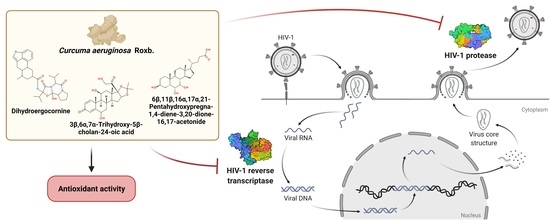
HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening
Abstract
:1. Introduction
2. Results
2.1. Plant Extract Preparation
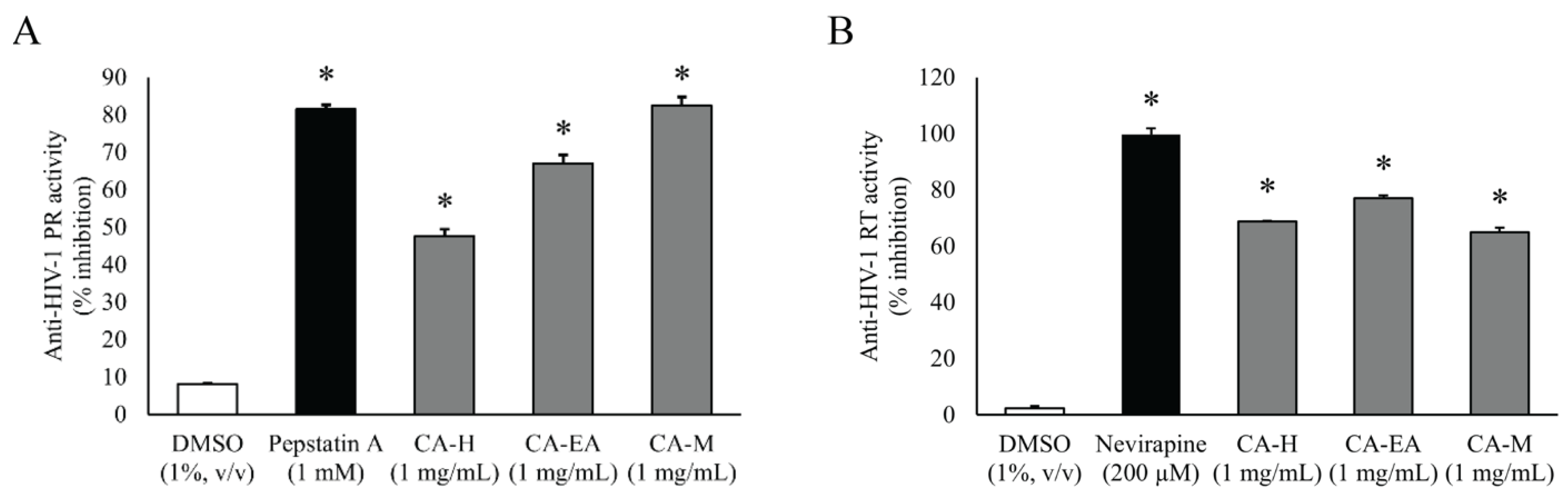
2.2. HIV-1 Protease Inhibitor Screening
2.3. HIV-1 Reverse Transcriptase Inhibitor Screening
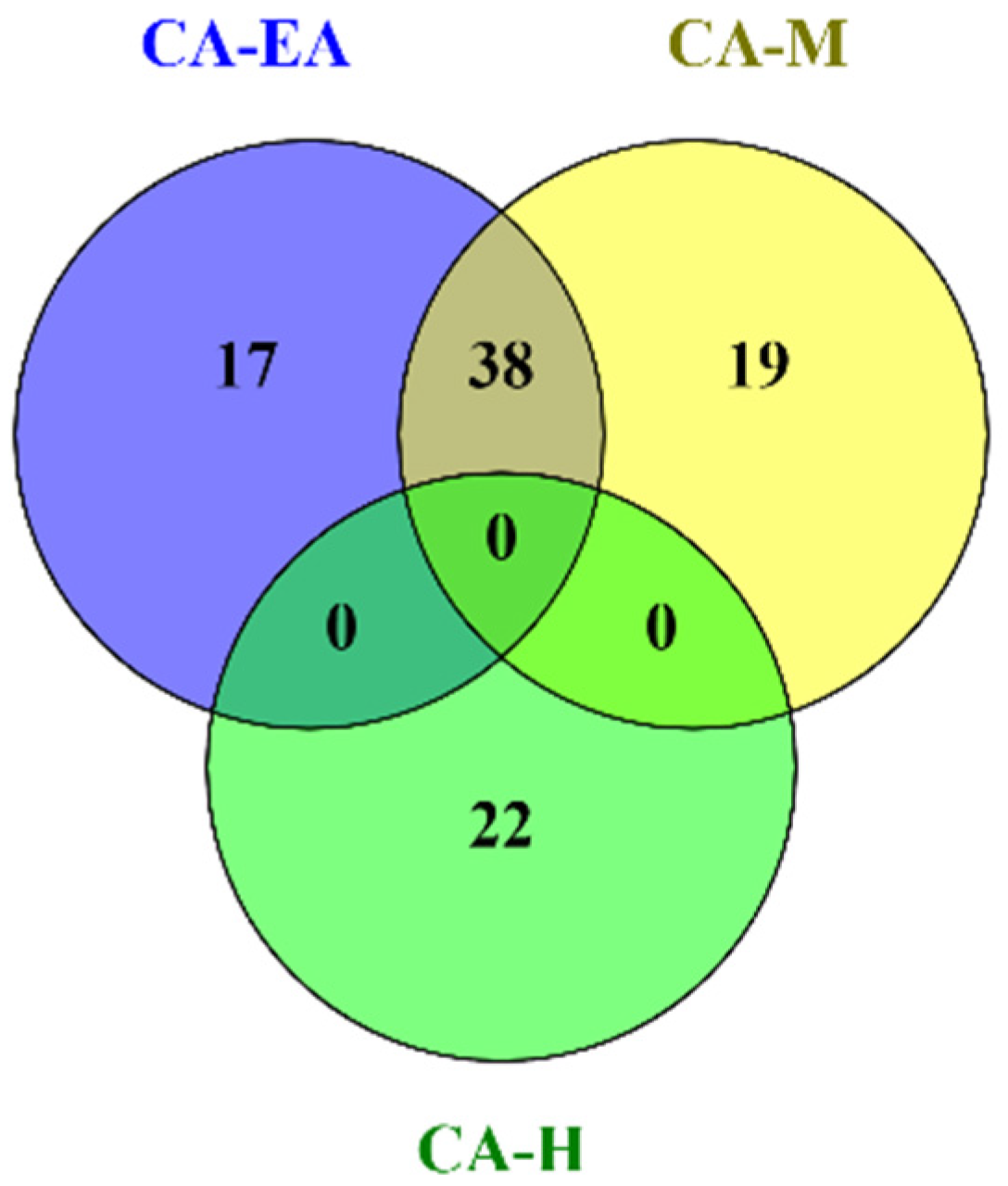
2.4. Phytochemical Profile Analysis
2.5. Evaluation of Lipinski′s Rule of Five and ADMET Parameters
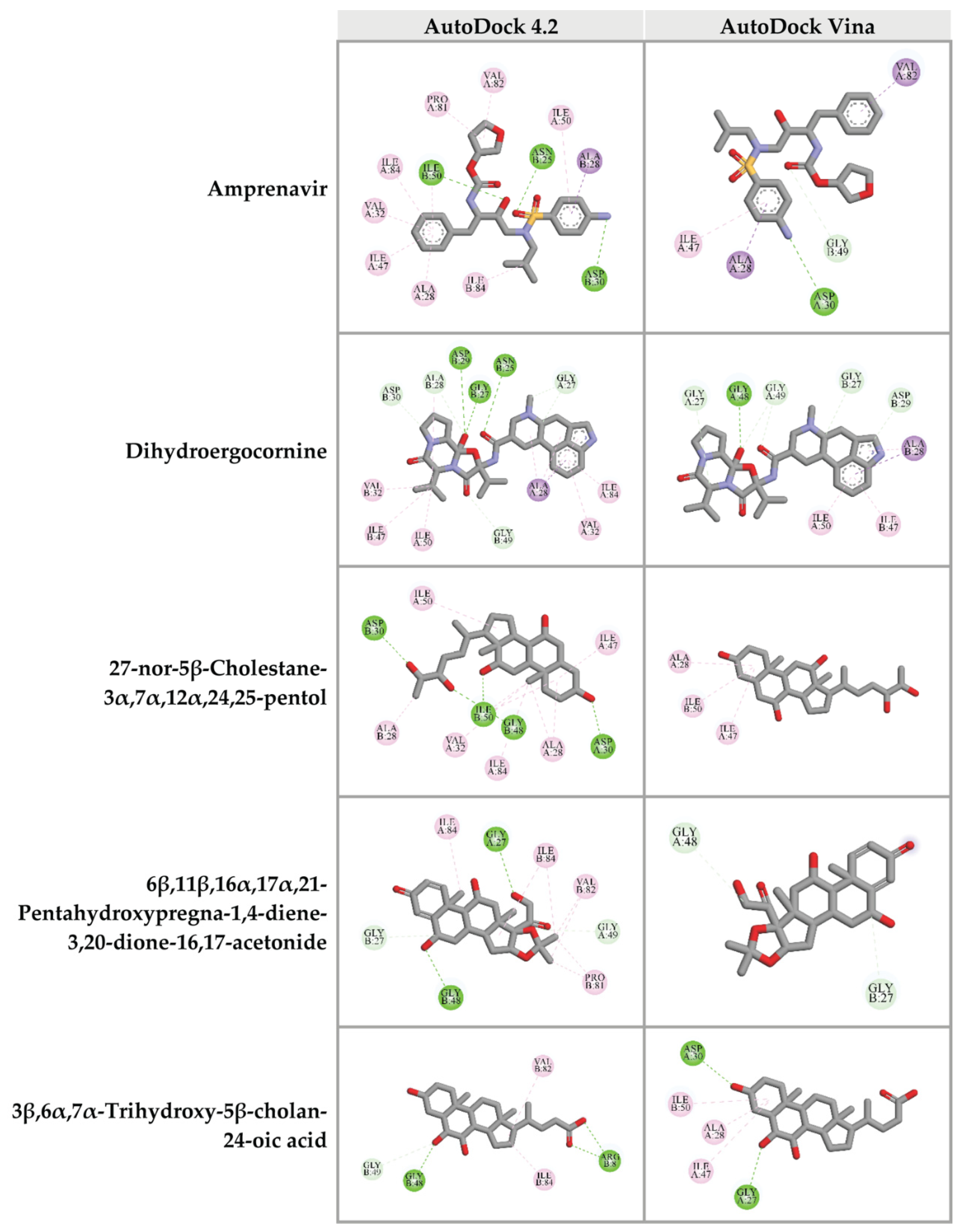
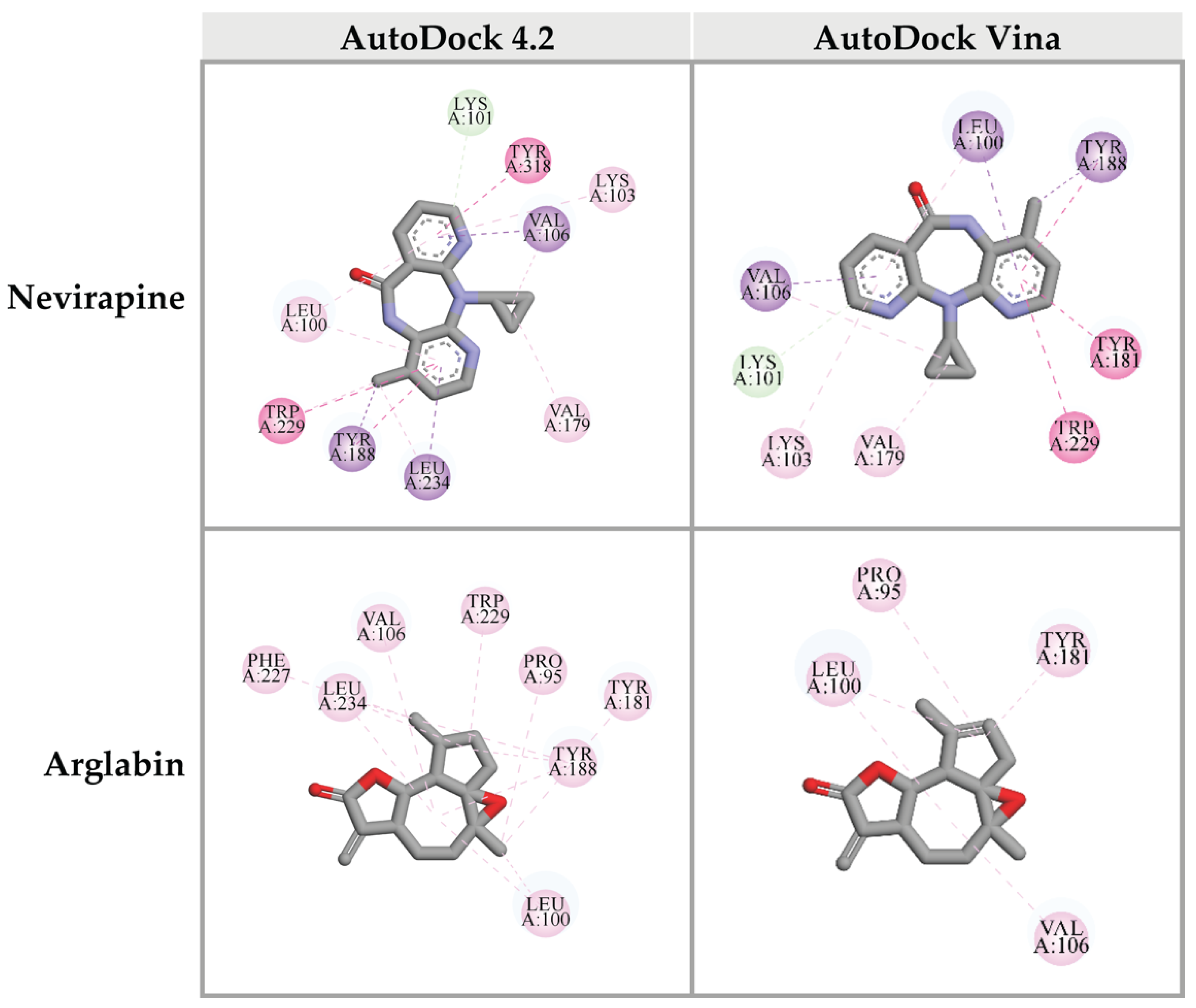
2.6. Prediction of Binding Affinity of Candidate Ligands against HIV-1 PR Active Site
2.7. Prediction of Binding Affinity of Candidate Ligands against DNA Polymerase Active Site of HIV-1 RT
2.8. Prediction of Binding Affinity of Candidate Ligands against RNase H Active Site of HIV-1 RT
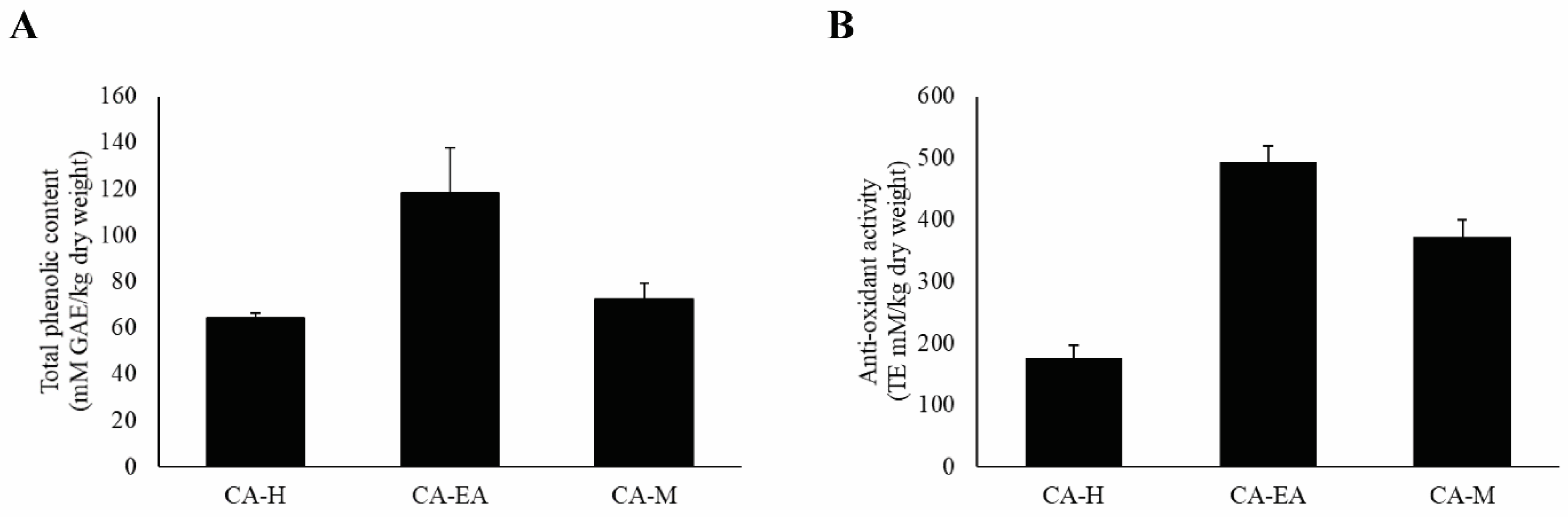
2.9. Investigations of Antioxidant Constituents and Antioxidant Activity
3. Discussion
4. Materials and Methods
4.1. Plant Material and Extract Preparation
4.2. HIV-1 Protease Inhibitor Screening
4.3. HIV-1 Reverse Transcriptase Inhibitor Screening
4.4. Phytochemical Profile Analysis
4.5. Evaluation of Lipinski’s Rule of Five and ADMET Parameters
4.6. Molecular Docking Analysis
4.7. Total Phenolic Content Measurement
4.8. Antioxidant Activity Measurement
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nyamweya, S.; Hegedus, A.; Jaye, A.; Rowland-Jones, S.; Flanagan, K.L.; Macallan, D.C. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 2013, 23, 221–240. [Google Scholar] [CrossRef]
- Kwan, C.K.; Ernst, J.D. HIV and tuberculosis: A deadly human syndemic. Clin. Microbiol. Rev. 2011, 24, 351–376. [Google Scholar] [CrossRef] [Green Version]
- Engelman, A.; Cherepanov, P. The structural biology of HIV-1: Mechanistic and therapeutic insights. Nat. Rev. Microbiol. 2012, 10, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS 2015, 7, 95–104. [Google Scholar]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and function of HIV-1 reverse transcriptase: Molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkman, K.; ter Hofstede, H.J.; Burger, D.M.; Smeitink, J.A.; Koopmans, P.P. Adverse effects of reverse transcriptase inhibitors: Mitochondrial toxicity as common pathway. Aids 1998, 12, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Ammassari, A.; Murri, R.; Pezzotti, P.; Trotta, M.P.; Ravasio, L.; De Longis, P.; Caputo, L.; Narciso, P.; Pauluzzi, S.; Carosi, G. Self-Reported symptoms and medication side effects influence adherence to highly active antiretroviral therapy in persons with HIV infection. J. Acquir. Immune Defic. Syndr. 