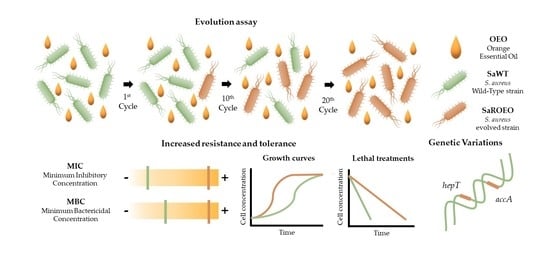
Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance
Abstract
1. Introduction
2. Results
2.1. Isolation of Resistant Strains by Evolution Assay with OEO
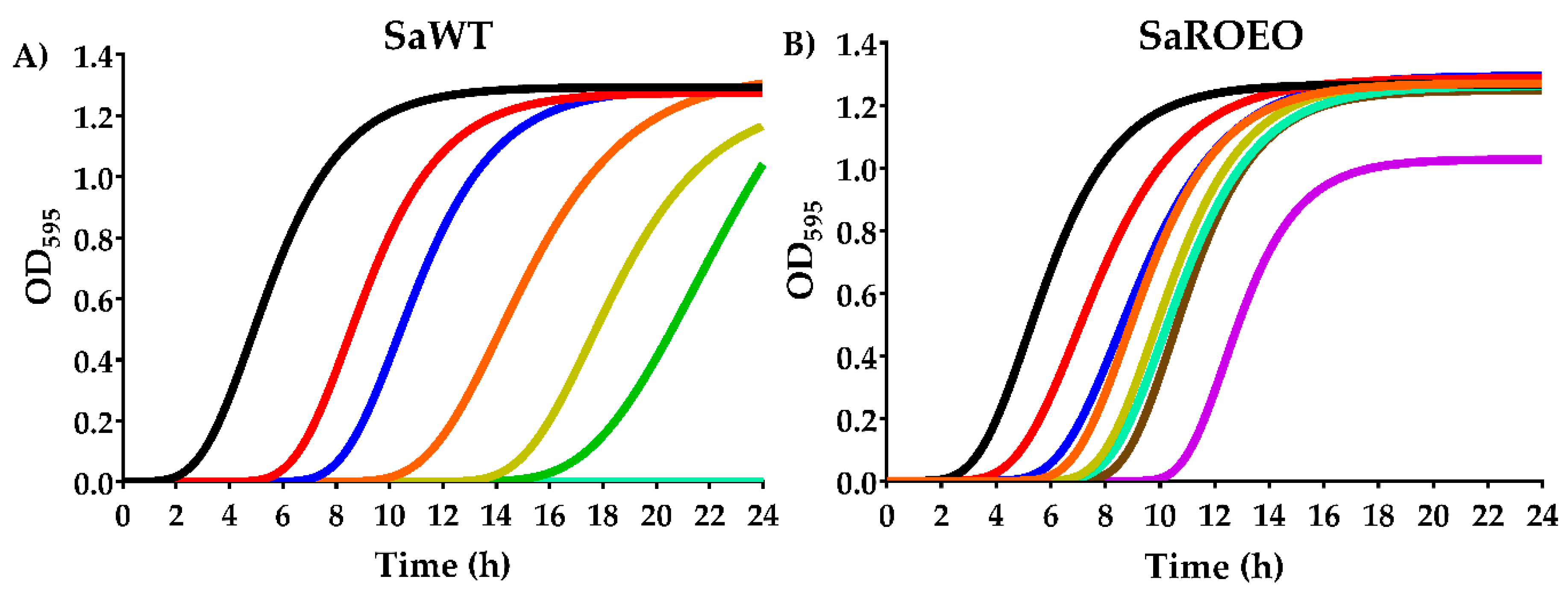
2.2. SaROEO Showed a Greater Fitness than SaWT in Presence of OEO
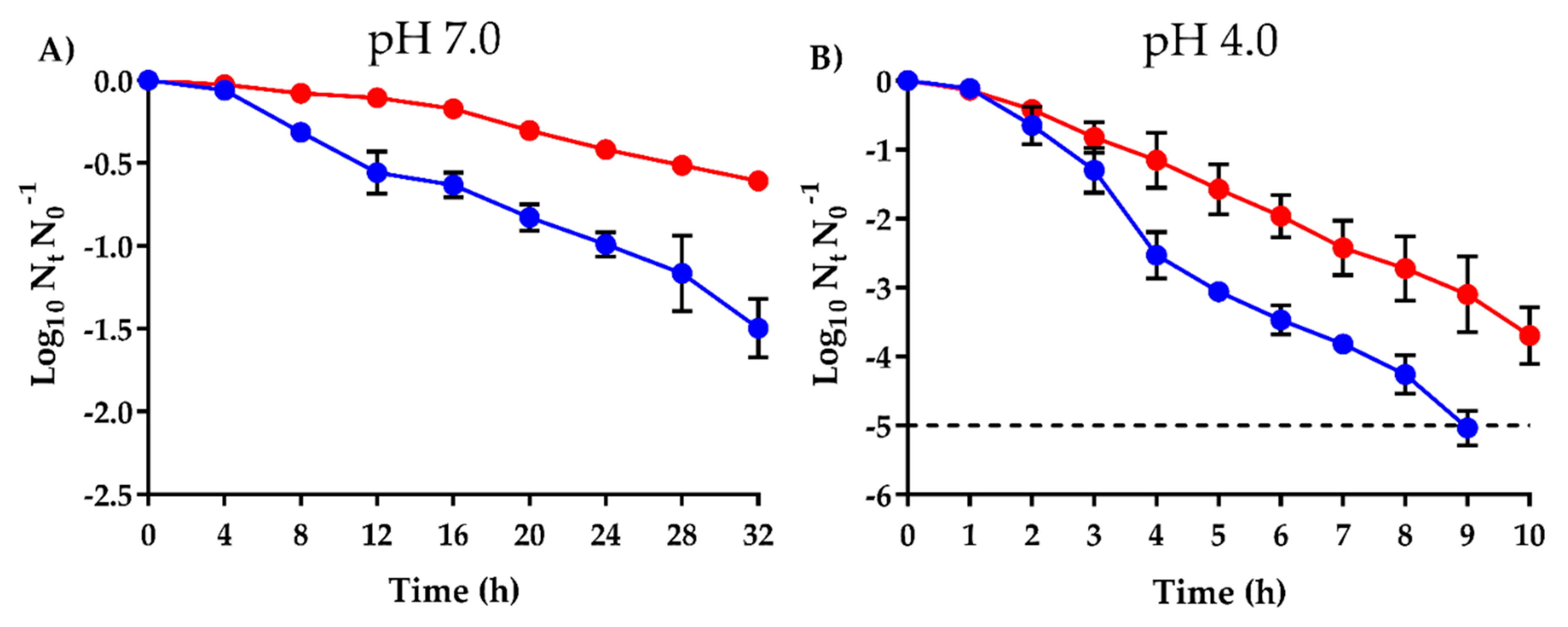
2.3. Higher Survival of SaROEO after OEO Treatments at both pH 7.0 and 4.0
2.4. SaROEO Displayed an Antibiotic Resistance Similar to SaWT
