Abstract
Triple negative breast cancer (TNBC) is one of the deadliest types of cancer for women of different age groups. Frequently this cancer does not respond to conservative treatment. Combinatorial RNAi can be suggested as an advanced approach to TNBC therapy. Due to the fact that TNBC cells overexpress chemokine receptor 4 we used modular L1 peptide-based nanoparticles modified with CXCR4 ligand for combinatorial delivery of siRNAs suppressing major transduction pathways. TNBC cell line MDA-MB-231 was used as a cellular model. Genes encoding the AQP3, CDC20, and COL4A2 proteins responsible for proliferative activity in TNBC cells were selected as RNAi targets. The siRNA binding ability of the carrier was studied at different charge ratios. The silencing specificity was demonstrated for all siRNAs studied. Alamar Blue proliferation assay has shown significant reduction in the anti-proliferative activity after combinatorial siRNA transfection compared to single siRNA delivery. The most significant synergistic effect has been demonstrated for combinatorial transfection of anti-COL4A2 and anti-CDC20 siRNAs what resulted in 1.5–2 fold inhibition of proliferation and migration of TNBC cells. Based on our findings, we have concluded that combinatorial treatment by CXCR4-ligand modified L1-polyplexes formed with AQP3, CDC20, and COL4A2 siRNAs effectively inhibits proliferation of TNBC cells and can be suggested as useful tool for RNAi-mediated cancer therapy.
1. Introduction
The mammary gland/breast is a hormone-dependent organ of the female reproductive system [1]. Mammary cancer is one of the most common types of cancers among women of any age, and for 30 years the number of detected breast cancer cases has increased by more than 40% [2]. Some successes were achieved in the mammary tumors treatment by negatively affecting estrogen (ER) and progesterone (PR) receptors as well as human epidermal growth factor receptor 2 (HER-2) when breast cancers displayed these receptors [3,4,5]. However, chemotherapy used in the mammary cancer treatment has a gonadotoxic effect and leads to a decrease or complete loss of fertility in women of reproductive age. It is worth noting that the triple negative breast cancer (TNBC) is characterized by the lack of ER- and PR-, as well as HER-2 gene expression [6]. Due to the absence of targeted therapy, TNBC remains a more aggressive among all mammary cancer subtypes. The lack of proper response to the chemotherapy results in the high recurrence rate and as a consequence short overall TNBC patients’ survival when compared to hormone-positive breast cancers [7,8]. TNBC is characterized by genomic and molecular aberrations that lead to dysregulation of signaling pathways [9,10]. A possible way for effective and targeted as well as fertility-preserving treatment of TNBC may be suppression of the signal transduction pathways or related proteins with the subsequent inhibition of the cancer cell proliferation and migration; this, in turn, can reduce the metastases.
TNBC progression and development is regulated by several pathways, including NF-kB, JAK/STAT3, PI3K–AKT–mTOR and PI3K/AKT, however only few of them are found to be useful for targeted therapy [11,12,13,14]. To address this issue an advanced therapy should be explored using novel approaches and/or a combination of already established strategies. Advanced technology such as RNAi in the combination with chemotherapy could be a promising approach to treat TNBC but some obstacles are present such as efficiency and specificity of siRNA delivery. The discovered phenomenon of RNA interference (RNAi) makes it possible to relatively easy silence the corresponding gene expression [15]. RNAi could become the targeted therapy for TNBC in order to downregulate the certain genes involved in the tumor development and progression. Previously it was shown, thatcancer cell proliferation, migration and metastasis are regulated by SDF-1 (stromal cell derived factor 1)/CXCR4 axis which stimulate downstream G-protein signaling leading to activation of protein kinase cascades [16]. CXCR4 receptor is abundantly overexpressed on surface of TNBC cells and can be used as specific target for siRNA therapeutics delivery [17,18]. Also, SDF-1-induced breast cancer signaling and migration requires aquaporin-3 (AQP3), which is a member of the aquaporin water channel family functioning as water and glycerol transporter [19]. It is also required for directional mammary cancer cell migration via SDF-1-induced cellular uptake of hydrogen peroxide [20]. AQP3 also may be contributed to ATP generation energizing cancer cells to proliferate and develop tumor [21]. AQP3 gene silencing was shown to significantly reduce proliferation and migration of TNBC cells [22]. An important cancer feature is the upregulation of cell division, due to the activation of proteins involved in cell cycle. One of these proteins is the cell division cycle protein 20 (CDC20), a participator of multiple cell cycle regulatory pathways that required for progression and completion of mitosis [23]. It activates a complex of proteins that regulate the anaphase promoting complex (APC), which initiate chromatid separation and the start of cell division at anaphase [24]. Thus, CDC20 suppression can be resulted to the arrest of the cell cycle and thereby reduction of the tumor progression [25]. CDC20 is highly expressed exactly in TNBC cells but not in hormonal breast cancer cell lines [26]. CDC20 downregulation in TNBC cells was shown to result in 40% inhibition of cell growth [27]. Another promising target for the breast cancer treatment is the proteins of the basement membrane components. One of such proteins is collagen type 4 alpha-2 (COL4A2), which is extremely important for functioning and stability of vascular basement membrane [28]. COL4A2 was demonstrated to be involved in the tumorigenesis of the reproductive system organs, prostate cancer, epithelial ovarian cancer, uterine leiomyoma, breast cancer and etc. [29,30,31,32]. It was also shown that COL4A2 is overexpressed in TNBC cells; therefore, it can be assumed that the COL4A2 protein plays an important role in the pathogenesis of this breast cancer subtype [33]. Suppression of COL4A2 gene in TNBC cells led to significant reduction in cell proliferation and migration level [34].
For RNAi to perform effectively, siRNA needs to be compacted with a vehicle to prevent extracellular degradation and to ensure its release to the cytoplasm [35]. To achieve targeted delivery to tumor cells we previously developed a new ligand peptide derived from N-terminus of chemokine SDF-1 for CXCR4 receptor specific binding and include it in the modular peptide vehicle [36,37,38]. The resulting L1 carrier was effective in siRNA delivery in vitro and in vivo studies [39,40]. Also, we demonstrated a 60–80% silencing of GFP expression after treatment of the triple negative breast cancer MDA-MB-231 cells with L1/siRNA complexes [41].
Here, a combinatorial approach based on CXCR4-specific delivery of TNBC-related siRNAs was studied. We used siRNA/L1 complexes containing siRNA against AQP3, CDC20 and COL4A2 for targeted inhibition of TNBC cell proliferation and migration. We analyzed the anti-tumor effects of AQP3, CDC20 and COL4A2 targeting alone and in combinations. Synergistic anti-tumor effects after combinatorial siRNA delivery were studied. We identified a more effective combination of siRNAs for therapeutic silencing that may provide a basis for targeted TNBC gene therapy.
2. Results and Discussion
The pharmaceutical attractiveness of siRNA for tumor gene therapy is based on its properties such as temporary effect due to lack of genome integration and low variation of chemicals because of controlled synthesis [42]. The development of siRNA-based cancer therapy has greatly relied on the effect of increased tumor permeability and retention [43]. The identification of specific target genes for suppression in TNBC opens up new possibilities for siRNA-based therapy of the disease. It could be interesting to study how the combinatorial silencing of the factors specific for TNBC can influence such important phases for the tumor progression as cell proliferation and migration.