2001, 28, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Montessori, V.; Press, N.; Harris, M.; Akagi, L.; Montaner, J.S. Adverse effects of antiretroviral therapy for HIV infection. Cmaj 2004, 170, 229–238. [Google Scholar]
- Yeni, P. Update on HAART in HIV. J. Hepatol. 2006, 44, S100–S103. [Google Scholar] [CrossRef]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D.; Investigators, H.O.S. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Corona, A.; Tramontano, E. HIV-1 reverse transcriptase still remains a new drug target: Structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions. Mol. Biol. Int. 2012, 2012, 586401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillapachaiyaporn, C.; Nilkhet, S.; Ung, A.T.; Chuchawankul, S. Anti-HIV-1 protease activity of the crude extracts and isolated compounds from Auricularia polytricha. BMC Complementary Altern. Med. 2019, 19, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sillapachaiyaporn, C.; Chuchawankul, S. HIV-1 protease and reverse transcriptase inhibition by tiger milk mushroom (Lignosus rhinocerus) sclerotium extracts: In vitro and in silico studies. J. Tradit. Complementary Med. 2020, 10, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; Carli, I.; Del Vecchio, C.; Xu, L.; Corona, A.; Grandi, N.; Piano, D.; Maccioni, E.; Distinto, S.; Parolin, C. Sennoside A, derived from the traditional chinese medicine plant Rheum L., is a new dual HIV-1 inhibitor effective on HIV-1 replication. Phytomedicine 2016, 23, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.S.; Maulana, S.; Widodo, A.; Mukaddas, A.; Pitopang, R. Docking study on anti-HIV-1 activity of secondary metabolites from Zingiberaceae plants. J. Pharm. Bioallied Sci. 2020, 12, S763–S767. [Google Scholar] [CrossRef] [PubMed]
- Yuandani, I.J.; Rohani, A.S.; Sumantri, I.B. Immunomodulatory effects and mechanisms of Curcuma species and their bioactive compounds: A review. Front. Pharmacol. 2021, 12, 643119. [Google Scholar] [CrossRef]
- Srivilai, J.; Khorana, N.; Waranuch, N.; Wisuitiprot, W.; Suphrom, N.; Suksamrarn, A.; Ingkaninan, K. Germacrene analogs are anti-androgenic on androgen-dependent cells. Nat. Prod. Commun. 2016, 11, 1225–1228. [Google Scholar] [CrossRef] [Green Version]