2.5. OEO does not Induce an Increased Mutagenesis
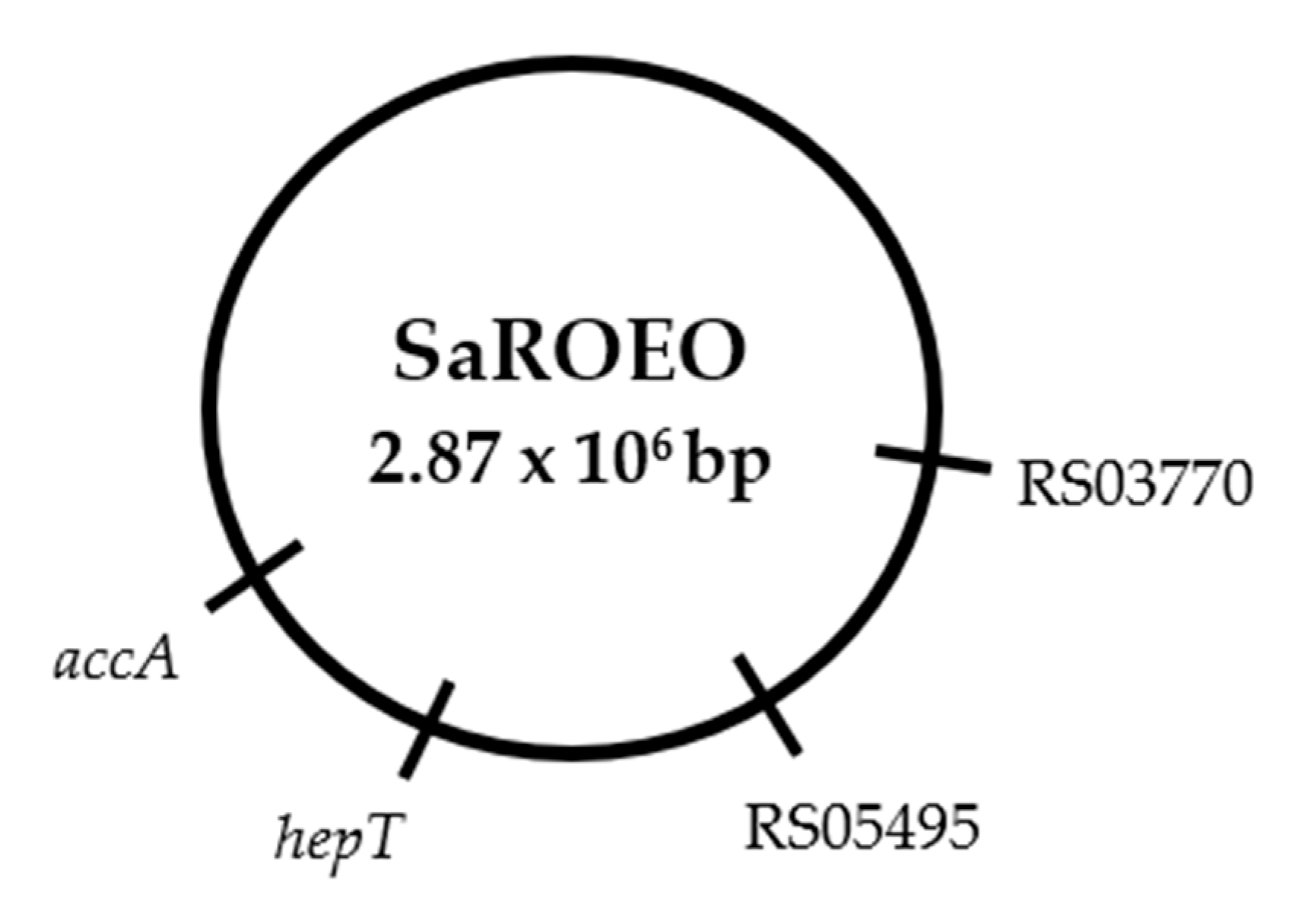
2.6. Four Missense Mutations Identified in SaROEO
- (1)
- A SNV was detected at position 993 bp in the SAUSA300_RS03770 locus resulting in a change of glutamic acid by asparagine in the enzyme allophanate hydrolase at position 331 amino acid.
- (2)
- A transversion from thymine to guanine was found at position 26 bp in SAUSA300_RS05495 locus coding a hypothetical protein in S. aureus USA300.
- (3)
- A replacement of cytosine by thymine was observed at position 272 bp in the hepT gene. This missense mutation resulted in a protein modification in the position 91 amino acid, from threonine to isoleucine, in the heptaprenyl diphosphate synthase subunit II.
- (4)
- A transition from cytosine to thymine was detected at position 481 bp in the accA gene. This mutation led to a protein change in the position 161 of proline to serine in the acetyl-CoA carboxylase carboxyl transferase subunit alpha.