2.1. In Vitro Transfection of MDA-MB-231 Cells
MDA-MB-231 breast cancer cells with constant gfp gene expression were used for in vitro transfection by L1/anti-GFP siRNA complexes (Figure 1). CXCR4 surface expression is known to be upregulated in TNBC cells and is an independent prognostic factor for disease relapse [44], so the L1 peptide carrier containing the ligand to CXCR4 receptor is a suitable vehicle for the marker and therapeutic siRNAs delivery into the cells. It should be noted that proliferation studies require low density to prevent contact inhibition of cell growth and thus require shorter time of incubation of cells with siRNA-polyplexes. On other hand, standard migration protocols employ high cell density and 4 h incubation [39]. Previously, effective inhibition of gfp expression was registered for MDA-MB-231 cells seeded in low density after their 2.5 h incubation with siRNA-polyplexes [41]. In order to harmonize proliferation and migration protocols we optimized L1-polyplexes mediated transfection: the siRNA-complexes were left for 2 and 4 h in a transfection medium and two siRNA concentrations were tested. As shown in Figure 1, control mock siRNA-containing polyplexes did not demonstrate gfp gene expression silencing, whereas, L1/anti-GFP-siRNA complexes delivery resulted in significant decrease in gfp expression up to 40–50%.
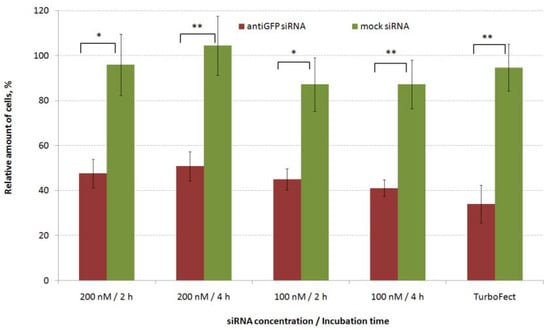
Figure 1.
Silencing of gfp expression after treatment of MDA-MB-231 cells with L1/antiGFP siRNA complexes at 8 to 1 of N/P ratio. * p < 0.05, ** p < 0.01 when compared with cells treated by mock siRNA-complexes.
These data were comparable to transfection efficiency of Turbofect/anti-GFP siRNA complexes which showed gfp silencing expression to 35%. We did not find any differences in transfection efficiency between 2 h and 4 h of incubation regarding the GFP protein level for both siRNA concentrations. Thus, lower incubation time was used in further transfection studies and functional tests.
2.2. Evaluation of siRNA-Complexes Cytotoxicity
Cytotoxicity of siRNA-complexes is one of the important factors that must be taken into account when developing nucleic acid delivery applications. It was reported that high positive charge density of polyplexes correlates with the lower cell viability [45]. L1/siRNA polyplexes were previously shown to have positive zeta potential [39]. For cytotoxicity studies MDA-MB-231 cells were seeded with high and low densities in accordance with the cell migration and proliferation protocols. The increase in length of transfection time is considered to affect a rise of cytotoxicity [46]. To this extent, we defined the cell viability after 2-h incubation with polyplexes at different siRNA concentration (100, 200 and 400 nM) (Figure 2).
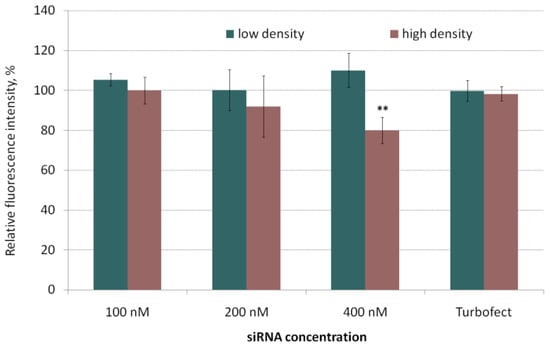
Figure 2.
Cytotoxicity of the siRNA-polyplexes at N/P ratio 8/1 in MDA-MB-231 cells seeded with low and high density. ** p < 0.01 when compared with intact cells.
In the most cases polyplexes added to cells showed no cytotoxicity. A slight decrease in the cell viability was observed at siRNA concentration of 400 nM for cells seeded at a high density. However, the relative number of the living cells remained at 80% compared to intact cells. Thus, the siRNA polyplexes used showed no serious damaging effects on the MDA-MB-231 cells and turned out to be suitable for in vitro studies aimed at specific suppression of cell migration and proliferation.
2.3. RNAi-Mediated Knockdown of TNBC-Related Genes
The lack of effective and readily available therapies that affect the proliferation and migration of TNBC cells stimulates the search for new targets and using of a combinatorial approach for targeting already known ones [47]. AQP3, CDC20 and COL4A2 genes involved in the tumor development and progression were chosen for analysis [22,27,33]. Before carrying out functional tests, antiAQP3, antiCDC20 and antiCOL4A2 siRNAs were used to confirm the specific silencing of the appropriate genes. The suppression efficacy was compared to the gene expression level after delivery of mock siRNA. siRNAs used in the experiments had a 200 nM concentration and formed polyplexes with L1 carrier at 8/1 N/P ratio. As shown in Figure 3, mock siRNA-complexes did not induce silencing of the target gene expression that confirms specificity of silencing mediated by experimental siRNAs. The cells treatment with L1/antiAQP3 siRNA complexes resulted in decrease of AQP3 expression to 47%. L1/antiCOL4A2 siRNA polyplexes was found to decrease COL4A2 expression to 32%. The most efficient gene suppression was shown using L1/antiCDC20 siRNA polyplexes which demonstrated CDC20 gene knockdown to 30%. The efficiency of L1/targeted siRNA polyplexes was comparable to or exceeded the efficacy of Turbofect/siRNA complexes as in the case of antiCDC20 siRNA. The results obtained suggested that the silencing of targeted genes in TNBC MDA-MB-231 cells was due to a specific RNAi but not from polyplex cytotoxicity.
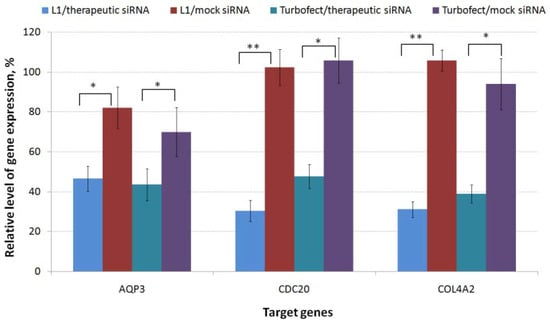
Figure 3.
Specific silencing of gene expression after MDA-MB-231 cells treatment by polyplexes formed with L1 and anti-AQP3, anti-CDC20, anti-COL4A2 siRNA or mock siRNA. * p < 0.05, ** p < 0.01 when compared with cells treated with mock siRNA-complexes.