- Suphrom, N.; Pumthong, G.; Khorana, N.; Waranuch, N.; Limpeanchob, N.; Ingkaninan, K. Anti-Androgenic effect of sesquiterpenes isolated from the rhizomes of Curcuma aeruginosa roxb. Fitoterapia 2012, 83, 864–871. [Google Scholar] [CrossRef]
- Hossain, C.F.; Al-Amin, M.; Sayem, A.S.M.; Siragee, I.H.; Tunan, A.M.; Hassan, F.; Kabir, M.M.; Sultana, G.N.N. Antinociceptive principle from Curcuma aeruginosa. BMC Complementary Altern. Med. 2015, 15, 191. [Google Scholar] [CrossRef] [Green Version]
- Kamazeri, T.S.A.T.; Abd Samah, O.; Taher, M.; Susanti, D.; Qaralleh, H. Antimicrobial activity and essential oils of Curcuma aeruginosa, Curcuma mangga, and Zingiber cassumunar from Malaysia. Asian Pac. J. Trop. Med. 2012, 5, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Otake, T.; Mori, H.; Morimoto, M.; Ueba, N.; Sutardjo, S.; Kusumoto, I.T.; Hattori, M.; Namba, T. Screening of Indonesian plant extracts for anti-human immunodeficiency virus—Type 1 (HIV-1) activity. Phytother. Res. 1995, 9, 6–10. [Google Scholar] [CrossRef]
- Daussy, C.F.; Galais, M.; Pradel, B.; Robert-Hebmann, V.; Sagnier, S.; Pattingre, S.; Biard-Piechaczyk, M.; Espert, L. HIV-1 Env induces pexophagy and an oxidative stress leading to uninfected CD4+ T cell death. Autophagy 2020, 17, 2465–2474. [Google Scholar] [CrossRef]
- Ezhilarasan, D.; Srilekha, M.; Raghu, R. HAART and hepatotoxicity. J. Appl. Pharm. Sci. 2017, 7, 220–226. [Google Scholar]
- Carr, A.; Cooper, D.A. Adverse effects of antiretroviral therapy. Lancet 2000, 356, 1423–1430. [Google Scholar] [CrossRef]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K. The worldwide trend of using botanical drugs and strategies for developing global drugs. BMB Rep. 2017, 50, 111. [Google Scholar] [CrossRef] [Green Version]
- Sookkongwaree, K.; Geitmann, M.; Roengsumran, S.; Petsom, A.; Danielson, U.H. Inhibition of viral proteases by Zingiberaceae extracts and flavones isolated from Kaempferia parviflora. Die Pharm.-Int. J. Pharm. Sci. 2006, 61, 717–721. [Google Scholar]
