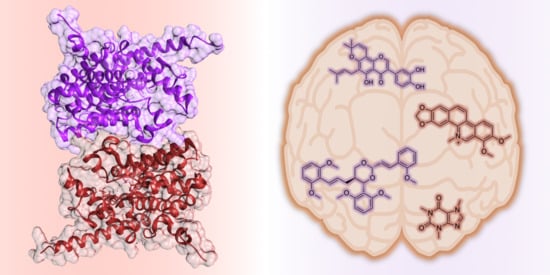
A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration
Abstract
1. Introduction
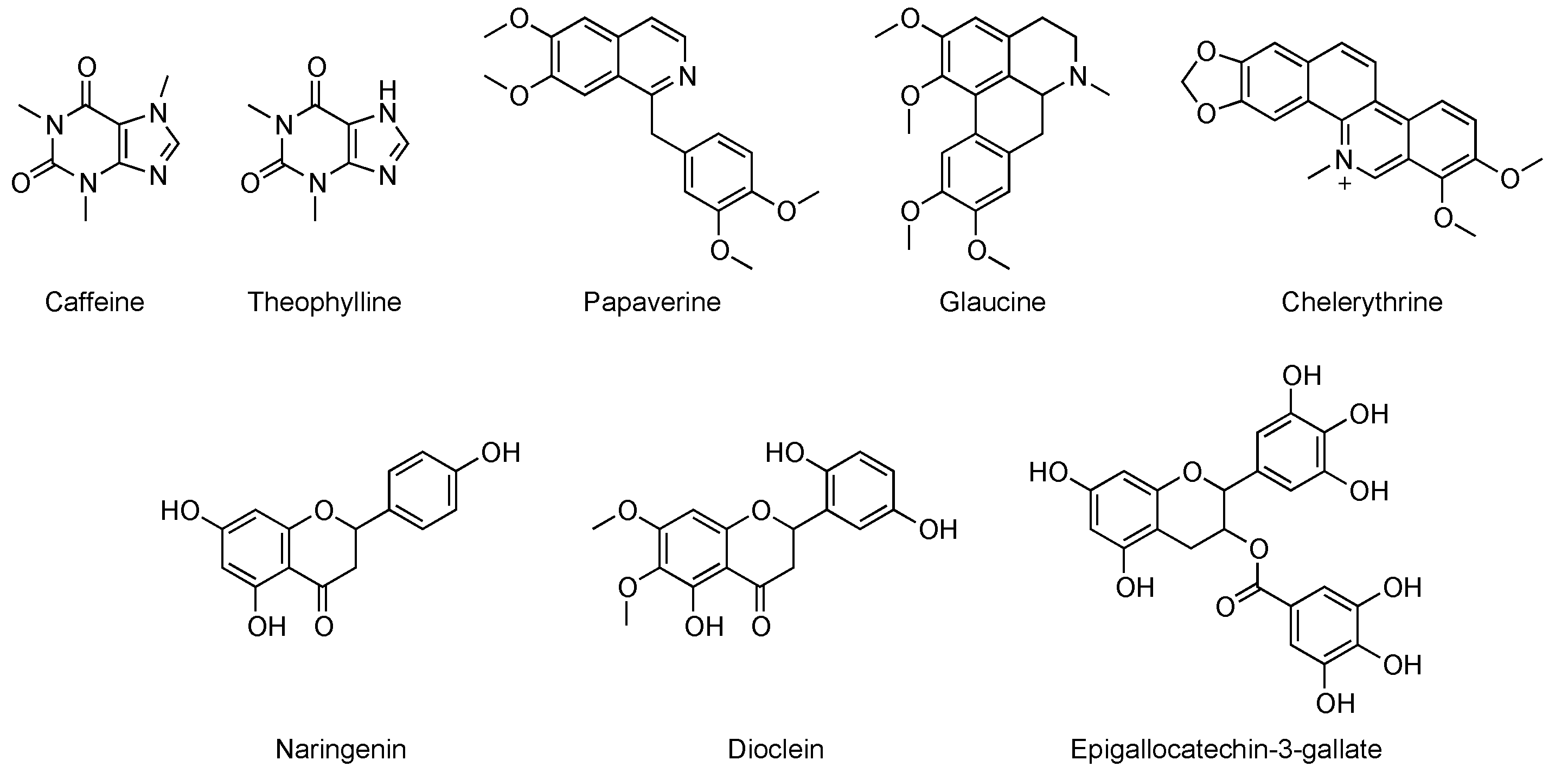
2. Chemical Classes of Natural Compounds Targeting PDEs
3. Natural and Nature-Inspired PDE9 Inhibitors
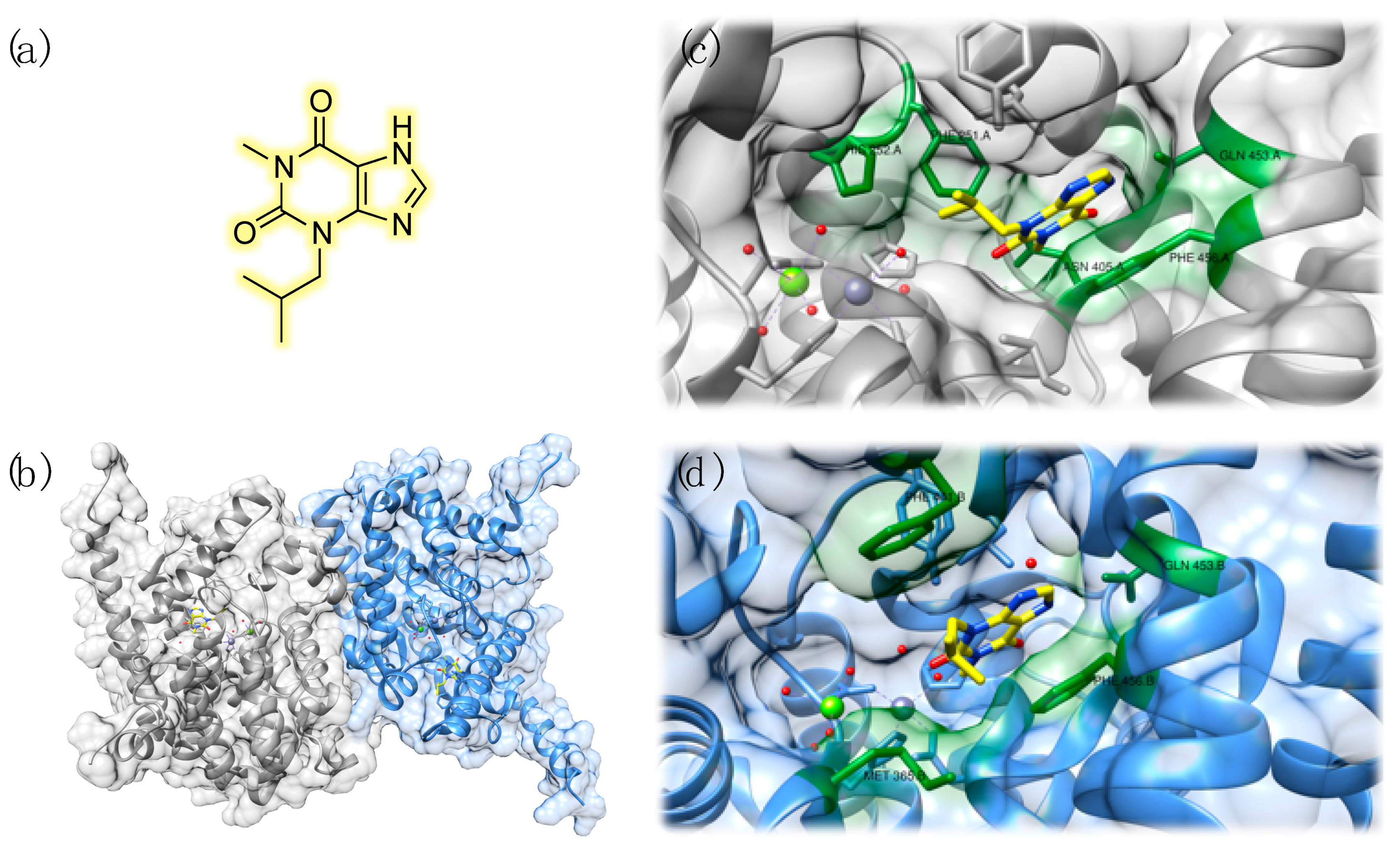
3.1. Structural Features of PDE9
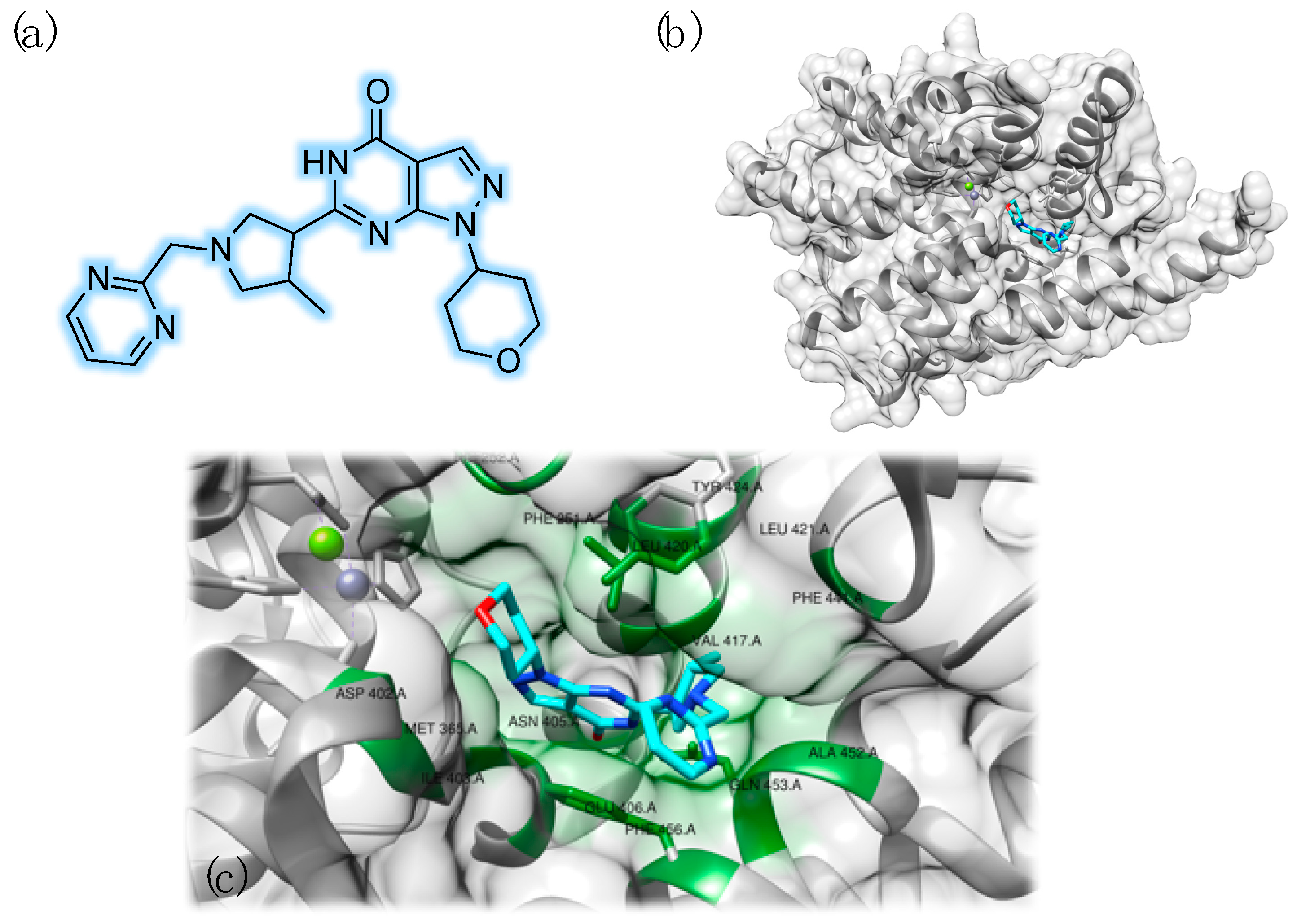
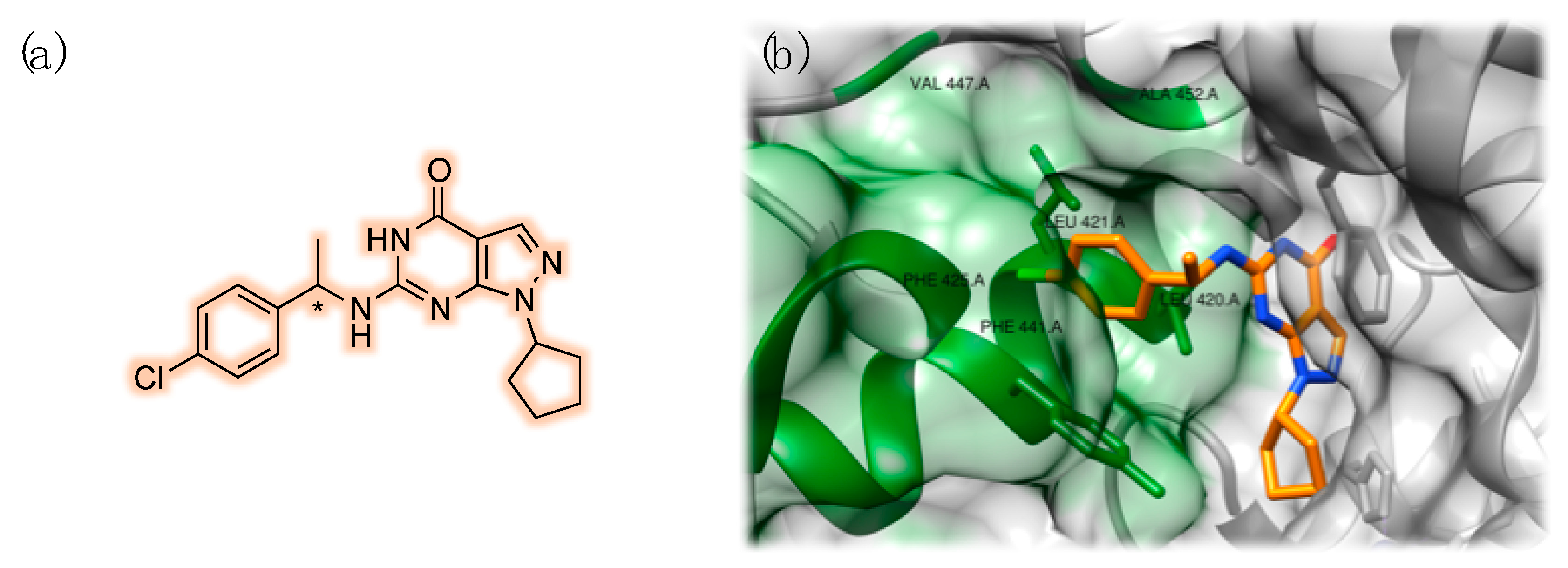
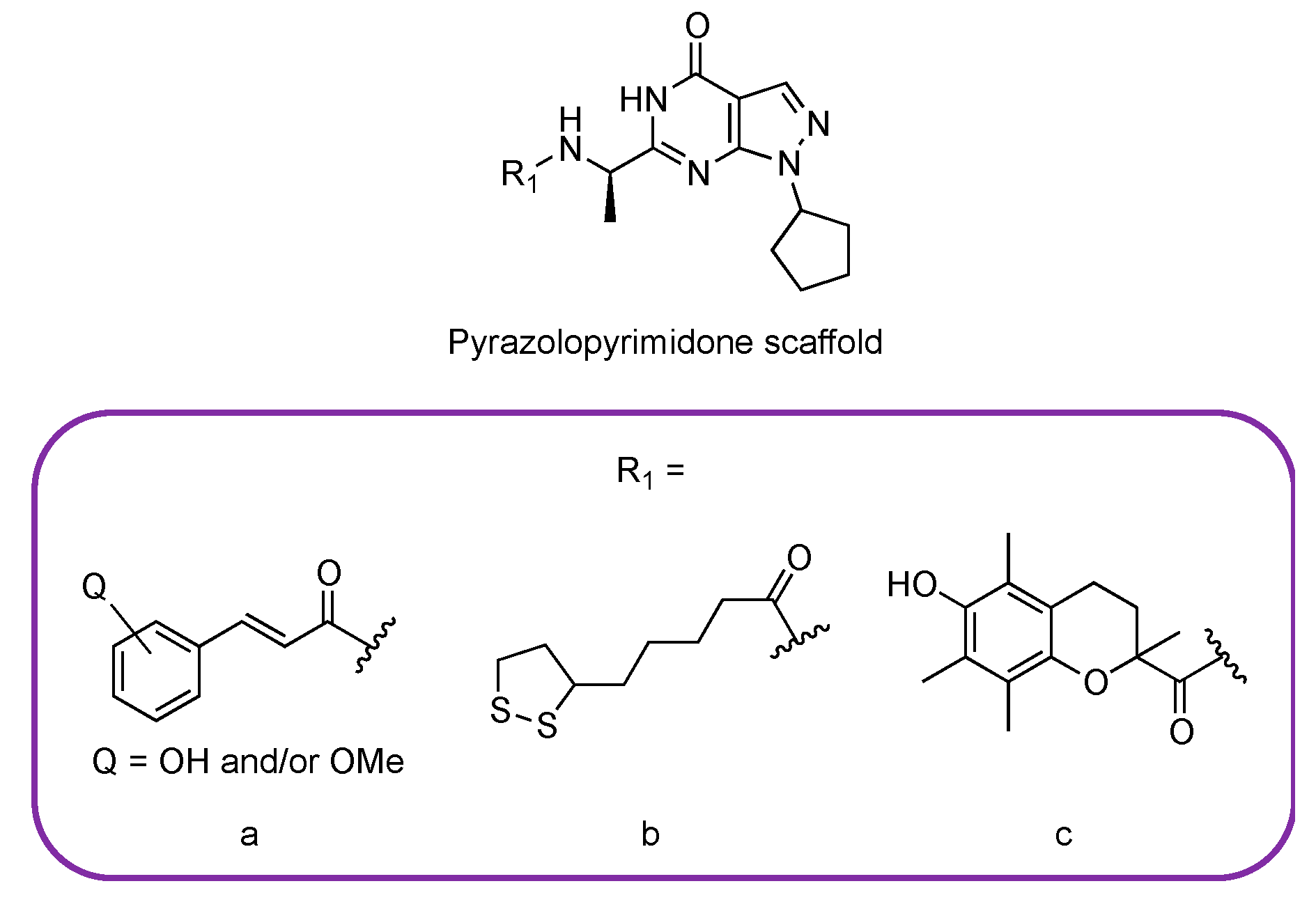
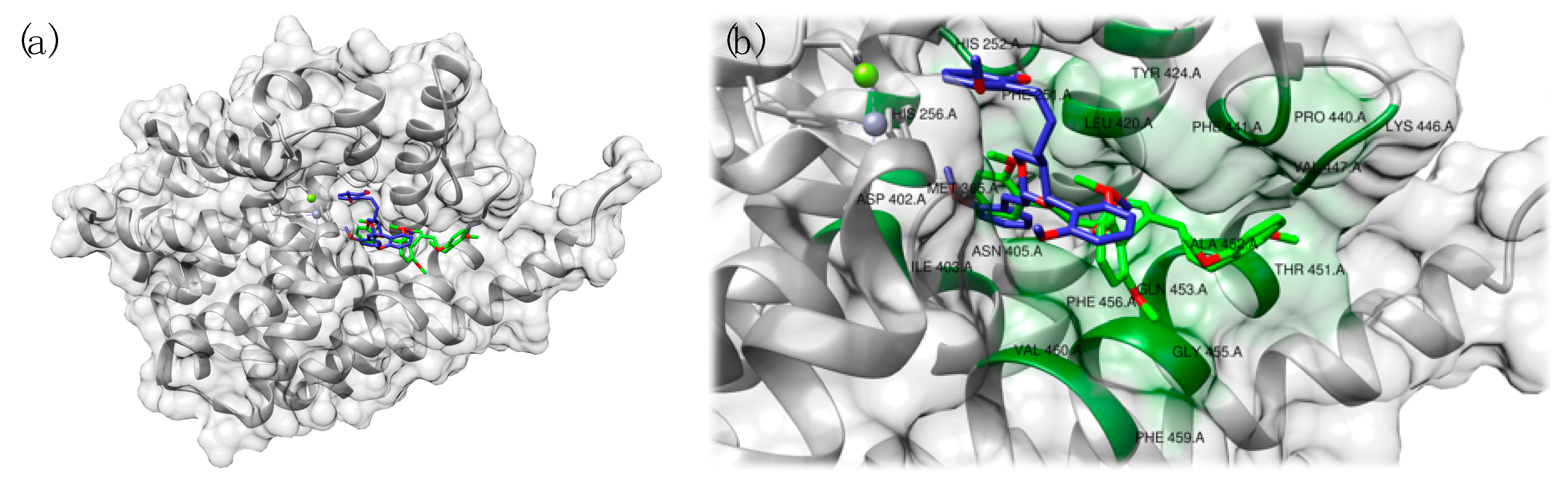
3.2. PDE9 Inhibitors Undergoing Clinical Studies
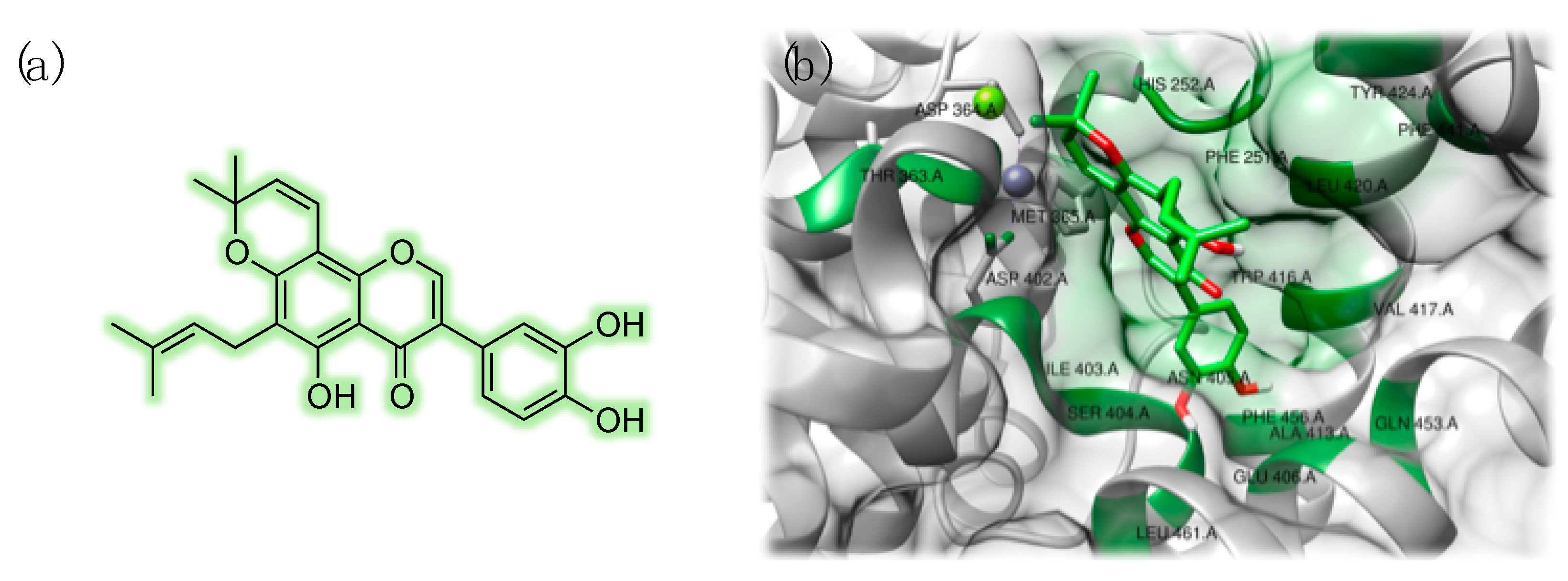
3.3. Studies on Natural and Nature-Inspired Compounds Targeting PDE9
4. Conclusions: The Perspective of the Medicinal Chemist
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lane, C.A.; Hardy, J.; Schott, J.M. Alzheimer’s disease. Eur. J. Neurol. 2018, 25, 59–70. [Google Scholar] [CrossRef]
- Hane, F.T.; Robinson, M.; Lee, B.Y.; Bai, O.; Leonenko, Z.; Albert, M.S. Recent Progress in Alzheimer’s Disease Research, Part 3: Diagnosis and Treatment. J. Alzheimer’s Dis. 2017, 57, 645–665. [Google Scholar] [CrossRef]
- Prickaerts, J.; Heckman, P.R.A.; Blokland, A. Investigational phosphodiesterase inhibitors in phase I and phase II clinical trials for Alzheimer’s disease. Expert Opin. Investig. Drugs 2017, 26, 1033–1048. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, V.; Singh, V.P.; Kaundal, M.; Gupta, M.K.; Bariwal, J.; Deshmukh, R. Herbs to curb cyclic nucleotide phosphodiesterase and their potential role in Alzheimer’s disease. Mech. Ageing Dev. 2015, 149, 75–87. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Therapeutic Potential of Phosphodiesterase (PDE) Inhibitors Against Neurodegeneration: The Perspective of the Medicinal Chemist. ACS Chem. Neurosci. 2020, 11, 1726–1739. [Google Scholar] [CrossRef]
- Bender, A.T.; Beavo, J.A. Cyclic nucleotide phosphodiesterases: Molecular regulation to clinical use. Pharmacol. Rev. 2006, 58, 488–520. [Google Scholar] [CrossRef]
- Ribaudo, G.; Pagano, M.A.; Bova, S.; Zagotto, G. New Therapeutic Applications of Phosphodiesterase 5 Inhibitors (PDE5-Is). Curr. Med. Chem. 2016, 23, 1239–1249. [Google Scholar] [CrossRef]