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Growth Conditions
4.2. Minimum Inhibitory Concentration (MIC)
4.3. Minimum Bactericidal Concentration (MBC)
4.4. Evolution Assay of OEO
4.5. Growth Curves in Presence of OEO
4.6. Survival Curves after Lethal OEO Treatments
4.7. Antibiotic Susceptibility Test
4.8. Mutagenesis Frequency Evaluation
4.9. Whole Genome Sequencing (WGS) and Identification of Genetic Variations
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Quinto, E.J.; Caro, I.; Villalobos-Delgado, L.H.; Mateo, J.; De-Mateo-Silleras, B.; Redondo-Del-Río, M.P. Food Safety through Natural Antimicrobials. Antibiotics 2019, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Espina, L.; Berdejo, D.; Alfonso, P.; García-Gonzalo, D.; Pagán, R. Potential use of carvacrol and citral to inactivate biofilm cells and eliminate biofouling. Food Control 2017, 82, 256–265. [Google Scholar] [CrossRef]
- Carocho, M.; Barreiro, M.F.; Morales, P.; Ferreira, I.C.F.R. Adding molecules to food, pros and cons: A review on synthetic and natural food additives. Compr. Rev. Food Sci. Food Saf. 2014, 13, 377–399. [Google Scholar] [CrossRef]
- Leite de Souza, E. The effects of sublethal doses of essential oils and their constituents on antimicrobial susceptibility and antibiotic resistance among food-related bacteria: A review. Trends Food Sci. Technol. 2016, 56, 1–12. [Google Scholar] [CrossRef]
- Vasireddy, L.; Bingle, L.E.H.; Davies, M.S. Antimicrobial activity of essential oils against multidrug-resistant clinical isolates of the Burkholderia cepacia complex. PLoS ONE 2018, 13, e0201835. [Google Scholar] [CrossRef] [PubMed]
- Yap, P.S.X.; Yiap, B.C.; Ping, H.C.; Lim, S.H.E. Essential oils, a new horizon in combating bacterial antibiotic resistance. Open Microbiol. J. 2014, 8, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, C.M.S.; Ikeda, N.Y.; Miano, A.C.; Saldaña, E.; Moreno, A.M.; Stashenko, E.; Contreras-Castillo, C.J.; Da Gloria, E.M. Unraveling the selective antibacterial activity and chemical composition of citrus essential oils. Sci. Rep. 2019, 9, 17719. [Google Scholar] [CrossRef]
- Hammer, K.A.; Carson, C.F.; Riley, T.V. Frequencies of resistance to Melaleuca alternifolia (tea tree) oil and rifampicin in Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2008, 32, 170–173. [Google Scholar] [CrossRef]
- Chueca, B.; Berdejo, D.; Gomes-Neto, N.J.; Pagán, R.; García-Gonzalo, D. Emergence of hyper-resistant Escherichia coli MG1655 derivative strains after applying sub-inhibitory doses of individual constituents of essential oils. Front. Microbiol. 2016, 7, 273. [Google Scholar] [CrossRef]
- Berdejo, D.; Chueca, B.; Pagan, E.; Renzoni, A.; Kelley, W.L.; Pagan, R.; Garcia-Gonzalo, D. Sub-inhibitory doses of individual constituents of essential oils can select for Staphylococcus aureus resistant mutants. Molecules 2019, 24, 170. [Google Scholar] [CrossRef]
- Hashemi, S.M.B.; Khorram, S.B.; Sohrabi, M. Antioxidant activity of essential oils in foods. In Essential Oils in Food Processing; Hashemi, S.M.B., Khaneghah, A.M., de Souza Sant’Ana, A., Eds.; Wiley Blackwell: New Hoboken, NJ, USA, 2017; pp. 247–265. [Google Scholar]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Berdejo, D.; Merino, N.; Pagán, E.; García-Gonzalo, D.; Pagán, R. Genetic variants and phenotypic characteristics of Salmonella Typhimurium-resistant mutants after exposure to carvacrol. Microorganisms 2020, 8, 937. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Lubbe, A.; Verpoorte, R. Cultivation of medicinal and aromatic plants for specialty industrial materials. Ind. Crop. Prod. 2011, 34, 785–801. [Google Scholar] [CrossRef]
- Cherrat, L.; Espina, L.; Bakkali, M.; Pagán, R.; Laglaoui, A. Chemical composition, antioxidant and antimicrobial properties of Mentha pulegium, Lavandula stoechas and Satureja calamintha Scheele essential oils and an evaluation of their bactericidal effect in combined processes. Innov. Food Sci. Emerg. Technol. 2014, 22, 221–229. [Google Scholar] [CrossRef]
