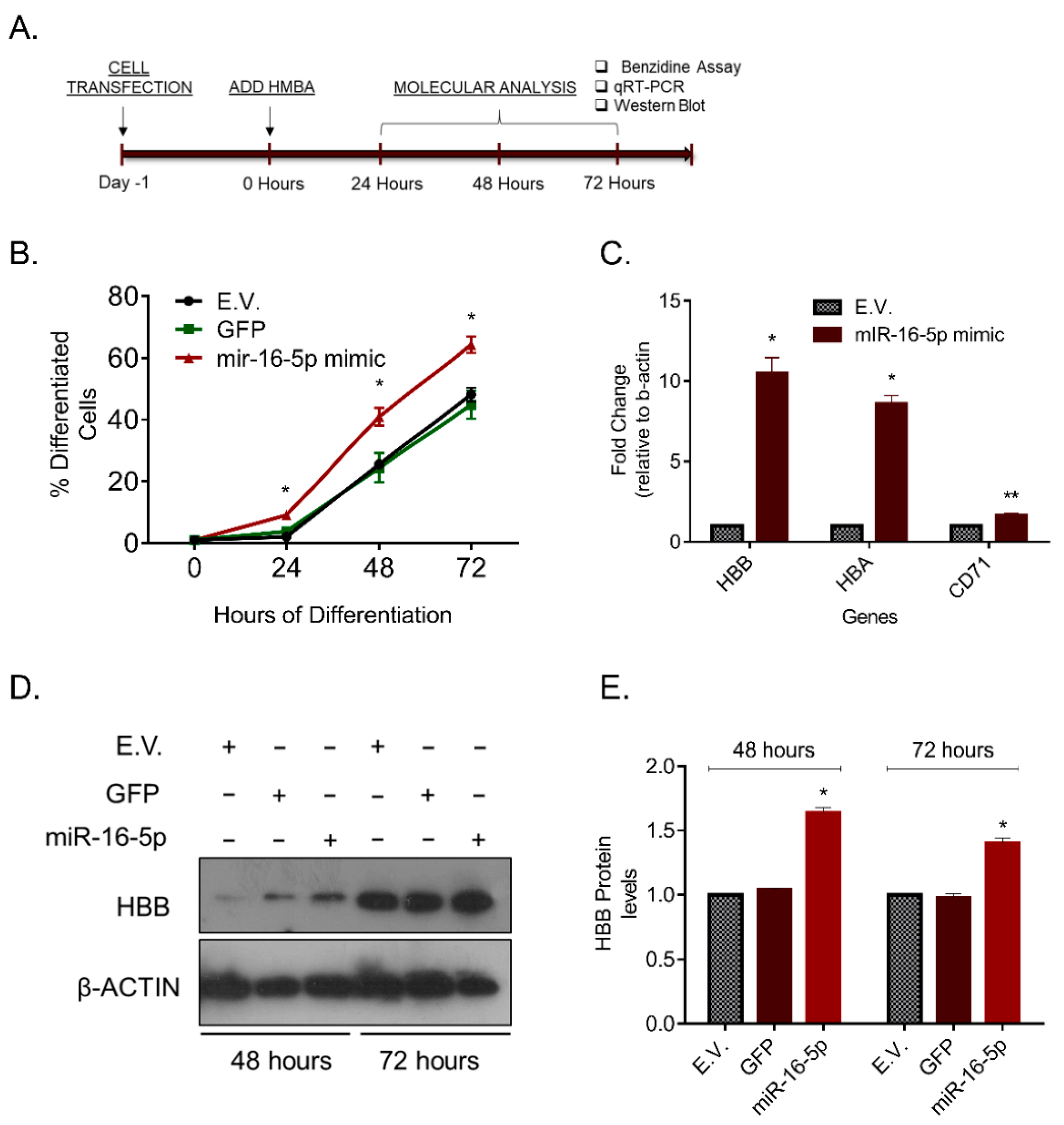
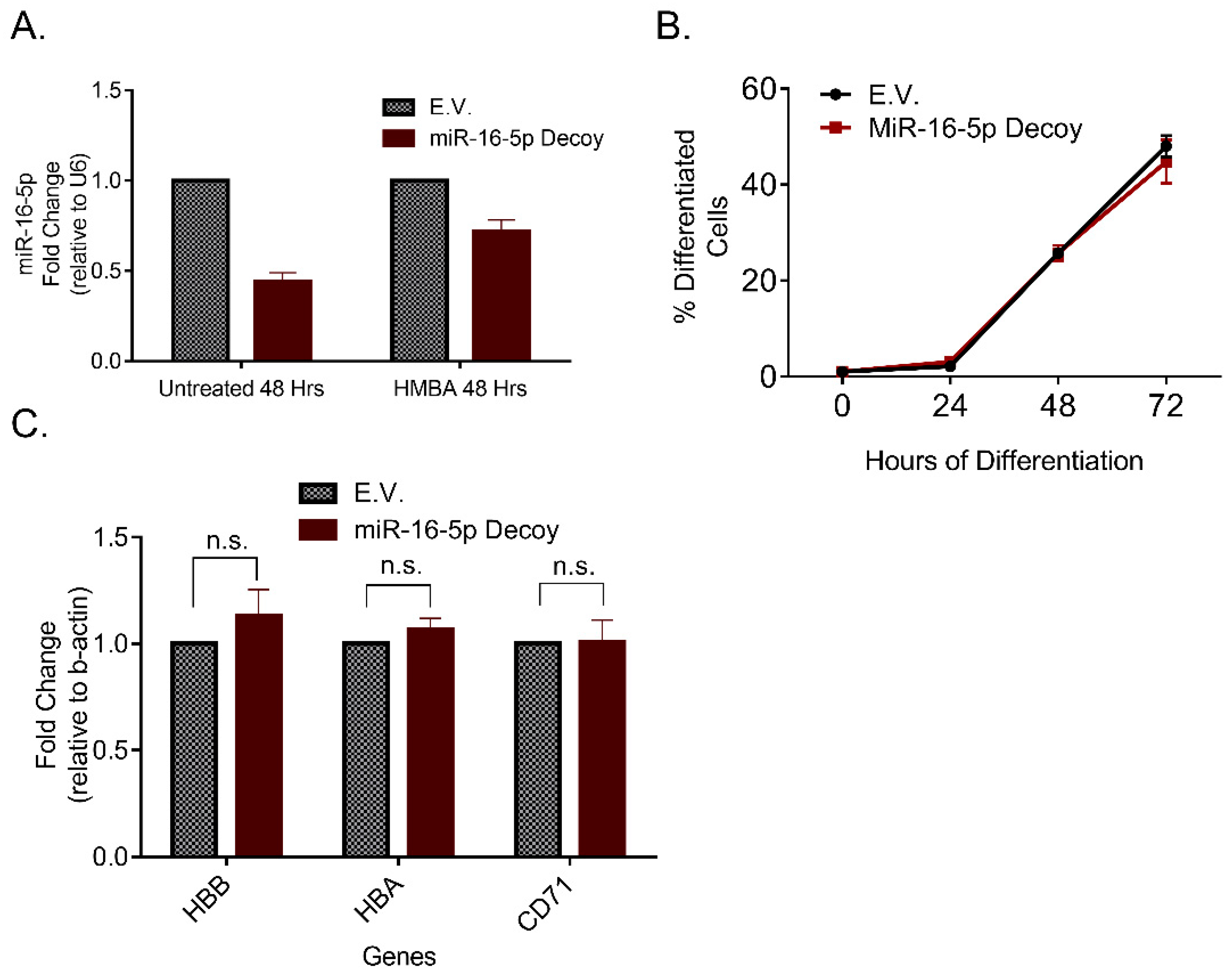
2.3. miR-16-5p Overexpression Increases Erythroid Differentiation of MEL Cells
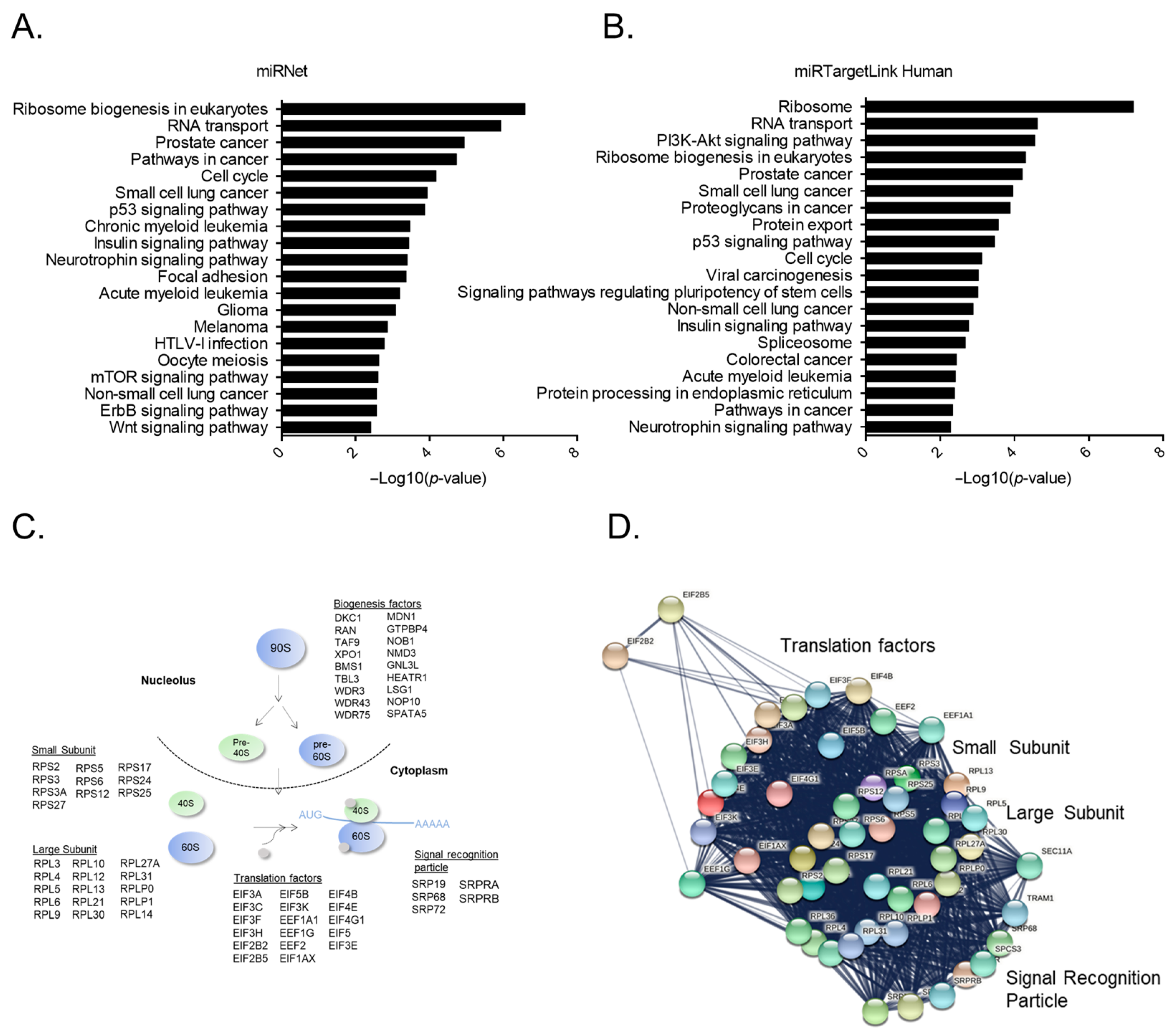
2.4. Bioinformatic Analysis Reveals Ribosome Constituents as the Main miR-16-5p Target Genes
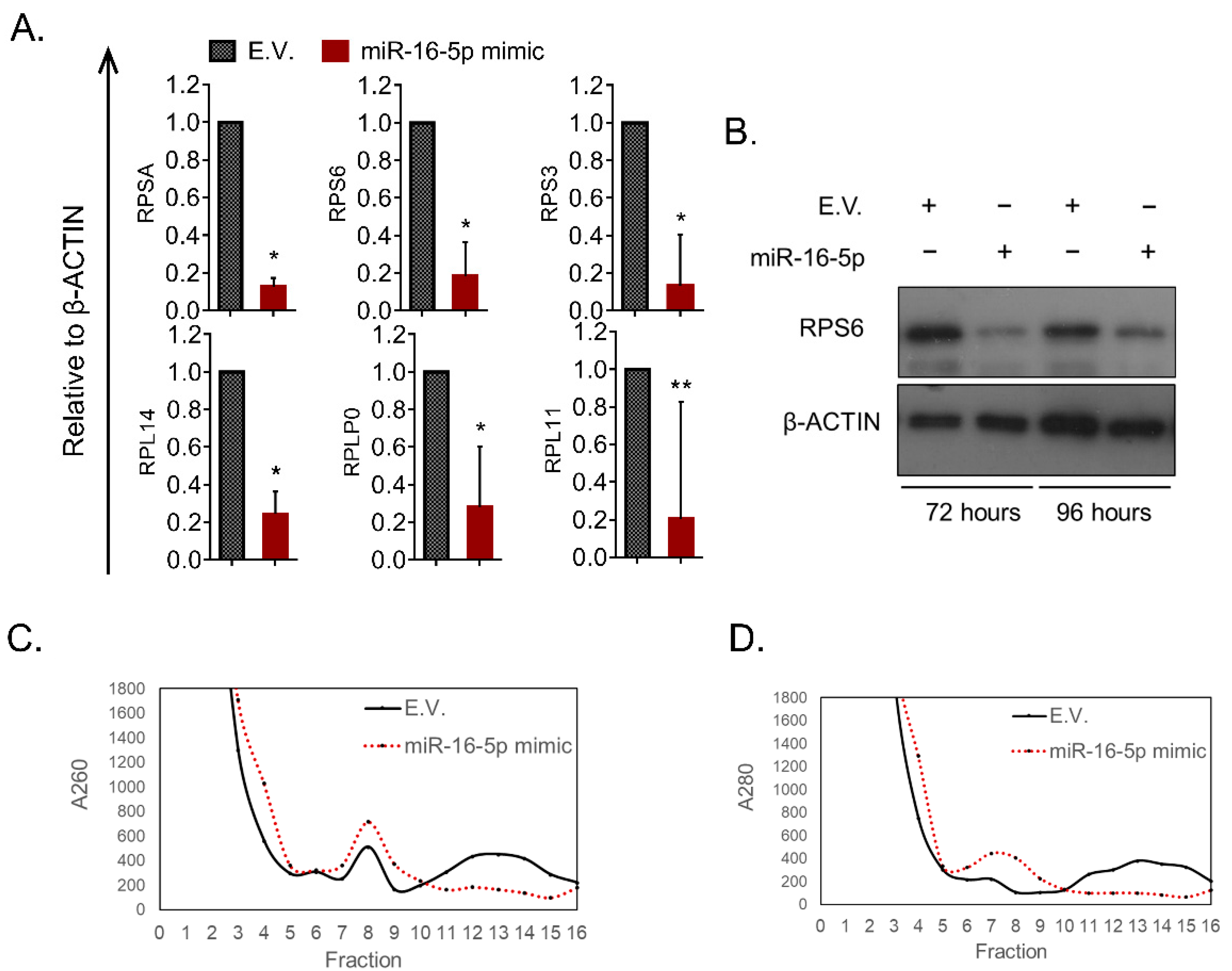
2.5. miR-16-5p Reduces the Overall Ribosomal Levels in MEL Cells
3. Discussion
4. Materials and Methods
4.1. MEL Cell Culture and Induction of Differentiation
4.2. RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) for miRNA Analysis
4.3. Cell Transfection
4.4. Cell Proliferation and Differentiation Assays
4.5. Western Blot Analysis
4.6. Bioinformatic Analysis of miR-16-5p Target Genes
4.7. Ribosome Fractionation Protocol
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Cho, W.C. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer 2007, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Ann. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, Z.X.; Wang, R. MicroRNA-21: A novel therapeutic target in human cancer. Cancer Biol. Ther. 2010, 10, 1224–1232. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The promising pole of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes. Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef]
- Tsiftsoglou, A.S.; Pappas, I.S.; Vizirianakis, I.S. The developmental program of murine erythroleukemia cells. Oncol. Res. 2003, 13, 339–346. [Google Scholar] [CrossRef]