- Rahimi, R.; Ghiasi, S.; Azimi, H.; Fakhari, S.; Abdollahi, M. A review of the herbal phosphodiesterase inhibitors; future perspective of new drugs. Cytokine 2010, 49, 123–129. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G. Natural Compounds Promoting Weight Loss: Mechanistic Insights from the Point of View of the Medicinal Chemist. Nat. Prod. J. 2019, 9, 78–85. [Google Scholar] [CrossRef]
- Kraus, M.M.; Prast, H. The nitric oxide system modulates the in vivo release of acetylcholine in the nucleus accumbens induced by stimulation of the hippocampal fornix/fimbria-projection. Eur. J. Neurosci. 2001, 14, 1105–1112. [Google Scholar] [CrossRef]
- Song, R.S.; Tolentino, R.; Sobie, E.A.; Neves-Zaph, S.R. Cross-regulation of Phosphodiesterase 1 and Phosphodiesterase 2 Activities Controls Dopamine-mediated Striatal α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Trafficking. J. Biol. Chem. 2016, 291, 23257–23267. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Kim, J.-E.; Lee, R.; Malberg, J.E.; Chen, J.; Steffen, C.; Zhang, Y.-J.; Nestler, E.J.; Duman, R.S. Regulation of neurogenesis in adult mouse hippocampus by cAMP and the cAMP response element-binding protein. J. Neurosci. 2002, 22, 3673–3682. [Google Scholar] [CrossRef] [PubMed]
- Argyrousi, E.K.; Heckman, P.R.A.; Prickaerts, J. Role of cyclic nucleotides and their downstream signaling cascades in memory function: Being at the right time at the right spot. Neurosci. Biobehav. Rev. 2020, 113, 12–38. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Irisarri, E.; Markerink-Van Ittersum, M.; Mengod, G.; de Vente, J. Expression of the cGMP-specific phosphodiesterases 2 and 9 in normal and Alzheimer’s disease human brains. Eur. J. Neurosci. 2007, 25, 3332–3338. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, C.S.; Chen, M.L.; Lynex, C.N.; Goh, D.L.M.; Brenner, S.; Tay, S.K.H. Changes in PDE4D isoforms in the hippocampus of a patient with advanced Alzheimer disease. Arch. Neurol. 2007, 64, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, A.; Gil-Bea, F.; García-Barroso, C.; Cedazo-Minguez, Á.; Ramírez, M.J.; Franco, R.; García-Osta, A.; Oyarzabal, J.; Cuadrado-Tejedor, M. Decreased levels of guanosine 3’, 5’-monophosphate (cGMP) in cerebrospinal fluid (CSF) are associated with cognitive decline and amyloid pathology