- Yu, Y.-B.; Miyashiro, H.; Nakamura, N.; Hattori, M.; Park, J.C. Effects of triterpenoids and flavonoids isolated from Alnus firma on HIV-1 viral enzymes. Arch. Pharmacal Res. 2007, 30, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Cheenpracha, S.; Karalai, C.; Ponglimanont, C.; Subhadhirasakul, S.; Tewtrakul, S. Anti-HIV-1 protease activity of compounds from Boesenbergia pandurata. Bioorganic Med. Chem. 2006, 14, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Safe, I.P.; Amaral, E.P.; Araújo-Pereira, M.; Lacerda, M.V.; Printes, V.S.; Souza, A.B.; Beraldi-Magalhães, F.; Monteiro, W.M.; Sampaio, V.S.; Barreto-Duarte, B. Adjunct N-acetylcysteine treatment in hospitalized patients with HIV-associated tuberculosis dampens the oxidative stress in peripheral blood: Results from the RIPENACTB Study trial. Front. Immunol. 2020, 11, 602589. [Google Scholar] [CrossRef]
- Moon-ai, W.; Niyomploy, P.; Boonsombat, R.; Sangvanich, P.; Karnchanatat, A. A superoxide dismutase purified from the rhizome of Curcuma aeruginosa Roxb. as inhibitor of nitric oxide production in the macrophage-like RAW 264.7 cell line. Appl. Biochem. Biotechnol. 2012, 166, 2138–2155. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.; Rautray, T.R. Estimation of trace elements, antioxidants, and antibacterial agents of regularly consumed Indian medicinal plants. Biol. Trace Elem. Res. 2021, 199, 1185–1193. [Google Scholar] [CrossRef]
- Tonsomboon, A.; Prasanth, M.I.; Plaingam, W.; Tencomnao, T. Kaempferia parviflora rhizome extract inhibits glutamate-induced toxicity in HT-22 mouse hippocampal neuronal cells and extends longevity in Caenorhabditis elegans. Biology 2021, 10, 264. [Google Scholar] [CrossRef]
- Simoh, S.; Zainal, A. Chemical profiling of Curcuma aeruginosa Roxb. rhizome using different techniques of solvent extraction. Asian Pac. J. Trop. Biomed. 2015, 5, 412–417. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangsinth, P.; Sillapachaiyaporn, C.; Nilkhet, S.; Tencomnao, T.; Ung, A.T.; Chuchawankul, S. Mushroom-Derived bioactive compounds potentially serve as the inhibitors of SARS-CoV-2 main protease: An in silico approach. J. Tradit. Complementary Med. 2021, 11, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Medina-O’Donnell, M.; Rivas, F.; Reyes-Zurita, F.J.; Cano-Muñoz, M.; Martinez, A.; Lupiañez, J.A.; Parra, A. Oleanolic acid derivatives as potential inhibitors of HIV-1 protease. J. Nat. Prod. 2019, 82, 2886–2896. [Google Scholar] [CrossRef]