2.4. Inhibition of MDA-MB-231 Cells Proliferation
The level of cell proliferative activity is known to determine the aggressiveness and malignancy of the tumor process [48]. AntiAQP3, antiCDC20 and antiCOL4A2 siRNA-polyplexes alone or in combination with each other were studied for their ability to inhibit MDA-MB-231 cell proliferation (Figure 4). Non-targeted mock siRNA-polyplexes were taken to demonstrate the cell growth suppression specificity by RNAi. siRNAs were used at 100 nM and 200 nM concentrations. We did not find significant differences between the antiAQP3 siRNA-polyplexes and mock siRNA-complexes (Figure 4). Previously, in several studies performed on different cancer cells involvement of AQP3 gene expression in cell proliferation was demonstrated [49,50]. Arif and colleagues have demonstrated a 28% reduction in amount of MDA-MB-231 cells for stably expressing AQP3 shRNA clone [22]. The efficiency of proliferation inhibiting may correlate with different factors such as stability and efficacy of the gene expression silencing, siRNA concentration, etc.
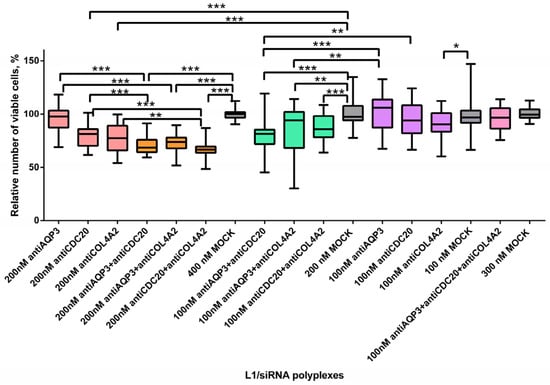
Figure 4.
Relative number of MDA-MB-231 cells after treatment with L1 formed with therapeutic siRNAs alone and in combinations. * p < 0.05, ** p < 0.01, *** p < 0.001 when compared with cells treated by mock siRNA. Coloring: single siRNA polyplexes at 100 nM (purple), at 200 nM (peach); dual siRNA polyplexes at 100 nM (green), at 200 nM (orange); triple siRNA polyplexes at 100 nM (pink); mock siRNA polyplexes at 100, 200, 300 and 400 nM (grey).
The results obtained demonstrated significant difference in relative cell number after their treatment with L1/antiCDC20 siRNA polyplexes at 200 nM concentration of siRNA and mock siRNA complexes. A 20% decrease in the proliferative activity of cells was found (Figure 4). Actually, silencing of CDC20 was previously shown to inhibit MDA-MB-231 cell proliferation [51]. CDC20 gene definitely plays a key role during mitosis of dividing cancer cells, so its silencing could decrease a tumor growth [27]. However, 100 nM concentration of antiCDC20 siRNA was not enough to decrease the cell proliferation level. The most efficient inhibition of MDA-MB-231 cell proliferation was shown using L1/antiCOL4A2 siRNA polyplexes which resulted in decrease in the cell growth by 23% at siRNA concentration of 200 nM (Figure 4). Song and colleagues using the integrated analysis showed that Col4A2 protein was the most significantly up-regulated in MDA-MB-231 cell line. Silencing of the gene resulted in inhibition in the TNBC cell proliferation [34]. Authors suppose that antiCOL4A2 siRNA treatment resulted in decrease of cell proliferation due to induction of apoptosis. Further improvement in therapeutic RNAi efficiency in TNBC cells is possible by means of alternate CXCR4-targeted siRNA delivery systems recently developed for cancer gene therapy [52,53].
To identify the synergistic effects of combinatorial siRNA delivery, several siRNAs were transfected using L1 carrier (Figure 4). AntiAQP3, antiCDC20 and antiCOL4A2 were mixed in equimolar concentrations of 100 nM and 200 nM. We also used triple siRNA-polyplexes consisted of L1 and each siRNA at the concentration of 100 nM. These polyplexes’ efficiency was compared with the efficacy of mock siRNA and each siRNA complexed alone by L1 carrier (Figure 4). The dual siRNA-polyplexes at both concentrations suppressed MDA-MB-321 cell proliferation as compared to appropriate L1/mock siRNA complexes. Moreover, complexes with a higher siRNA concentration suppressed the proliferation more efficiently (Figure 4). On the over hand, triple polyplexes with siRNA concentration of 100 nM did not decrease MDA-MB-231 cell number. We assume that several factors influencing cell proliferation could exist as the efficiency of siRNAs for gene silencing, siRNA concentration, and the amount of AGO2. An increase in the number of siRNA could lead to a self-inhibition effect caused by AGO2 depletion. Previously it was shown that a multi-knockdown siRNA system may not always effectively compete with cellular miRNAs for the AGO2 complex thus reducing RNAi efficacy [54]. Alternate combinatorial approach is an encapsulation of siRNA and chemotherapeutic drug in a single delivery system, however, this way seems very difficult due to difference in their physicochemical properties [11]. For instance, Su et al. reported a photothermal system for co-delivery of indocyanine green, paclitaxel, and survivin siRNAto TNBC cells [55]. The combined treatment resulted in significant improvement in antitumor efficacy, which opens perspectives for combinatorial application of other TNBC-related siRNAs including those described here.
We compared the efficacy of dual polyplexes with single ones formed at the same concentrations. As it can be seen in Figure 4 L1/antiAQP3 siRNA complexes did not decrease cell proliferation, whereas the addition of antiCDC20 or antiCOL4A2 resulted in a significant cell growth inhibition. The treatment by antiAQP3 and antiCDC20 or antiCOL4A2 siRNAs led to the decrease in the relative cell number to 70% or 74% at siRNA concentration 200 nM and to 81% or 82% at the concentration 100 nM, respectively (Figure 4). When compared to L1/antiCDC20 siRNA polyplexes, the addition of antiAQP3 promoted a decrease in the cell proliferation level by 10% and 15% at 200 nM and 100 nM of siRNAs, accordingly (Figure 4). Thus, due to the combinatorial targeting, we managed to demonstrate the antiAQP3 siRNA contribution to cell proliferation suppression. The combinatorial treatment with anti-CDC20 and antiCOL4A2 siRNAs-polyplexes resulted in significant inhibition of cell proliferation as compared to L1/antiCDC20 siRNA complexes only at siRNA concentration of 200 nM. However, this siRNAs combination was the most successful and led to a decrease in the cell proliferation intensity by 11% compared with anti-CDC20 siRNA only to a level of 66% (Figure 4). As compared to L1/antiCOL4A2 siRNA complexes significant differences were registered only for combinatorial treatment by antiCDC20 and antiCOL4A2 siRNAs at 200 nM of siRNA concentration. To sum up, the combinatorial delivery of antiCDC20 and antiCOL4A2 siRNAs seems to be more effective for the inhibition of cell growth.
2.5. Inhibition of MDA-MB-231 Cells Migration
Active migration of cancer cells is a prerequisite for their invasion and metastasis [56]. A scratch assay was performed to determine whether MDA-MB-231 cell migration was affected by antiAQP3, antiCDC20, or antiCOL4A2 siRNAs as well as their dual and triple combinations. L1/mock siRNA polyplexes were used as a negative control and to demonstrate the specificity of RNAi functional effects. MDA-MB-231 cells were treated with L1/siRNA-polyplexes with different siRNA concentrations. The cell monolayer was damaged and the number of cells that migrated into the cell-free area was counted; the number of migrated untreated cells was taken as 100% (Figure 5).