in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2015, 41, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Torres, S.; Cortés, R.; Tolnay, M.; Probst, A.; Palacios, J.; Mengod, G. Alterations on phosphodiesterase type 7 and 8 isozyme mRNA expression in Alzheimer’s disease brains examined by in situ hybridization. Exp. Neurol. 2003, 182, 322–334. [Google Scholar] [CrossRef]
- Rosenbrock, H.; Giovannini, R.; Schänzle, G.; Koros, E.; Runge, F.; Fuchs, H.; Marti, A.; Reymann, K.G.; Schröder, U.H.; Fedele, E.; et al. The Novel Phosphodiesterase 9A Inhibitor BI 409306 Increases Cyclic Guanosine Monophosphate Levels in the Brain, Promotes Synaptic Plasticity, and Enhances Memory Function in Rodents. J. Pharmacol. Exp. Ther. 2019, 371, 633–641. [Google Scholar] [CrossRef]
- Fisher, D.A.; Smith, J.F.; Pillar, J.S.; St Denis, S.H.; Cheng, J.B. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J. Biol. Chem. 1998, 273, 15559–15564. [Google Scholar] [CrossRef]
- Huai, Q.; Wang, H.; Zhang, W.; Colman, R.W.; Robinson, H.; Ke, H. Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor 3-isobutyl-1-methylxanthine binding. Proc. Natl. Acad. Sci. USA 2004, 101, 9624–9629. [Google Scholar] [CrossRef]
- Lakics, V.; Karran, E.H.; Boess, F.G. Quantitative comparison of phosphodiesterase mRNA distribution in human brain and peripheral tissues. Neuropharmacology 2010, 59, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.S.; Klett, J.; Pilarzyk, K.; Lee, D.I.; Kass, D.; Menniti, F.S.; Kelly, M.P. Identification of new PDE9A isoforms and how their expression and subcellular compartmentalization in the brain change across the life span. Neurobiol. Aging 2018, 65, 217–234. [Google Scholar] [CrossRef] [PubMed]
- Abusnina, A.; Lugnier, C. Therapeutic potentials of natural compounds acting on cyclic nucleotide phosphodiesterase families. Cell. Signal. 2017, 39, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Zanforlin, E.; Zagotto, G.; Ribaudo, G. An Overview of New Possible Treatments of Alzheimer’s Disease, Based on Natural Products and Semi-Synthetic Compounds. Curr. Med. Chem. 2017, 24, 3749–3773. [Google Scholar] [CrossRef]