- Xiao, S.; Tian, Z.; Wang, Y.; Si, L.; Zhang, L.; Zhou, D. Recent progress in the antiviral activity and mechanism study of pentacyclic triterpenoids and their derivatives. Med. Res. Rev. 2018, 38, 951–976. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.W.; Ayeni, C.; Breuer, S.; Torbett, B.E. Virtual screening for HIV protease inhibitors: A comparison of AutoDock 4 and Vina. PLoS ONE 2010, 5, e11955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock vina adopts more accurate binding poses but autodock4 forms better binding affinity. J. Chem. Inf. Modeling 2019, 60, 204–211. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freis, E.D.; Stanton, J.R.; Litter, J.; Culbertson, J.W.; Halperin, M.H.; Moister, F.C.; Wilkins, R.W. The hemodynamic effects of hypotensive drugs in man. II. Dihydroergocornine. J. Clin. Investig. 1949, 28, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Ravi, S.; Shanmugam, B.; Subbaiah, G.V.; Prasad, S.H.; Reddy, K.S. Identification of food preservative, stress relief compounds by GC–MS and HR-LC/Q-TOF/MS; evaluation of antioxidant activity of Acalypha indica leaves methanolic extract (in vitro) and polyphenolic fraction (in vivo). J. Food Sci. Technol. 2017, 54, 1585–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumbhar, S.T.; Patil, S.P.; Une, H.D. Phytochemical analysis of Canna indica L. roots and rhizomes extract. Biochem. Biophys. Rep. 2018, 16, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Pellicciari, R.; Passeri, D.; De Franco, F.; Mostarda, S.; Filipponi, P.; Colliva, C.; Gadaleta, R.M.; Franco, P.; Carotti, A.; Macchiarulo, A. Discovery of 3α, 7α, 11β-trihydroxy-6α-ethyl-5β-cholan-24-oic acid (TC-100), a novel bile acid as potent and highly selective FXR agonist for enterohepatic disorders. J. Med. Chem. 2016, 59, 9201–9214. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, X.; Hu, L.; Pham, L.; Poole, K.M.; Tang, Y.; Mahon, B.P.; Tang, W.; Li, K.; Goldfarb, N.E. Effects of hinge-region natural polymorphisms on human immunodeficiency virus-type 1 protease structure, dynamics, and drug pressure evolution. J. Biol. Chem. 2016, 291, 22741–22756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lansdon, E.B.; Liu, Q.; Leavitt, S.A.; Balakrishnan, M.; Perry, J.K.; Lancaster-Moyer, C.; Kutty, N.; Liu, X.; Squires, N.H.; Watkins, W.J. Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 2011, 55, 2905–2915. [Google Scholar] [CrossRef] [Green Version]