- Tsiftsoglou, A.S.; Vizirianakis, I.S.; Strouboulis, J. Erythropoiesis: Model systems, molecular regulators, and developmental programs. IUBMB Life 2009, 61, 800–830. [Google Scholar] [CrossRef] [PubMed]
- Vizirianakis, I.S.; Wong, W.; Tsiftsoglou, A.S. Analysis of the inhibition of commitment of murine erythroleukemia (MEL) cells to terminal maturation by N6-methyladenosine. Biochem. Pharmacol. 1992, 44, 927–936. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Tsiftsoglou, A.S. Blockade of murine erythroleukemia cell differentiation by hypomethylating agents causes accumulation of discrete small poly(A)-RNAs hybridized to 3’-end flanking sequences of beta(major) globin gene. Biochim. Biophys. Acta 2005, 1743, 101–114. [Google Scholar] [CrossRef][Green Version]
- Vizirianakis, I.S.; Papachristou, E.T.; Andreadis, P.; Zopounidou, E.; Matragkou, C.N.; Tsiftsoglou, A.S. Genetic manipulation of RPS5 gene expression modulates the initiation of commitment of MEL cells to erythroid maturation: Implications in understanding ribosomopathies. Int. J. Oncol. 2015, 47, 303–314. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vizirianakis, I.S.; Tsiftsoglou, A.S. Induction of murine erythroleukemia cell differentiation is associated with methylation and differential stability of poly(A)+ RNA transcripts. Biochim. Biophys. Acta 1996, 1312, 8–20. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Tsiftsoglou, A.S. N6-methyladenosine inhibits murine erythroleukemia cell maturation by blocking methylation of RNA and memory via conversion to S-(N6-methyl)-adenosylhomocysteine. Biochem. Pharmacol. 1995, 50, 1807–1814. [Google Scholar] [CrossRef]