- Zanforlin, E.; Zagotto, G.; Ribaudo, G. The Medicinal Chemistry of Natural and Semisynthetic Compounds against Parkinson’s and Huntington’s Diseases. ACS Chem. Neurosci. 2017, 8, 2356–2368. [Google Scholar] [CrossRef]
- Temkitthawon, P.; Viyoch, J.; Limpeanchob, N.; Pongamornkul, W.; Sirikul, C.; Kumpila, A.; Suwanborirux, K.; Ingkaninan, K. Screening for phosphodiesterase inhibitory activity of Thai medicinal plants. J. Ethnopharmacol. 2008, 119, 214–217. [Google Scholar] [CrossRef]
- Ribaudo, G.; Zanforlin, E.; Canton, M.; Bova, S.; Zagotto, G. Preliminary studies of berberine and its semi-synthetic derivatives as a promising class of multi-target anti-parkinson agents. Nat. Prod. Res. 2018, 32, 1395–1401. [Google Scholar] [CrossRef]
- Pavan, V.; Mucignat-Caretta, C.; Redaelli, M.; Ribaudo, G.; Zagotto, G. The Old Made New: Natural Compounds against Erectile Dysfunction. Arch. Pharm. 2015, 348, 607–614. [Google Scholar] [CrossRef]
- Reneerkens, O.A.H.; Rutten, K.; Steinbusch, H.W.M.; Blokland, A.; Prickaerts, J. Selective phosphodiesterase inhibitors: A promising target for cognition enhancement. Psychopharmacology 2009, 202, 419–443. [Google Scholar] [CrossRef]
- Lugnier, C.; Schini, V.B. Characterization of cyclic nucleotide phosphodiesterases from cultured bovine aortic endothelial cells. Biochem. Pharmacol. 1990, 39, 75–84. [Google Scholar] [CrossRef]
- Ribaudo, G.; Pagano, M.A.; Pavan, V.; Redaelli, M.; Zorzan, M.; Pezzani, R.; Mucignat-Caretta, C.; Vendrame, T.; Bova, S.; Zagotto, G. Semi-synthetic derivatives of natural isoflavones from Maclura pomifera as a novel class of PDE-5A inhibitors. Fitoterapia 2015, 105, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Vendrame, T.; Bova, S. Isoflavones from Maclura pomifera: Structural elucidation and in silico evaluation of their interaction with PDE5. Nat. Prod. Res. 2017, 31, 1988–1994. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Ongaro, A.; Zagotto, G. 5-Hydroxy-3-(4-hydroxyphenyl)-8,8-dimethyl-6-(3-methylbut-2-enyl)pyrano[2,3-h]chromen-4-one. Molbank 2018, 2018, M1004. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Ongaro, A.; Zagotto, G.; Memo, M.; Ribaudo, G. 2-(3,4-Dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol. Molbank 2020, 2020, M1144. [Google Scholar] [CrossRef]