- Sillapachaiyaporn, C.; Rangsinth, P.; Nilkhet, S.; Ung, A.T.; Chuchawankul, S.; Tencomnao, T. Neuroprotective effects against Glutamate-Induced HT-22 hippocampal cell damage and Caenorhabditis elegans lifespan/healthspan enhancing activity of Auricularia polytricha mushroom extracts. Pharmaceuticals 2021, 14, 1001. [Google Scholar] [CrossRef]
- Panthong, S.; Boonsathorn, N.; Chuchawankul, S. Antioxidant activity, anti-proliferative activity, and amino acid profiles of ethanolic extracts of edible mushrooms. Genet. Mol. Res. 2016, 15, gmr15048886. [Google Scholar] [CrossRef] [PubMed]



| Identified Compound | Extract |
|---|---|
| (22S)-1α,25-Dihydroxy-22-methoxy-26,27-dimethyl-23,24-tetradehydro-20-epivitamin D3 | CA-EA |
| (4Z)-4-(6,6-Dimethyl-2-methylidenecyclohex-3-en-1-ylidene) pentan-2-ol | CA-H |
| (6R)-Vitamin D3 6,19-(4-phenyl-1,2,4-triazoline-3,5-dione) adduct/(6R)-cholecalciferol 6,19-(4-phenyl-1,2,4-triazoline-3,5-dione) adduct | CA-M |
| (E)-2-Methylglutaconic acid | CA-EA, CA-M |
| 10-keto Tridecanoic acid | CA-EA |
| 12-Hydroxy-10-octadecynoic acid | CA-EA, CA-M |
| 13-Hydroxy-tridecanoic acid | CA-M |
| 1α-Hydroxy-24-(dimethylphosphoryl)-25,26,27-trinorvitamin D3 | CA-M |
| 2,2,4-Trimethyl-3-[(3E,7E,11E)-3,8,12,16-tetramethylheptadeca-3,7,11,15-tetraenyl] cyclohexan-1-ol | CA-H |
| 2,4-Dimethyl-2-eicosenoic acid | CA-EA |
| 2-[3-Carboxy-3-(methylammonio)propyl]-L-histidine | CA-EA, CA-M |
| 27-nor-5β-Cholestane-3α,7α,12α,24,25-pentol | CA-EA, CA-M |
| 2-Hydroxyethanesulfonate | CA-EA |
| 2-oxo-Dodecanoic acid | CA-EA |
| 3-(3,3,8,8-Tetramethyl-5-tricyclo [5.1.0.02,5] oct-5-enyl) propanoic acid | CA-H |
| 3-Dodecynoic acid | CA-M |
| 3-n-Decyl acrylic acid | CA-EA, CA-M |
| 3-oxo-Tridecanoic acid | CA-EA, CA-M |
| 3-Tridecynoic acid | CA-M |
| 3β,6α,7α -Trihydroxy-5β-cholan-24-oic acid | CA-EA, CA-M |
| 4-(2-Hydroxy-3isopropylaminoproxy) benzyloxy acetic acid | CA-EA |
| 4-(3,3-dimethylbut-1-ynyl)-4-hydroxy-2,6,6-trimethylcyclohex-2-en-1-one | CA-H |
| 4,7,7-Trimethyl-4-(2-methylallyl) tricyclo [3.3.0.02,8] octane-3,6-dione | CA-H |
| 4-Heptanone | CA-EA, CA-M |
| 4-Hydroxy capric acid | CA-EA |
| 4-Methylpentanal | CA-EA, CA-M |
| 4Z-Decenedioic acid | CA-EA |
| 6-(3-Hydroxyprop-1-en-2-yl)-4,8a-dimethyl-1,3,5,6,7,8-hexahydronaphthalen-2-one | CA-H |
| 6E-Nonenoic acid | CA-EA, CA-M |
| 6β,11β,16α,17α,21-Pentahydroxypregna-1,4-diene-3,20-dione-16,17-acetonide | CA-EA, CA-M |
| 7E,9Z-Dodecadien-1-ol | CA-EA |
| 7-Hydroxymethotrexate | CA-EA, CA-M |
| 9-Dodecen-1-ol | CA-EA, CA-M |