- Tsiftsoglou, A.S.; Pappas, I.S.; Vizirianakis, I.S. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol. Ther. 2003, 100, 257–290. [Google Scholar] [CrossRef]
- Bruchova-Votavova, H.; Yoon, D.; Prchal, J.T. miR-451 enhances erythroid differentiation in K562 cells. Leuk. Lymphoma 2010, 51, 686–693. [Google Scholar] [CrossRef]
- Pase, L.; Layton, J.E.; Kloosterman, W.P.; Carradice, D.; Waterhouse, P.M.; Lieschke, G.J. miR-451 regulates zebrafish erythroid maturation in vivo via its target GATA2. Blood 2009, 113, 1794–1804. [Google Scholar] [CrossRef]
- Yu, D.; dos Santos, C.O.; Zhao, G.; Jiang, J.; Amigo, J.D.; Khandros, E.; Dore, L.C.; Yao, Y.; D’Souza, J.; Zhang, Z.; et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010, 24, 1620–1633. [Google Scholar] [CrossRef]
- Grabher, C.; Payne, E.M.; Johnston, A.B.; Bolli, N.; Lechman, E.; Dick, J.E.; Kanki, J.P.; Look, A.T. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia 2011, 25, 506–514. [Google Scholar] [CrossRef]
- Zhao, H.; Kalota, A.; Jin, S.; Gewirtz, A.M. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 2009, 113, 505–516. [Google Scholar] [CrossRef]
- Felli, N.; Pedini, F.; Romania, P.; Biffoni, M.; Morsilli, O.; Castelli, G.; Santoro, S.; Chicarella, S.; Sorrentino, A.; Peschle, C.; et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica 2009, 94, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Liu, H.; Lv, X.; Liu, Y.; Wang, X.; Zhang, M.; Zhang, X.; Li, Y.; Lou, Q.; Li, S.; et al. MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget 2017, 8, 72400–72410. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L.; Qian, X. MiR-16-5p inhibits breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci. Rep. 2019, 39, BSR20191611. [Google Scholar] [CrossRef] [PubMed]
- Haghi, M.; Taha, M.F.; Javeri, A. Suppressive effect of exogenous miR-16 and miR-34a on tumorigenesis of breast cancer cells. J. Cell. Biochem. 2019, 120, 13342–13353. [Google Scholar] [CrossRef]
- Zhan, M.; Miller, C.P.; Papayannopoulou, T.; Stamatoyannopoulos, G.; Song, C.Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp. Hematol. 2007, 35, 1015–1025. [Google Scholar] [CrossRef]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet—Dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef]
- Hamberg, M.; Backes, C.; Fehlmann, T.; Hart, M.; Meder, B.; Meese, E.; Keller, A. MiRTargetLink-miRNAs, gGenes and interaction networks. Int. J. Mol. Sci. 2016, 17, 564. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk-database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Woo, P.; Venkataraman, K.; Karzai, A.W. Ribosome purification approaches for studying interactions of regulatory proteins and RNAs with the ribosome. Methods Mol. Biol. 2012, 905, 273–289. [Google Scholar] [CrossRef]
- Gautier, E.F.; Leduc, M.; Ladli, M.; Schulz, V.P.; Lefèvre, C.; Boussaid, I.; Fontenay, M.; Lacombe, C.; Verdier, F.; Guillonneau, F.; et al. Comprehensive proteomic analysis of murine terminal erythroid differentiation. Blood Adv. 2020, 4, 1464–1477. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef]