- Orallo, F.; Alvarez, E.; Basaran, H.; Lugnier, C. Comparative study of the vasorelaxant activity, superoxide-scavenging ability and cyclic nucleotide phosphodiesterase-inhibitory effects of hesperetin and hesperidin. Naunyn Schmiedebergs Arch. Pharmacol. 2004, 370, 452–463. [Google Scholar] [CrossRef]
- Orallo, F.; Camiña, M.; Alvarez, E.; Basaran, H.; Lugnier, C. Implication of cyclic nucleotide phosphodiesterase inhibition in the vasorelaxant activity of the citrus-fruits flavonoid (+/-)-naringenin. Planta Med. 2005, 71, 99–107. [Google Scholar] [CrossRef]
- Gonçalves, R.L.; Lugnier, C.; Keravis, T.; Lopes, M.J.; Fantini, F.A.; Schmitt, M.; Cortes, S.F.; Lemos, V.S. The flavonoid dioclein is a selective inhibitor of cyclic nucleotide phosphodiesterase type 1 (PDE1) and a cGMP-dependent protein kinase (PKG) vasorelaxant in human vascular tissue. Eur. J. Pharmacol. 2009, 620, 78–83. [Google Scholar] [CrossRef]
- Alvarez, E.; Campos-Toimil, M.; Justiniano-Basaran, H.; Lugnier, C.; Orallo, F. Study of the mechanisms involved in the vasorelaxation induced by (-)-epigallocatechin-3-gallate in rat aorta. Br. J. Pharmacol. 2006, 147, 269–280. [Google Scholar] [CrossRef]
- Hutson, P.H.; Finger, E.N.; Magliaro, B.C.; Smith, S.M.; Converso, A.; Sanderson, P.E.; Mullins, D.; Hyde, L.A.; Eschle, B.K.; Turnbull, Z.; et al. The selective phosphodiesterase 9 (PDE9) inhibitor PF-04447943 (6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) enhances synaptic plasticity and cognitive function in rodents. Neuropharmacology 2011, 61, 665–676. [Google Scholar]
- Hershey, L.A.; Coleman-Jackson, R. Pharmacological Management of Dementia with Lewy Bodies. Drugs Aging 2019, 36, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Oselladore, E.; Ongaro, A.; Zagotto, G.; Memo, M.; Ribaudo, G.; Gianoncelli, A. Combinatorial library generation, molecular docking and molecular dynamics simulations for enhancing the isoflavone scaffold in phosphodiesterase inhibition. New J. Chem. 2020, 44, 19472–19488. [Google Scholar] [CrossRef]
- Cameron, R.T.; Coleman, R.G.; Day, J.P.; Yalla, K.C.; Houslay, M.D.; Adams, D.R.; Shoichet, B.K.; Baillie, G.S. Chemical informatics uncovers a new role for moexipril as a novel inhibitor of cAMP phosphodiesterase-4 (PDE4). Biochem. Pharmacol. 2013, 85, 1297–1305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Singh, N.; Saravanan, P.; Thakur, M.S.; Patra, S. Development of Xanthine Based Inhibitors Targeting Phosphodiesterase 9A. Lett. Drug Des. Discov. 2017, 14, 1122–1137. [Google Scholar] [CrossRef]