| 9-Isopropyl-1-methyl-2-methylene-5-oxatricyclo [5.4.0.03,8] undecane | CA-H |
| Ala Glu His | CA-M |
| Amiloxate | CA-EA, CA-M |
| Arglabin | CA-H |
| Benzenehexanoic acid, 2,5-dihydroxy-3,4-dimethoxy-6-methyl- | CA-EA, CA-M |
| Betaine | CA-M |
| Cadinol T | CA-H |
| Citronellic acid | CA-EA, CA-M |
| Cycloisolongifolene,8,9-dehydro-9-formyl- | CA-H |
| Cyclopropanebutanoic acid, 2-[[2-[[2-[(2-pentylcyclopropyl) methyl] cyclopropyl] methyl] | CA-H |
| Deoxyribose | CA-EA, CA-M |
| Deoxysappanone B 7,3′- dimethyl ether acetate | CA-EA |
| Dihydrocostunolide | CA-H |
| Dihydroergocornine | CA-M |
| Dihydrojasmonic acid, methyl ester | CA-EA, CA-M |
| Dihydrosphingosine | CA-EA, CA-M |
| Elephantopin | CA-M |
| Ethyl Oxalacetate | CA-M |
| Gemfibrozil | CA-EA, CA-M |
| Gemfibrozil M1 | CA-EA, CA-M |
| Gemfibrozil M3 | CA-EA, CA-M |
| Gln Lys Arg | CA-EA, CA-M |
| GPEtn(12:0/0:0) | CA-EA, CA-M |
| Hexadecasphinganine | CA-EA, CA-M |
| Hydroxycyclohexanecarboxylic acid | CA-EA, CA-M |
| Hydroxyibuprofen | CA-EA, CA-M |
| Ibutilide | CA-M |
| Ile Asp | CA-EA |
| Ile Leu Leu | CA-EA, CA-M |
| Ile Thr | CA-EA |
| Isoaromadendrene epoxide | CA-H |
| Lactone of PGF-MUM | CA-EA, CA-M |
| Leucine | CA-M |
| Linoleic acid | CA-H |
| Linoleic acid, methyl ester | CA-H |
| Methyl jasmonate | CA-EA, CA-M |
| Methyldopexamine sulfate | CA-M |
| N-(2-fluro-ethyl)-eicosanoyl amine | CA-M |
| N-(2-hydroxyethyl) icosanamide | CA-M |
| Octanal | CA-EA, CA-M |
| Oleic Acid | CA-H |
| Palmitic acid | CA-H |
| Pantoic acid | CA-EA |
| Phe Ala Arg | CA-M |
| Phe Ala Pro | CA-EA |
| Phe Gln Arg | CA-EA, CA-M |
| Phytosphingosine | CA-EA, CA-M |
| Pro Glu | CA-EA |
| Prostaglandin F1a alcohol | CA-M |
| Prostaglandin H1 | CA-EA, CA-M |
| Punctaporin B | CA-EA, CA-M |
| QH2 | CA-EA, CA-M |
| Swietenine | CA-M |
| Taurine | CA-EA, CA-M |
| Trp Gln Trp | CA-EA |
| Undecanal | CA-EA |
| Val Glu | CA-EA, CA-M |
| Val Val | CA-M |
| Xanthumin | CA-H |
| α-Cadinol | CA-H |
| α-Terpineol | CA-H |
| β-Elemene | CA-H |
| β-Levantenolide | CA-H |
| Compound | Binding Energy (kcal/mol) | |
|---|---|---|
| AutoDock 4.2 | AutoDock Vina | |
| Amprenavir (original inhibitor) | −9.73 | −9.0 |
| Dihydroergocornine | −12.65 | −11.3 |
| 27-nor-5β-Cholestane-3α,7α,12α,24,25-pentol | −11.53 | −9.8 |
| 3β,6α,7α-Trihydroxy-5β-cholan-24-oic acid | −10.92 | −9.3 |
| 6β,11β,16α,17α,21-Pentahydroxypregna-1,4-diene-3,20-dione-16,17-acetonide | −10.71 | −9.7 |