- Isman, M.B. Bridging the gap: Moving botanical insecticides from the laboratory to the farm. Ind. Crop. Prod. 2017, 110, 10–14. [Google Scholar] [CrossRef]
- McClements, D.J. Nanoemulsion-based oral delivery systems for lipophilic bioactive components: Nutraceuticals and pharmaceuticals. Ther. Deliv. 2013, 4, 841–857. [Google Scholar] [CrossRef]
- Verzera, A.; Trozzi, A.; Dugo, G.; Di Bella, G.; Cotroneo, A. Biological lemon and sweet orange essential oil composition. Flavour Fragr. J. 2004, 19, 544–548. [Google Scholar] [CrossRef]
- Bento, R.; Pagán, E.; Berdejo, D.; de Carvalho, R.J.; García-Embid, S.; Maggi, F.; Magnani, M.; de Souza, E.L.; García-Gonzalo, D.; Pagán, R. Chitosan nanoemulsions of cold-pressed orange essential oil to preserve fruit juices. Int. J. Food Microbiol. 2020, 331, 108786. [Google Scholar] [CrossRef]
- Espina, L.; Somolinos, M.; Lorán, S.; Conchello, P.; García, D.; Pagán, R. Chemical composition of commercial citrus fruit essential oils and evaluation of their antimicrobial activity acting alone or in combined processes. Food Control 2011, 22, 896–902. [Google Scholar] [CrossRef]
- O’Bryan, C.A.; Crandall, P.G.; Chalova, V.I.; Ricke, S.C. Orange essential oils antimicrobial activities against Salmonella spp. J. Food Sci. 2008, 73, M264–M267. [Google Scholar] [CrossRef] [PubMed]
- Espina, L.; Gelaw, T.K.; de Lamo-Castellvi, S.; Pagan, R.; Garcia-Gonzalo, D. Mechanism of bacterial inactivation by (+)-limonene and its potential use in food preservation combined processes. PLoS ONE 2013, 8, e56769. [Google Scholar] [CrossRef] [PubMed]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Differential mechanism of Escherichia coli inactivation by (+)-limonene as a function of cell physiological state and drug’s concentration. PLoS ONE 2014, 9, e94072. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Y.; Chen, Y.-W.; Hou, C.-Y. Antioxidant and antibacterial activity of seven predominant terpenoids. Int. J. Food Prop. 2019, 22, 230–238. [Google Scholar] [CrossRef]
- Lingan, K. A review on major constituents of various essential oils and its application. J. Transl. Med. 2018, 08, 201. [Google Scholar] [CrossRef]
- Gomes Neto, N.J.; Luz Ida, S.; Tavares, A.G.; Honorio, V.G.; Magnani, M.; de Souza, E.L. Rosmarinus officinalis L. essential oil and its majority compound 1,8-cineole at sublethal amounts induce no direct and cross protection in Staphylococcus aureus ATCC 6538. Foodborne Pathog. Dis. 2012, 9, 1071–1076. [Google Scholar] [CrossRef]
- Gomes-Neto, N.J.; Luz, I.S.; Franco, O.L.; Magnani, M.; Souza, E.L. Tolerance evaluation in Salmonella enterica serovar Typhimurium challenged with sublethal amounts of Rosmarinus officinalis L. essential oil or 1,8-cineole in meat model. Int. J. Food Sci. Tech. 2014, 49, 1912–1917. [Google Scholar] [CrossRef]
- Silva da Luz, I.; Gomes Neto, N.J.; Tavares, A.G.; Nunes, P.C.; Magnani, M.; de Souza, E.L. Lack of induction of direct protection or cross-protection in Staphylococcus aureus by sublethal concentrations of Origanum vulgare L. essential oil and carvacrol in a meat-based medium. Arch. Microbiol. 2013, 195, 587–593. [Google Scholar] [CrossRef]
- Chueca, B.; Renzoni, A.; Berdejo, D.; Pagan, R.; Kelley, W.L.; Garcia-Gonzalo, D. Whole-genome sequencing and genetic analysis reveal novel stress responses to individual constituents of essential oils in Escherichia coli. Appl. Environ. Microbiol. 2018, 84, e02538-17. [Google Scholar] [CrossRef]
- Bridges, M.A.; Mattice, M.R. Over two thousand estimations of the pH of representative foods. J. Dig. Dis. 1939, 6, 440–449. [Google Scholar] [CrossRef]