- Huang, M.; Shao, Y.; Hou, J.; Cui, W.; Liang, B.; Huang, Y.; Li, Z.; Wu, Y.; Zhu, X.; Liu, P.; et al. Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor. Mol. Pharmacol. 2015, 88, 836–845. [Google Scholar] [CrossRef]
- Maurice, D.H.; Ke, H.; Ahmad, F.; Wang, Y.; Chung, J.; Manganiello, V.C. Advances in targeting cyclic nucleotide phosphodiesterases. Nat. Rev. Drug Discov. 2014, 13, 290–314. [Google Scholar] [CrossRef]
- van der Staay, F.J.; Rutten, K.; Bärfacker, L.; Devry, J.; Erb, C.; Heckroth, H.; Karthaus, D.; Tersteegen, A.; van Kampen, M.; Blokland, A.; et al. The novel selective PDE9 inhibitor BAY 73-6691 improves learning and memory in rodents. Neuropharmacology 2008, 55, 908–918. [Google Scholar] [CrossRef]
- Li, J.; Liu, C.-N.; Wei, N.; Li, X.-D.; Liu, Y.-Y.; Yang, R.; Jia, Y.-J. Protective effects of BAY 73-6691, a selective inhibitor of phosphodiesterase 9, on amyloid-β peptides-induced oxidative stress in in-vivo and in-vitro models of Alzheimer’s disease. Brain Res. 2016, 1642, 327–335. [Google Scholar] [CrossRef]
- Schwam, E.M.; Nicholas, T.; Chew, R.; Billing, C.B.; Davidson, W.; Ambrose, D.; Altstiel, L.D. A multicenter, double-blind, placebo-controlled trial of the PDE9A inhibitor, PF-04447943, in Alzheimer’s disease. Curr. Alzheimer Res. 2014, 11, 413–421. [Google Scholar] [CrossRef]
- Moschetti, V.; Boland, K.; Feifel, U.; Hoch, A.; Zimdahl-Gelling, H.; Sand, M. First-in-human study assessing safety, tolerability and pharmacokinetics of BI 409306, a selective phosphodiesterase 9A inhibitor, in healthy males. Br. J. Clin. Pharmacol. 2016, 82, 1315–1324. [Google Scholar] [CrossRef]
- Höllerhage, M.; Moebius, C.; Melms, J.; Chiu, W.-H.; Goebel, J.N.; Chakroun, T.; Koeglsperger, T.; Oertel, W.H.; Rösler, T.W.; Bickle, M.; et al. Protective efficacy of phosphodiesterase-1 inhibition against alpha-synuclein toxicity revealed by compound screening in LUHMES cells. Sci. Rep. 2017, 7, 11469. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Kotani, S.; Watanabe, N.; Fukushima, T. PRECLINICAL CHARACTERIZATION OF E2027, A NOVEL PHOSPHODIESTERASE 9 INHIBITOR. Alzheimer’s Dement. 2017, 13, P946. [Google Scholar] [CrossRef]