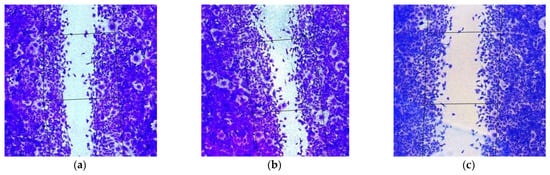
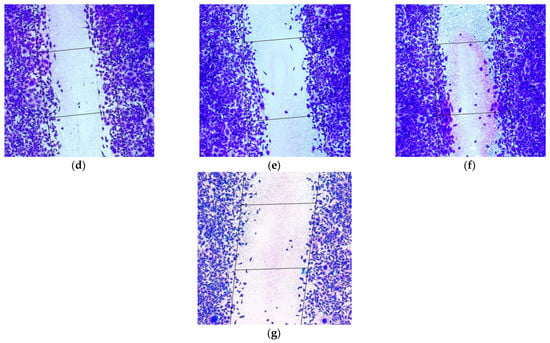
Figure 5.
Appearance of migrated MDA-MB-231 intact cells (magnification ×100) (a) and after their treatment with (b) L1/mock siRNA (200 nM), (c) L1/mock siRNA (400 nM), (d) L1/anti-AQP3 siRNA (200 nM), (e) L1/anti-CDC20 siRNA (200 nM), (f) L1/anti-COL4A2 siRNA (200 nM) and (g) L1/anti-CDC20 siRNA+anti-COL4A2 siRNA (200 + 200 nM) complexes.
Previous results suggested that AQP3 is one of the key regulators of MDA-MB-231 migration [22]. After L1/antiAQP3 siRNA polyplexes cell treatment decrease of cell migration level was found to be to 73% and 63% with 100 nM and 200 nM of siRNA, respectively, in comparison with corresponding mock siRNA-complexes (Figure 6). Satooka and Hara-Chikuma demonstrated the importance of AQP3 for SDF-1/CXCR4-dependant migration of the breast cancer cells [20]. Moreover, AQP3 was shown to be a necessary component for fibroblasts growth factor 2-induced cell migration [57].
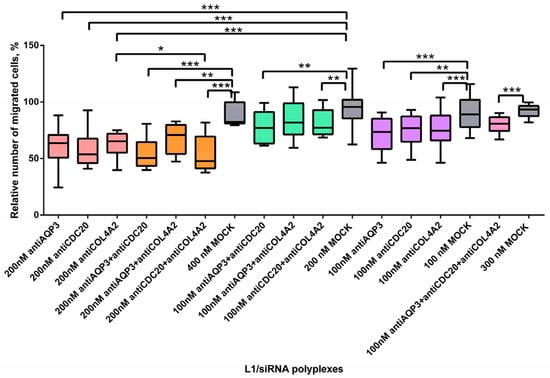
Figure 6.
Relative number of migrated MDA-MB-231 cells (%) after treatment with L1 formed with therapeutic siRNAs aloneand in combinations. * p < 0.05, ** p < 0.01, *** p < 0.001 when compared with cells treated by mock siRNA. Coloring: single siRNA polyplexes at 100 nM (purple), at 200 nM (peach); dual siRNA polyplexes at 100 nM (green), at 200 nM (orange); triple siRNA polyplexes at 100 nM (pink); mock siRNA polyplexes at 100, 200, 300 and 400 nM (grey).
Aberrant CDC20 expression could play a role in the aggressive metastasizing of TNBC [26]. Under antiCDC20 siRNA-treatment, MDA-MB-231 cells showed a significantly reduced level of migration to 77% and 54% at siRNA concentrations of 100 nM and 200 nM, respectively, as compared to L1/mock siRNA complexes (Figure 6). In fact, CDC20 is one of the main factors for cancer cell migration and its overexpression promotes the metastasizing ability of the breast cancer cells [58]. CDC20 overexpression may be associated with an aggressive course of TNBC [26]. Abnormal CDC20 expression leads to mitotic errors which are the main reasons for chromosome instability (CIN). Bakhoum and colleagues demonstrated that CIN facilitated the cell metastasis through cGAS–STING-dependent manner (cyclic GMP-AMP synthase–stimulator of interferon genes) [59]. Paul and colleagues previously showed CDC20-mediated proteasomal degradation of SMAR1 (the tumor suppressor scaffold matrix attachment region binding protein 1) facilitated the cancer cell migration [60].
For L1/antiCOL4A2 siRNA polyplexes we also demonstrated significant differences in the MDA-MB-231 cell migration efficacy compared to mock siRNA-complexes. The treatment with antiCOL4A2 siRNA-polyplexes promoted a decrease in relative migration level to 75% and 65% (Figure 6). The important role of the COL4A2 gene in TNBC cell migration was already demonstrated previously [34]. The authors suppose the COL4A2 gene targets the Jak-STAT signaling pathway, which in turn is one of the TNBC-specific regulatory feedback programs [34,61].
Thus, treatment of TNBC MDA-MB-231 cells with antiAQP3, antiCDC20, or antiCOL4A2 siRNA-polyplexes resulted in strongly reduced cell migration ability. The combinatorial siRNAs delivery can allow us to determine more effective therapeutic combinations to prevent breast cancer metastasis.
We found that using triple siRNAs-polyplexes resulted in a significant decrease of the cell migration rate in comparison with L1/mock siRNA complexes (Figure 6). The level of MDA-MB-231 cell migration dropped to 80%. However, no significant difference was found when compared with single siRNA-polyplexes treatment at 100 nM of concentration. Most of dual polyplexes at both concentrations effectively suppressed MDA-MB-321 cell migration as compared to corresponding mock siRNA complexes (Figure 6). Significant increase of cell migration was demonstrated when using antiCDC20 and antiCOL4A2 siRNAs in comparison with single antiCOL4A2 siRNA treatment. Transfection with antiCDC20 siRNA and antiCOL4A2 siRNA-polyplexes facilitated a decrease in the cell migration level to 48% at 200 nM of each siRNA (Figure 6). It can be suggested that combinatorial delivery in most cases did not lead to a cumulative effect due to significantly reduced level of the cell migration caused by single siRNA-polyplexes (Figure 6).
The data obtained are summarized in Table 1. The most profound synergistic anti-proliferative and migration inhibiting activity was found for co-delivery of antiCDC20 and antiCOL4A2 siRNAs. In other cases we demonstrated that silencing of several targets is not always beneficial for effective anti-cancer therapy. However, the positive results of the previous study on combinatorial delivery of antiCD20 and anti-phosphatase siRNAs confirm our findings on revealed synergistic effects [27]. Thus, it can be suggested that combinatorial siRNA delivery against CDC20 and COL4A2 could be a promising strategy for RNAi-based TNBC therapy.

Table 1.
Summary of registered anti-proliferative effects after antiAQP3, antiCDC20, or antiCOL4A2 siRNA delivery and co-delivery mediated by an L1 carrier (↓ means decrease of migration/proliferation; - means no effect; ↓↓ means synergistic effect; ↓↓↓ means the most profound synergistic effect).