- Blázquez, J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 2003, 37, 1201–1209. [Google Scholar] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Kubicek-Sutherland, J.Z.; Lofton, H.; Vestergaard, M.; Hjort, K.; Ingmer, H.; Andersson, D.I. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J. Antimicrob. Chemother. 2017, 72, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Bouyahya, A.; Abrini, J.; Dakka, N.; Bakri, Y. Essential oils of Origanum compactum increase membrane permeability, disturb cell membrane integrity, and suppress quorum-sensing phenotype in bacteria. J. Pharm. Anal. 2019, 9, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Sánchez, D.; Cabo, M.L.; Rodríguez-Herrera, J.J. Antimicrobial activity of essential oils against Staphylococcus aureus biofilms. Food Sci. Technol. Int. 2015, 21, 559–570. [Google Scholar] [CrossRef]
- Kavanaugh, N.L.; Ribbeck, K. Selected antimicrobial essential oils eradicate Pseudomonas spp. and Staphylococcus aureus biofilms. Appl. Environ. Microbiol. 2012, 78, 4057–4061. [Google Scholar] [CrossRef]
- Ragno, R.; Papa, R.; Patsilinakos, A.; Vrenna, G.; Garzoli, S.; Tuccio, V.; Fiscarelli, E.; Selan, L.; Artini, M. Essential oils against bacterial isolates from cystic fibrosis patients by means of antimicrobial and unsupervised machine learning approaches. Sci. Rep. 2020, 10, 2653. [Google Scholar] [CrossRef]
- Fan, C.; Li, Z.; Yin, H.; Xiang, S. Structure and function of allophanate hydrolase. J. Biol. Chem. 2013, 288, 21422–21432. [Google Scholar] [CrossRef]
- Lin, Y.; St Maurice, M. The structure of allophanate hydrolase from Granulibacter bethesdensis provides insights into substrate specificity in the amidase signature family. Biochemistry (Moscow) 2013, 52, 690–700. [Google Scholar] [CrossRef]
- Shapir, N.; Sadowsky, M.J.; Wackett, L.P. Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J. Bacteriol. 2005, 187, 3731–3738. [Google Scholar] [CrossRef]
- Cheng, G.; Shapir, N.; Sadowsky, M.J.; Wackett, L.P. Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl. Environ. Microbiol. 2005, 71, 4437–4445. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Juttukonda, L.J.; Chazin, W.J.; Skaar, E.P. Acinetobacter baumannii coordinates urea metabolism with metal import to resist host-mediated metal limitation. MBio 2016, 7, e01475-16. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.; Liu, Y.-L.; Wei, H.; Liu, W.; Ko, T.-P.; Guo, R.-T.; Oldfield, E. Structure, function, and inhibition of Staphylococcus aureus heptaprenyl diphosphate synthase. ChemMedChem 2016, 11, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Chueca, B.; Pagán, R.; García-Gonzalo, D. Oxygenated monoterpenes citral and carvacrol cause oxidative damage in Escherichia coli without the involvement of tricarboxylic acid cycle and Fenton reaction. Int. J. Food Microbiol. 2014, 189, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-K.; Yusoff, K.; Thomas, W.; Akseer, R.; Alhosani, M.S.; Abushelaibi, A.; Lim, S.-H.-E.; Lai, K.-S. Lavender essential oil induces oxidative stress which modifies the bacterial membrane permeability of carbapenemase producing Klebsiella pneumoniae. Sci. Rep. 2020, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Berti, A.D.; Shukla, N.; Rottier, A.D.; McCrone, J.S.; Turner, H.M.; Monk, I.R.; Baines, S.L.; Howden, B.P.; Proctor, R.A.; Rose, W.E. Daptomycin selects for genetic and phenotypic adaptations leading to antibiotic tolerance in MRSA. J. Antimicrob. Chemother. 2018, 73, 2030–2033. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.S.; Solbiati, J.; Cronan, J.E., Jr. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 2000, 275, 28593–28598. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C.; Curtiss, R. Escherichia coli and Salmonella: Cellular and Molecular Biology, 1st ed.; American Society for Microbiology: Washington DC, USA, 1988; Volume 2, pp. 463–464. [Google Scholar]