- Zheng, C.; Zheng, Z.; Sun, J.; Zhang, Y.; Wei, C.; Ke, X.; Liu, Y.; Deng, L.; Wang, H. MiR-16-5p mediates a positive feedback loop in EV71-induced apoptosis and suppresses virus replication. Sci. Rep. 2017, 7, 16422. [Google Scholar] [CrossRef] [PubMed]
- Munson, P.B.; Hall, E.M.; Farina, N.H.; Pass, H.I.; Shukla, A. Exosomal miR-16-5p as a target for malignant mesothelioma. Sci. Rep. 2019, 9, 11688. [Google Scholar] [CrossRef] [PubMed]
- Passetti, F.; Jorge, N.A.; Durham, A. Using bioinformatics tools to study the role of microRNA in cancer. Methods Mol. Biol. 2014, 1168, 99–116. [Google Scholar] [CrossRef]
- Chen, W.; Gao, C.; Liu, Y.; Wen, Y.; Hong, X.; Huang, Z. Bioinformatics analysis of prognostic miRNA signature and potential critical genes in colon cancer. Biol. Front. Genet. 2020, 11, 478. [Google Scholar] [CrossRef] [PubMed]
- Banwait, J.K.; Bastola, D.R. Contribution of bioinformatics prediction in microRNA-based cancer therapeutics. Adv. Drug Delivery Rev. 2015, 81, 94–103. [Google Scholar] [CrossRef]
- Falzone, L.; Grimaldi, M.; Celentano, E.; Augustin, L.S.A.; Libra, M. Identification of modulated microRNAs associated with breast cancer, diet, and physical activity. Cancers 2020, 12, 2555. [Google Scholar] [CrossRef]
- Crimi, S. Droplet digital PCR analysis of liquid biopsy samples unveils the diagnostic role of HSA-miR-133a-3p and HSA-miR-375-3p in oral cancer. Biology 2020, 9, 379. [Google Scholar] [CrossRef]