3. Materials and Methods
3.1. Cell Lines
GFP-positive triple negative breast cancer cells MDA-MB 231 were cultivated under mycoplasma-free conditions in DMEM (Biolot LLC, Saint Petersburg, Russia) supplemented with 10% St-Biol—heat-inactivated fetal calf serum (Biolot LLC), 2 mM L-glutamine with addition of 0.012% of gentamicin as described previously [62].
3.2. Peptide Synthesis and Design
L1 peptide was synthesized by NPF Verta, LLC (Saint Petersburg, Russia) using solid-phase Boc-chemistry as described previously [41]. A dry powder of the peptide was stored at −70 °C. A dry sample (1–2 mg) was dissolved in 0.1% TFA at 2 mg/mL and stored at −20 °C. A high-performance liquid chromatography was used to determine the peptide purity which was in the range 90–95%. L1 peptide consists of the KPVSLSYRSPSRFFESH ligand part linked with DNA-binding motif (CHRRRRRRHC) by two ε-aminocaproic acids (Ahx) [38].
3.3. siRNA, Preparation of Peptide/siRNA Complexes
The sense sequence of anti-AQP3 siRNA 5′-GGG UCG UCA CUC CUU UAA Utt-3′ targets human AQP3 mRNA [63]. The sense sequence of anti-CDC20 siRNA 5′-GGG AAU AUA UAU CCU CUG Utt-3′ targets human CDC20 mRNA [64]. The sense sequence of anti-COL4A2 siRNA 5′-GGC AGA AAG GUG AGC CUU Att-3′ targets human COL4A2 mRNA [65]. The sense sequence of anti-GFP siRNA 5′-CAA GCU GAC CCU GAA GUU Ctt-3′ targets gfp mRNA [66]. A non-silencing siRNA 5′-UUC UCC GAA CGU GUC ACG U-3′ served as a mock siRNA [67]. siRNAs were synthesized by Syntol JSC, Moscow, Russia. The peptide/siRNA complexes were prepared at 8 to 1 peptide nitrogen/RNA phosphorus ratio (N/P ratios). siRNA solution was obtained by dilution of required amount of siRNA to 100 μg/m Lin Hepes-buffered mannitol (HBM) (5% w/v mannitol, 5 mM Hepes, pH 7.5). The L1 peptide at a certain N/P ration diluted in HBM was added to the siRNA solution in equal volume and mixed. Complexes were kept at room temperature for 30 min.
3.4. Cytotoxicity Assay
Cytotoxicity of peptide/siRNA complexes added to cells with low and high density was evaluated using Alamar blue assay (BioSource Intl., Camarillo, CA, USA). 0.6 × 104 (low density) and 2 × 104 (high density) of MDA-MB-231 cells were seeded in 96-well plates a day before experiment. The cytotoxicity experiments were carried out as described previously [38]. The cell viability was studied after 16 h of cell incubation with AlamarBlue. The fluorescence was measured on a Wallac 1420D scanning multilabel counter (Thermo Fisher Scientific Oyj, Vantaa, Finland) using excitation and emission wavelength at 544 nm and at 590 nm, consequently. Relative fluorescence intensity was counted by (F − Ff)/(Fb − Ff) ∗ 100%, where Fb is a fluorescence intensity in untreated control and Ff is a fluorescence intensity without cells. The results are presented as mean ± S.E.M from three independent experiments with three samples.
3.5. siRNA Transfer to MDA-MB-231 Cells
5.2 × 104 or 10.5 × 104 cells at high density were seeded in 48-well or 24-well plates, respectively, and incubated overnight. Before transfection the medium was replaced with serum-free one.Anti-GFP siRNA and mock siRNA-containing polyplexes were added in 48-well plates and incubated with cells for 2 and 4 h. L1 complexed with anti-AQP3, anti-CDC20, anti-COL4A2 and mock siRNAs were added in 24-well plates and incubated with cells for 2 h. The final concentrations of siRNAs were 100 or 200 nM and volumes of medium were 500 and 1000 µL for 48- and 24-well plates, accordingly. After incubation in full culture medium for the next 48 h, cells from 48-well plates were permeabilized for further GFP fluorescence measurement with a Wallac 1420D scanning multilabel counter (Thermo Fisher Scientific Oyj) at 485 nm excitation and 535 nm emission wavelengths as described previously [41]. The GFP fluorescence was normalized by the total protein concentration, measured with Bradford reagent (Helicon, Moscow, Russia). The results are presented as mean ± S.E.M from three independent experiments with three samples. After 48 h of incubation cells from 24-well plates were prepared for RNA extraction with Trizol reagent (Qiagen, Düsseldorf, Germany) and subsequent cDNA synthesis using an OT-1 Reverse Transcription kit (Syntol JSC) according to the manufacturer’s instructions.
3.6. Quantitative RT-PCR
Quantitative PCRs were carried out with EvaGreen PCR kit (Syntol JSC) using AQP3 forward primer 5′-GCAGCCTGTCCATCTGTG-3′, reverse primer 5′-ACCCTACTTCCCAAAAGCC-3′; CDC20 forward primer 5′-CGCTATATCCCCCATCGCAG-3′, reverse primer 5′-GATGTTCCTTCTTGGTGGGC-3′; COL4A2 forward primer 5′-ATTCCTTCCTCATGCACACG-3′, reverse primer 5′-ACTTGTTGGCGTAGTAGTGG-3′, and β-actin gene as endogenous reference was detected using forward 5′-TGCCGACAGGATGCAGAAG-3′, reverse primer 5′-GCCGATCCACACGGAGTACT-3′. 1.25 µg of cDNA was added to the reaction [39,63,64,65]. As previously, PCR was performed in Rotor-Gene 3000 thermocycler (Corbett Life Science, Sydney, Australia) with the following conditions:94 °C for 10 min, followed by 40 cycles of 95 °C for 20 s and 60 °C for 1 min; subsequent qPCR analysis was carried out using the Rotor-Gene version 6.1.71 software [39]. A standard curve of Ct value vs. log amount of standard was generated for each gene using 5-times dilutions of cDNA (1.6; 0.8; 0.4; 0.2; 0.1 µg) prepared from untreated cells. The values are presented as mean ± S.E.M from three independent experiments with two samples.
3.7. Proliferation Assay
0.6 × 104 of MDA-MB-231 cells were seeded in a 96-well plate a day before experiments. Cell culture medium was replaced with 50 µL of serum-free medium. L1 complexed with siRNAs in concentrations of 100, 200 and 300 nM were added and incubated with cells for 2 h. Then cell culture medium was replaced with 100 µL of medium containing 2.5% FBS for the next 72 h for further cell proliferation to be analyzed using Alamar blue assay [41]. The fluorescence was measured and relative fluorescence intensity was counted as described above (see Section 3.4). The results are shown as mean ± S.E.M from five independent experiments with four samples.