- Lee, G.; Cummings, J.; Decourt, B.; Leverenz, J.B.; Sabbagh, M.N. Clinical drug development for dementia with Lewy bodies: Past and present. Expert Opin. Investig. Drugs 2019, 28, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A.; Ribaudo, G. Natural phosphodiesterase 5 (PDE5) inhibitors: A computational approach. Nat. Prod. Res. 2019, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Hou, J.; Shao, Y.-X.; Wu, P.-Y.; Huang, M.; Zhu, X.; Cai, Y.; Li, Z.; Xu, J.; Liu, P.; et al. Structure-based discovery of highly selective phosphodiesterase-9A inhibitors and implications for inhibitor design. J. Med. Chem. 2012, 55, 8549–8558. [Google Scholar] [CrossRef]
- Shao, Y.; Huang, M.; Cui, W.; Feng, L.-J.; Wu, Y.; Cai, Y.; Li, Z.; Zhu, X.; Liu, P.; Wan, Y.; et al. Discovery of a phosphodiesterase 9A inhibitor as a potential hypoglycemic agent. J. Med. Chem. 2014, 57, 10304–10313. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, Q.; Wu, X.-N.; Huang, Y.-D.; Zhou, J.; Lai, Z.; Wu, Y.; Luo, H.-B. Discovery of novel PDE9A inhibitors with antioxidant activities for treatment of Alzheimer’s disease. J. Enzyme Inhib. Med. Chem. 2018, 33, 260–270. [Google Scholar] [CrossRef]
- Cheng, Z.-B.; Lu, X.; Bao, J.-M.; Han, Q.-H.; Dong, Z.; Tang, G.-H.; Gan, L.-S.; Luo, H.-B.; Yin, S. (±)-Torreyunlignans A–D, Rare 8–9′ Linked Neolignan Enantiomers as Phosphodiesterase-9A Inhibitors from Torreya yunnanensis. J. Nat. Prod. 2014, 77, 2651–2657. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Ribaudo, G.; Coghi, P.; Zanforlin, E.; Law, B.Y.K.; Wu, Y.Y.J.; Han, Y.; Qiu, A.C.; Qu, Y.Q.; Zagotto, G.; Wong, V.K.W. Semi-synthetic isoflavones as BACE-1 inhibitors against Alzheimer’s disease. Bioorg. Chem. 2019, 87, 474–483. [Google Scholar] [CrossRef]
- Chit, K.; Myint, W.; Thein, K.; Maw, W.W.; Myint, M.M.; Than, A.; Khin, M. Cyclic AMP Phosphodiesterase Inhibitory Activity and Chemical Screening of Four Medicinal Plants. Pharm. Biol. 2001, 39, 181–183. [Google Scholar] [CrossRef]