| β-Levantenolide | −9.52 | −8.2 |
| Deoxysappanone B 7,3′-dimethyl ether acetate | −7.96 | −7.8 |
| Xanthumin | −7.84 | −7.4 |
| Punctaporin B | −7.67 | −7.1 |
| Dihydrocostunolide | −7.51 | −7.6 |
| Prostaglandin H1 | −7.45 | −6.4 |
| Compound | Binding Energy (kcal/mol) | |
|---|---|---|
| AutoDock 4.2 | AutoDock Vina | |
| Nevirapine (original inhibitor) | −9.31 | −11.7 |
| 3β,6α,7α-Trihydroxy-5β-cholan-24-oic acid | −10.39 | −5.4 |
| 27-nor-5β-Cholestane-3α,7α,12α,24,25-pentol | −9.99 | −2.8 |
| Xanthumin | −9.73 | −8.9 |
| Prostaglandin F1a alcohol | −9.72 | −8.4 |
| QH2 | −9.69 | −8.6 |
| Prostaglandin H1 | −9.50 | −7.9 |
| Lactone of PGF-MUM | −9.48 | −8.7 |
| Dihydrocostunolide | −9.24 | −9.9 |
| Cadinol T | −9.22 | −9.7 |
| Arglabin | −9.22 | −10.8 |
| Compound | Binding Energy (kcal/mol) | |
|---|---|---|
| AutoDock 4.2 | AutoDock Vina | |
| P4Y (original inhibitor) | −5.29 | −6.0 |
| 3β,6α,7α-Trihydroxy-5β-cholan-24-oic acid | −6.77 | −6.4 |
| 27-nor-5β-Cholestane-3α,7α,12α,24,25-pentol | −6.58 | −5.9 |
| Hydroxyibuprofen | −5.92 | −4.3 |
| 6β,11β,16α,17α,21-Pentahydroxypregna-1,4-diene-3,20-dione-16,17-acetonide | −5.90 | −6.3 |
| Dihydroergocornine | −5.80 | −6.5 |
| Lactone of PGF-MUM | −5.57 | −4.7 |
| Pro Glu | −5.44 | −4.2 |
| β-Levantenolide | −5.38 | −6.4 |
| Val Val | −5.34 | −4.1 |
| 6-(3-Hydroxyprop-1-en-2-yl)-4,8a-dimethyl-1,3,5,6,7,8-hexahydronaphthalen-2-one | −5.25 | −4.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sillapachaiyaporn, C.; Rangsinth, P.; Nilkhet, S.; Moungkote, N.; Chuchawankul, S. HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening. Pharmaceuticals 2021, 14, 1115. https://doi.org/10.3390/ph14111115
Sillapachaiyaporn C, Rangsinth P, Nilkhet S, Moungkote N, Chuchawankul S. HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening. Pharmaceuticals. 2021; 14(11):1115. https://doi.org/10.3390/ph14111115
Chicago/Turabian StyleSillapachaiyaporn, Chanin, Panthakarn Rangsinth, Sunita Nilkhet, Nuntanat Moungkote, and Siriporn Chuchawankul. 2021. "HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening" Pharmaceuticals 14, no. 11: 1115. https://doi.org/10.3390/ph14111115
APA StyleSillapachaiyaporn, C., Rangsinth, P., Nilkhet, S., Moungkote, N., & Chuchawankul, S. (2021). HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening. Pharmaceuticals, 14(11), 1115. https://doi.org/10.3390/ph14111115