3.8. Scratch Migration Assay
2 × 104 of MDA-MB-231 cells were seeded into a 96-well plate 24 h before transfection. Transfection was performed as described above (see Section 3.5) in 175 mkl of serum-free medium. The final concentrations of siRNAs were 100, 200 and 300 nM. After 2 h incubation with L1/siRNA polyplexes, cells were washed by medium, the monolayer was scratched with a 300 μL pipette tip (Biohit Oyj, Helsinki, Finland) with subsequent cell wash by Hanks’ solution and photographing of wound area as described previously [39]. After cell incubation in FBS-containing medium (10%) for the 24 h cells were washed, stained in 100 µL of 0.2% crystal violet in 5% ethanol, dried at 37 °C overnight and photographed by MIBR microscope (LOMO, Saint Petersburg, Russia). Number of cells (N) migrated to wound area was counted using ImagePro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA). Relative number of migrated cells was calculated by (N/N′) ∗ (ρ′/ρ), where N′ is number of migrated cells in untreated control and ρ′ is cell density in untreated control. The results are presented as mean ± S.E.M from five independent experiments with four samples.
3.9. Statistical Analysis
Statistical analysis was performed by Student’s t-test using GraphPad Prism5 Software (GraphPad, McIntosh, CA, USA). Statistical significance was defined as * p < 0.05, ** p < 0.01 and *** p < 0.001.
4. Conclusions
Targeting of triple negative breast cancer-related genes by means of RNA interference is a very promising approach for treatment of this deadly disease. It should be noted that an efficient and TNBC-specific siRNA delivery system is a prerequisite for the development of RNAi-based medicines. The efficiency of an anti-proliferative treatment can be further improved via the combinatorial delivery of siRNAs against multiple targets responsible for proliferative activity in TNBC cells. Here, we have demonstrated the efficient down-regulation of TNBC cells migration and proliferation by antiAQP3, antiCDC20, and antiCOL4A2 siRNA delivery mediated by peptide-based vector L1. Several types of L1-based siRNA polyplexes have been tested and the most efficient combination of siRNAs has been found. Particularly, co-delivery of antiCDC20 and antiCOL4A2 siRNAs synergistically inhibited migration and proliferation of MDA-MB 231 cells. Therefore, the combinatorial strategy based on co-delivery of antiCDC20 and antiCOL4A2 siRNAs by means of CXCR4-targeted non-viral vehicle could be a promising approach to further development of RNAi-based therapy of TNBC.
Author Contributions
A.K. and A.E. designed the work; I.P. and M.M. performed the cytotoxicity and transfection experiments; A.E. and I.P. performed the cell function assays; I.P., A.K. and A.E. analyzed the data, V.B. and A.K. supervised the work, A.E. prepared original draft, A.K. performed review and editing of the manuscript, all other authors revised it critically. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 19-73-10045 (gene expression silencing study), and by Ministry of Science and Higher Education of the Russian Federation, project number AAAA-A19-119021290033-1 (cell functional study).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. Authors can confirm that all relevant data are included in the article.
Acknowledgments
The authors are grateful to Jessica Rosenholm (Abo Academy, Turku, Finland) for providing with GFP-positive triple negative breast cancer cell line MDA-MB 231.
Conflicts of Interest
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Patel, S. Breast cancer: Lesser-known facets and hypotheses. Biomed. Pharmacother. 2018, 98, 499–506. [Google Scholar] [CrossRef]
- Miller, K.D.; Fidler-Benaoudia, M.; Keegan, T.H.; Hipp, H.S.; Jemal, A.; Siegel, R.L. Cancer statistics for adolescents and young adults, 2020. CA Cancer J. Clin. 2020, 70, 443–459. [Google Scholar] [CrossRef]
- Ligresti, G.; Libra, M.; Militello, L.; Clementi, S.; Donia, M.; Imbesi, R.; Malaponte, G.; Cappellani, A.; McCubrey, J.A.; Stivala, F. Breast cancer: Molecular basis and therapeutic strategies (Review). Mol. Med. Rep. 2008, 1, 451–458. [Google Scholar] [CrossRef]
- Canine, B.F.; Wang, Y.; Hatefi, A. Biosynthesis and characterization of a novel genetically engineered polymer for targeted gene transfer to cancer cells. J. Control. Release 2009, 138, 188–196. [Google Scholar] [CrossRef]
- Nuciforo, P.; Radosevic-Robin, N.; Ng, T.; Scaltriti, M. Quantification of HER family receptors in breast cancer. Breast Cancer Res. 2015, 17, 53. [Google Scholar] [CrossRef]
- Masoud, V.; Pagès, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef]
- Rhee, J.; Han, S.-W.; Oh, D.-Y.; Kim, J.H.; Im, S.-A.; Han, W.; Ae Park, I.; Noh, D.-Y.; Bang, Y.-J.; Kim, T.-Y. The clinicopathologic characteristics and prognostic significance of triple-negativity in node-negative breast cancer. BMC Cancer 2008, 8, 307. [Google Scholar] [CrossRef]
- Vici, P.; Pizzuti, L.; Natoli, C.; Gamucci, T.; Di Lauro, L.; Barba, M.; Sergi, D.; Botti, C.; Michelotti, A.; Moscetti, L.; et al. Triple positive breast cancer: A distinct subtype? Cancer Treat. Rev. 2015, 41, 69–76. [Google Scholar] [CrossRef]
- Jiang, X.; Shapiro, D.J. The immune system and inflammation in breast cancer. Mol. Cell. Endocrinol. 2014, 382, 673–682. [Google Scholar] [CrossRef]
- Hon, J.D.C.; Singh, B.; Sahin, A.; Du, G.; Wang, J.; Wang, V.Y.; Deng, F.-M.; Zhang, D.Y.; Monaco, M.E.; Lee, P. Breast cancer molecular subtypes: From TNBC to QNBC. Am. J. Cancer Res. 2016, 6, 1864–1872. [Google Scholar]
- Chadar, R.; Afsana; Kesharwani, P. Nanotechnology-based siRNA delivery strategies for treatment of triple negative breast cancer. Int. J. Pharm. 2021, 605, 120835. [Google Scholar] [CrossRef]
- Amjad, M.W.; Kesharwani, P.; Mohd Amin, M.C.I.; Iyer, A.K. Recent advances in the design, development, and targeting mechanisms of polymeric micelles for delivery of siRNA in cancer therapy. Prog. Polym. Sci. 2017, 64, 154–181. [Google Scholar] [CrossRef]
- Ossovskaya, V.; Wang, Y.; Budoff, A.; Xu, Q.; Lituev, A.; Potapova, O.; Vansant, G.; Monforte, J.; Daraselia, N. Exploring Molecular Pathways of Triple-Negative Breast Cancer. Genes Cancer 2011, 2, 870–879. [Google Scholar] [CrossRef]