- Alcaro, S.; Bolognesi, M.L.; García-Sosa, A.T.; Rapposelli, S. Editorial: Multi-Target-Directed Ligands (MTDL) as Challenging Research Tools in Drug Discovery: From Design to Pharmacological Evaluation. Front. Chem. 2019, 7, 71. [Google Scholar] [CrossRef]
- Povolo, C.; Foschini, A.; Ribaudo, G. Optimization of the extraction of bioactive molecules from Lycium barbarum fruits and evaluation of the antioxidant activity: A combined study. Nat. Prod. Res. 2018, 33, 2694–2698. [Google Scholar] [CrossRef] [PubMed]
- Bortoli, M.; Dalla Tiezza, M.; Muraro, C.; Pavan, C.; Ribaudo, G.; Rodighiero, A.; Tubaro, C.; Zagotto, G.; Orian, L. Psychiatric Disorders and Oxidative Injury: Antioxidant Effects of Zolpidem Therapy disclosed In Silico. Comput. Struct. Biotechnol. J. 2019, 17, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Muraro, C.; Dalla Tiezza, M.; Pavan, C.; Ribaudo, G.; Zagotto, G.; Orian, L. Major Depressive Disorder and Oxidative Stress: In Silico Investigation of Fluoxetine Activity against ROS. Appl. Sci. 2019, 9, 3631. [Google Scholar] [CrossRef]
- Ribaudo, G.; Bortoli, M.; Pavan, C.; Zagotto, G.; Orian, L. Antioxidant Potential of Psychotropic Drugs: From Clinical Evidence to In Vitro and In Vivo Assessment and toward a New Challenge for in Silico Molecular Design. Antioxidants 2020, 9, 714. [Google Scholar] [CrossRef]
- García-Barroso, C.; Ugarte, A.; Martínez, M.; Rico, A.J.; Lanciego, J.L.; Franco, R.; Oyarzabal, J.; Cuadrado-Tejedor, M.; García-Osta, A. Phosphodiesterase inhibition in cognitive decline. J. Alzheimers. Dis. 2014, 42 (Suppl. S4), S561–S573. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribaudo, G.; Memo, M.; Gianoncelli, A. A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration. Pharmaceuticals 2021, 14, 58. https://doi.org/10.3390/ph14010058
Ribaudo G, Memo M, Gianoncelli A. A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration. Pharmaceuticals. 2021; 14(1):58. https://doi.org/10.3390/ph14010058
Chicago/Turabian StyleRibaudo, Giovanni, Maurizio Memo, and Alessandra Gianoncelli. 2021. "A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration" Pharmaceuticals 14, no. 1: 58. https://doi.org/10.3390/ph14010058
APA StyleRibaudo, G., Memo, M., & Gianoncelli, A. (2021). A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration. Pharmaceuticals, 14(1), 58. https://doi.org/10.3390/ph14010058