- Pelletier, J.; Thomas, G.; Volarević, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2018, 18, 51–63. [Google Scholar] [CrossRef]
- van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef]
- Vizirianakis, I.S.; Pappas, I.S.; Gougoumas, D.; Tsiftsoglou, A.S. Expression of ribosomal protein S5 cloned gene during differentiation and apoptosis in murine erythroleukemia (MEL) cells. Oncol. Res. 1999, 11, 409–419. [Google Scholar]
- Yelick, P.C.; Trainor, P.A. Ribosomopathies: Global process, tissue specific defects. Rare Dis. 2015, 3, e1025185. [Google Scholar] [CrossRef]
- Amanatiadou, E.P.; Papadopoulos, G.L.; Strouboulis, J.; Vizirianakis, I.S. GATA1 and PU.1 bind to ribosomal protein genes in erythroid cells: Implications for ribosomopathies. PLoS ONE 2015, 10, e0140077. [Google Scholar] [CrossRef]
- Hensold, J.O.; Barth-Baus, D.; Stratton, C.A. Inducers of erythroleukemic differentiation cause messenger RNAs that lack poly(A)-binding protein to accumulate in translationally inactive, salt-labile 80 S ribosomal complexes. J. Biol. Chem. 1996, 271, 23246–23254. [Google Scholar] [CrossRef]
- Tagliaferri, P.; Rossi, M.; Di Martino, M.T.; Amodio, N.; Leone, E.; Gulla, A.; Neri, A.; Tassone, P. Promises and challenges of microRNA-based treatment of multiple myeloma. Curr. Cancer Drug Targets 2012, 12, 838–846. [Google Scholar] [CrossRef]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Deliv. 2020, 15, 987–992. [Google Scholar] [CrossRef]