- Soni, N.; Soni, N.; Pandey, H.; Maheshwari, R.; Kesharwani, P.; Tekade, R.K. Augmented delivery of gemcitabine in lung cancer cells exploring mannose anchored solid lipid nanoparticles. J. Colloid Interface Sci. 2016, 481, 107–116. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef]
- Yang, M.; Zeng, C.; Li, P.; Qian, L.; Ding, B.; Huang, L.; Li, G.; Jiang, H.; Gong, N.; Wu, W. Impact of CXCR4 and CXCR7 knockout by CRISPR/Cas9 on the function of triple-negative breast cancer cells. OncoTargets Ther. 2019, 12, 3849–3858. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Ma, T.; Verkman, A.S. Aquaporin water channels in gastrointestinal physiology. J. Physiol. 1999, 517, 317–326. [Google Scholar] [CrossRef]
- Satooka, H.; Hara-Chikuma, M. Aquaporin-3 controls breast cancer cell migration by regulating hydrogen peroxide transport and its downstream cell signaling. Mol. Cell. Biol. 2016, 36, 1206–1218. [Google Scholar] [CrossRef]
- Verkman, A.S.; Hara-Chikuma, M.; Papadopoulos, M.C. Aquaporins—New players in cancer biology. J. Mol. Med. 2008, 86, 523–529. [Google Scholar] [CrossRef]
- Arif, M.; Kitchen, P.; Conner, M.T.; Hill, E.J.; Nagel, D.; Bill, R.M.; Dunmore, S.J.; Armesilla, A.L.; Gross, S.; Carmichael, A.R.; et al. Downregulation of aquaporin 3 inhibits cellular proliferation, migration and invasion in the MDA-MB-231 breast cancer cell line. Oncol. Lett. 2018, 16, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Yu, H. Cdc20: A WD40 Activator for a Cell Cycle Degradation Machine. Mol. Cell 2007, 27, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Baxter, J.E.; Fry, A.M.; Yamano, H. A Role for the Fizzy/Cdc20 Family of Proteins in Activation of the APC/C Distinct from Substrate Recruitment. Mol. Cell 2008, 32, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.B.; Aliabadi, H.M.; Mahdipoor, P.; Kucharski, C.; Maranchuk, R.; Hugh, J.C.; Uludag, H. Targeting Cell Cycle Proteins in Breast Cancer Cells with siRNA by Using Lipid-Substituted Polyethylenimines. Front. Bioeng. Biotechnol. 2015, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Lowe, V.J.; Lee, S. Inhibition of Cdc20 suppresses the metastasis in triple negative breast cancer (TNBC). Breast Cancer 2021, 28, 1073–1086. [Google Scholar] [CrossRef]
- Parmar, M.B.; Remant Bahadur, K.C.; Löbenberg, R.; Uludag, H. Additive Polyplexes to Undertake siRNA Therapy against CDC20 and Survivin in Breast Cancer Cells. Biomacromolecules 2018, 19, 4193–4206. [Google Scholar] [CrossRef]
- Jeanne, M.; Jorgensen, J.; Gould, D.B. Molecular and genetic analyses of collagen type IV mutant mouse models of spontaneous intracerebral hemorrhage identify mechanisms for stroke prevention. Circulation 2015, 131, 1555–1565. [Google Scholar] [CrossRef]
- Zhu, J.; Pan, C.; Jiang, J.; Deng, M.; Gao, H.; Men, B.; McClelland, M.; Mercola, D.; Zhong, W.-D.; Jia, Z. Six stroma-based RNA markers diagnostic for prostate cancer in European-Americans validated at the RNA and protein levels in patients in China. Oncotarget 2015, 6, 16757–16765. [Google Scholar] [CrossRef][Green Version]
- Brown, C.W.; Brodsky, A.S.; Freiman, R.N. Notch3 Overexpression Promotes Anoikis Resistance in Epithelial Ovarian Cancer via Upregulation of COL4A2. Mol. Cancer Res. 2015, 13, 78–85. [Google Scholar] [CrossRef]
- Maekawa, R.; Sato, S.; Yamagata, Y.; Asada, H.; Tamura, I.; Lee, L.; Okada, M.; Tamura, H.; Takaki, E.; Nakai, A.; et al. Genome-Wide DNA Methylation Analysis Reveals a Potential Mechanism for the Pathogenesis and Development of Uterine Leiomyomas. PLoS ONE 2013, 8, e66632. [Google Scholar] [CrossRef]
- Wang, C.; Gao, C.; Zhuang, J.-L.; Ding, C.; Wang, Y. A combined approach identifies three mRNAs that are down-regulated by microRNA-29b and promote invasion ability in the breast cancer cell line MCF-7. J. Cancer Res. Clin. Oncol. 2012, 138, 2127–2136. [Google Scholar] [CrossRef]
- He, J.; Yang, J.; Chen, W.; Wu, H.; Yuan, Z.; Wang, K.; Li, G.; Sun, J.; Yu, L. Molecular features of triple negative breast cancer: Microarray evidence and further integrated analysis. PLoS ONE 2015, 10, e0129842. [Google Scholar] [CrossRef]
- He, J.; Hong, G.; Yang, J.; Zheng, D.; Li, F.; Chen, W.; Luo, X.; Mao, Y.; OuYang, Y.; Pan, Y.; et al. siRNA-Mediated suppression of collagen type iv alpha 2 (COL4A2) mRNA inhibits triple-negative breast cancer cell proliferation and migration. Oncotarget 2017, 8, 2585–2593. [Google Scholar] [CrossRef]
- David, S.; Pitard, B.; Benoît, J.-P.; Passirani, C. Non-viral nanosystems for systemic siRNA delivery. Pharmacol. Res. 2010, 62, 100–114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Egorova, A.; Kiselev, A.; Hakli, M.; Ruponen, M.; Baranov, V.; Urtti, A. Chemokine-derived peptides as carriers for gene delivery to CXCR4 expressing cells. J. Gene Med. 2009, 11, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, A.; Egorova, A.; Laukkanen, A.; Baranov, V.; Urtti, A. Characterization of reducible peptide oligomers as carriers for gene delivery. Int. J. Pharm. 2013, 441, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Egorova, A.; Bogacheva, M.; Shubina, A.; Baranov, V.; Kiselev, A. Development of a receptor-targeted gene delivery system using CXCR4 ligand-conjugated cross-linking peptides. J. Gene Med. 2014, 16, 336–351. [Google Scholar] [CrossRef]
- Egorova, A.; Shubina, A.; Sokolov, D.; Selkov, S.; Baranov, V.; Kiselev, A. CXCR4-targeted modular peptide carriers for efficient anti-VEGF siRNA delivery. Int. J. Pharm. 2016, 515, 431–440. [Google Scholar] [CrossRef]
- Egorova, A.; Petrosyan, M.; Maretina, M.; Balashova, N.; Polyanskih, L.; Baranov, V.; Kiselev, A. Anti-angiogenic treatment of endometriosis via anti-VEGFA siRNA delivery by means of peptide-based carrier in a rat subcutaneous model. Gene Ther. 2018, 25, 548–555. [Google Scholar] [CrossRef]
- Egorova, A.A.; Shtykalova, S.V.; Maretina, M.A.; Sokolov, D.I.; Selkov, S.A.; Baranov, V.S.; Kiselev, A.V. Synergistic Anti-Angiogenic Effects Using Peptide-Based Combinatorial Delivery of siRNAs Targeting VEGFA, VEGFR1, and Endoglin Genes. Pharmaceutics 2019, 11, 261. [Google Scholar] [CrossRef]
- Wang, J.; Mi, P.; Lin, G.; Wáng, Y.X.J.; Liu, G.; Chen, X. Imaging-guided delivery of RNAi for anticancer treatment. Adv. Drug Deliv. Rev. 2016, 104, 44–60. [Google Scholar] [CrossRef]
- Peer, D.; Lieberman, J. Special delivery: Targeted therapy with small RNAs. Gene Ther. 2011, 18, 1127–1133. [Google Scholar] [CrossRef]
- Liang, S.; Peng, X.; Li, X.; Yang, P.; Xie, L.; Li, Y.; Du, C.; Zhang, G. Silencing of CXCR4 sensitizes triple-negative breast cancer cells to cisplatin. Oncotarget 2015, 6, 1020–1030. [Google Scholar] [CrossRef]
- Ren, Y.; Jiang, X.; Pan, D.; Mao, H.-Q. Charge density and molecular weight of polyphosphoramidate gene carrier are key parameters influencing its DNA compaction ability and transfection efficiency. Biomacromolecules 2010, 11, 3432–3439. [Google Scholar] [CrossRef]
- Nikcevic, G.; Kovacevic-Grujicic, N.; Stevanovic, M. Improved transfection efficiency of cultured human cells. Cell Biol. Int. 2003, 27, 735–737. [Google Scholar] [CrossRef]
- Mayer, I.A.; Abramson, V.G.; Lehmann, B.D.; Pietenpol, J.A. New strategies for triple-negative breast cancer-deciphering the heterogeneity. Clin. Cancer Res. 2014, 20, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.; Custodio, S.; Maestro De Las Casas, M.-L.; García-Sáenz, J.-A.; De La Torre, J.-C.; Bellón-Cano, J.-M.; López-Tarruella, S.; Vidaurreta-Lazaro, M.; De La Orden, V.; Jerez, Y.; et al. Circulating tumor cells following first chemotherapy cycle: An early and strong predictor of outcome in patients with metastatic breast cancer. Oncologist 2013, 18, 917–923. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhu, Z.; Sun, M.; Wang, J.; Guo, R.; Shen, L.; Wu, W. Critical role of aquaporin-3 in the human epidermal growth factor-induced migration and proliferation in the human gastric adenocarcinoma cells. Cancer Biol. Ther. 2010, 9, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Ma, Y.-F.; Yu, C.-H.; Li, Y.-J.; Tang, J.; Li, J.-B.; Zhao, Y.-N.; Liu, Y. Aquaporin 3 knockdown suppresses tumour growth and angiogenesis in experimental non-small cell lung cancer. Exp. Physiol. 2014, 99, 974–984. [Google Scholar] [CrossRef]
- Tang, J.; Lu, M.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Overexpression of ASPM, CDC20, and TTK Confer a Poorer Prognosis in Breast Cancer Identified by Gene Co-expression Network Analysis. Front. Oncol. 2019, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xie, Y.; Williams, J.; Hang, Y.; Richter, L.; Becker, M.; Amador, C.; Oupický, D.; Hyde, R.K. Use of polymeric CXCR4 inhibitors as siRNA delivery vehicles for the treatment of acute myeloid leukemia. Cancer Gene Ther. 2020, 27, 45–55. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Li, J.; Hang, Y.; Oupický, D. Promise of chemokine network-targeted nanoparticles in combination nucleic acid therapies of metastatic cancer. WIREs Nanomed. Nanobiotechnol. 2019, 11, e1528. [Google Scholar] [CrossRef]
- Castanotto, D.; Sakurai, K.; Lingeman, R.; Li, H.; Shively, L.; Aagaard, L.; Soifer, H.; Gatignol, A.; Riggs, A.; Rossi, J.J. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007, 35, 5154–5164. [Google Scholar] [CrossRef]
- Su, S.; Tian, Y.; Li, Y.; Ding, Y.; Ji, T.; Wu, M.; Wu, Y.; Nie, G. “Triple-Punch” Strategy for Triple Negative Breast Cancer Therapy with Minimized Drug Dosage and Improved Antitumor Efficacy. ACS Nano 2015, 9, 1367–1378. [Google Scholar] [CrossRef]
- Entschladen, F.; Drell, T.L., IV; Lang, K.; Joseph, J.; Zaenker, K.S. Tumour-cell migration, invasion, and metastasis: Navigation by neurotransmitters. Lancet Oncol. 2004, 5, 254–258. [Google Scholar] [CrossRef]
- Cao, X.-C.; Zhang, W.-R.; Cao, W.-F.; Liu, B.-W.; Zhang, F.; Zhao, H.-M.; Meng, R.; Zhang, L.; Niu, R.-F.; Hao, X.-S.; et al. Aquaporin3 Is Required for FGF-2-Induced Migration of Human Breast Cancers. PLoS ONE 2013, 8, e56735. [Google Scholar] [CrossRef]
- Cheng, S.; Castillo, V.; Sliva, D. CDC20 associated with cancer metastasis and novel mushroom-derived CDC20 inhibitors with antimetastatic activity. Int. J. Oncol. 2019, 54, 2250–2256. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Cantley, L.C. The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 2018, 174, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Ghorai, S.; Dinesh, U.S.; Shetty, P.; Chattopadhyay, S.; Santra, M.K. Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017, 8, e2882. [Google Scholar] [CrossRef] [PubMed]
- Poage, G.M.; Hartman, Z.C.; Brown, P.H. Revealing targeted therapeutic opportunities in triple-negative breast cancers: A new strategy. Cell Cycle 2013, 12, 2705–2706. [Google Scholar] [CrossRef]
- Slita, A.; Egorova, A.; Casals, E.; Kiselev, A.; Rosenholm, J.M. Characterization of modified mesoporous silica nanoparticles as vectors for siRNA delivery. Asian J. Pharm. Sci. 2018, 13, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, Y.; Chen, H.; Guo, Z. MicroRNA-488 inhibits proliferation, invasion and EMT in osteosarcoma cell lines by targeting aquaporin 3. Int. J. Oncol. 2018, 53, 1493–1504. [Google Scholar] [CrossRef]
- Hemati, M.; Haghiralsadat, F.; Jafary, F.; Moosavizadeh, S.; Moradi, A. Targeting cell cycle protein in gastric cancer with CDC20siRNA and anticancer drugs (Doxorubicin and quercetin) co-loaded cationic PEGylated nanoniosomes. Int. J. Nanomed. 2019, 14, 6575–6585. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Yang, H.; Wu, J.; Wang, A.; Chen, X.; Hu, S.; Zhang, Y.; Bai, D.; Jin, Z. COL4A2 in the tissue-specific extracellular matrix plays important role on osteogenic differentiation of periodontal ligament stem cells. Theranostics 2019, 9, 4265–4286. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, L.; Yeh, J.; Wu, X.; Cao, Z.; Wang, Y.A.; Zhang, M.; Yang, L.; Mao, H. Reducing non-specific binding and uptake of nanoparticles and improving cell targeting with an antifouling PEO-b-PγMPS copolymer coating. Biomaterials 2010, 31, 5397–5407. [Google Scholar] [CrossRef]
- Wu, J.; Qu, L.; Meng, L.; Zeng, Y.; Shou, C.; Xu, H.; Jiang, B.; Ren, T. N-α-Acetyltransferase 10 protein inhibits apoptosis through RelA/p65-regulated MCL1 expression. Carcinogenesis 2012, 33, 1193–1202. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).