- Giassafaki, L.N.; Siqueira, S.; Panteris, E.; Psatha, K.; Chatzopoulou, F.; Aivaliotis, M.; Tzimagiorgis, G.; Müllertz, A.; Fatouros, D.G.; Vizirianakis, I.S. Towards analyzing the potential of exosomes to deliver microRNA therapeutics. J. Cell Physiol. 2021, 236, 1529–1544. [Google Scholar] [CrossRef]
- Yoon, H.; Belmonte, K.C.; Kasten, T.; Bateman, R.; Kim, J. Intra- and Iinter-individual variability of microRNA levels in human cerebrospinal fluid: Critical implications for biomarker discovery. Sci. Rep. 2017, 7, 12720. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papagiannopoulos, C.I.; Theodoroula, N.F.; Vizirianakis, I.S. miR-16-5p Promotes Erythroid Maturation of Erythroleukemia Cells by Regulating Ribosome Biogenesis. Pharmaceuticals 2021, 14, 137. https://doi.org/10.3390/ph14020137
Papagiannopoulos CI, Theodoroula NF, Vizirianakis IS. miR-16-5p Promotes Erythroid Maturation of Erythroleukemia Cells by Regulating Ribosome Biogenesis. Pharmaceuticals. 2021; 14(2):137. https://doi.org/10.3390/ph14020137
Chicago/Turabian StylePapagiannopoulos, Christos I., Nikoleta F. Theodoroula, and Ioannis S. Vizirianakis. 2021. "miR-16-5p Promotes Erythroid Maturation of Erythroleukemia Cells by Regulating Ribosome Biogenesis" Pharmaceuticals 14, no. 2: 137. https://doi.org/10.3390/ph14020137
APA StylePapagiannopoulos, C. I., Theodoroula, N. F., & Vizirianakis, I. S. (2021). miR-16-5p Promotes Erythroid Maturation of Erythroleukemia Cells by Regulating Ribosome Biogenesis. Pharmaceuticals, 14(2), 137. https://doi.org/10.3390/ph14020137