- Freiberg, C.; Brunner, N.A.; Schiffer, G.; Lampe, T.; Pohlmann, J.; Brands, M.; Raabe, M.; Häbich, D.; Ziegelbauer, K. Identification and characterization of the first class of potent bacterial acetyl-coa carboxylase inhibitors with antibacterial activity. J. Biol. Chem. 2004, 279, 26066–26073. [Google Scholar] [CrossRef]
- Meades, G., Jr.; Henken, R.L.; Waldrop, G.L.; Rahman, M.M.; Gilman, S.D.; Kamatou, G.P.; Viljoen, A.M.; Gibbons, S. Constituents of cinnamon inhibit bacterial acetyl CoA carboxylase. Planta Med. 2010, 76, 1570–1575. [Google Scholar] [CrossRef]
- Sullivan, G.J.; Delgado, N.N.; Maharjan, R.; Cain, A.K. How antibiotics work together: Molecular mechanisms behind combination therapy. Curr. Opin. Microbiol. 2020, 57, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically Approved Standard—Tenth Edition; CLSI Doc. M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Lallemand, E.A.; Lacroix, M.Z.; Toutain, P.-L.; Boullier, S.; Ferran, A.A.; Bousquet-Melou, A. In vitro degradation of antimicrobials during use of broth microdilution method can increase the measured minimal inhibitory and minimal bactericidal concentrations. Front. Microbiol. 2016, 7, 2051. [Google Scholar] [CrossRef] [PubMed]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fourth Informational Supplement; CLSI Doc. M100-S24; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014. [Google Scholar]
- Aubry-Damon, H.; Soussy, C.J.; Courvalin, P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 1998, 42, 2590–2594. [Google Scholar] [CrossRef]
- Rosche, W.A.; Foster, P.L. Determining mutation rates in bacterial populations. Methods 2000, 20, 4–17. [Google Scholar] [CrossRef]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet (Lond. Engl.) 2006, 367, 731–739. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxf. Engl.) 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics (Oxf. Engl.) 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef]

| Strains | Carvacrol Concentration (µL/L) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SaWT | 0 | 250 | 500 | 750 | 1000 | 1250 | 1500 | 1750 | 2000 | 2500 | 3000 | 4000 | 5000 |
| SaROEO1–5 | 0 | 250 | 500 | 750 | 1000 | 1250 | 1500 | 1750 | 2000 | 2500 | 3000 | 4000 | 5000 |
| OEO (µL/L) | A (OD595) | µm (OD595/h) | λ (h) | |||
|---|---|---|---|---|---|---|
| SaWT | SaROEO | SaWT | SaROEO | SaWT | SaROEO | |
| 0 | 1.294 ± 0.012 a | 1.268 ± 0.010 A | 0.248 ± 0.019 a | 0.254 ± 0.018 A | 2.902 ± 0.228 a | 3.319 ± 0.191 A |
| 250 | 1.276 ± 0.014 a | 1.287 ± 0.013 A | 0.238 ± 0.015 a | 0.217 ± 0.014 AB | 6.468 ± 0.215 b | 4.776 ± 0.215 B* |
| 500 | 1.293 ± 0.013 a | 1.295 ± 0.019 A | 0.229 ± 0.011 a | 0.218 ± 0.015 AB | 8.239 ± 0.162 c | 6.311 ± 0.251 C* |
| 750 | 1.324 ± 0.030 ab | 1.271 ± 0.018 A | 0.181 ± 0.008 b | 0.244 ± 0.021 AB* | 11.410 ± 0.170 d | 6.876 ± 0.271D* |
| 1000 | 1.262 ± 0.032 ab | 1.280 ± 0.018 A | 0.183 ± 0.006 b | 0.251 ± 0.020 A* | 15.000 ± 0.099 e | 7.884 ± 0.243 E* |
| 1250 | 1.185 ± 0.038 b | 1.280 ± 0.017 A | 0.167 ± 0.008 b | 0.240 ± 0.017 AB* | 17.430 ± 0.149 f | 7.761 ± 0.226 F* |
| 1500 | / | 1.272 ± 0.019 A | / | 0.210 ± 0.014 AB | / | 8.062 ± 0.237 G |
| 1750 | / | 1.254 ± 0.019 AB | / | 0.237 ± 0.019 AB | / | 8.447 ± 0.236 H |
| 2000 | / | 1.246 ± 0.018 ABC | / | 0.244 ± 0.019 AB | / | 8.563 ± 0.225 I |
| 2500 | / | 1.212 ± 0.023 BC | / | 0.250 ± 0.019 A | / | 9.482 ± 0.240 J |
| 3000 | / | 1.198 ± 0.019 C | / | 0.227 ± 0.017 AB | / | 9.516 ± 0.204 K |
| 4000 | / | 1.112 ± 0.020 D | / | 0.208 ± 0.015 AB | / | 10.300 ± 0.196 L |
| 5000 | / | 1.052 ± 0.014 E | / | 0.193 ± 0.010 B | / | 10.850 ± 0.139 M |
| Antibiotics | Strains | |
|---|---|---|
| SaWT | SaROEO | |
| Tetracycline | 28.07 ± 1.11 | 28.93 ± 2.03 |
| Chloramphenicol | 22.23 ± 1.32 | 23.37 ± 1.25 |
| Nalidixic acid | 15.88 ± 1.24 | 15.45 ± 1.27 |
| Rifampicin | 30.80 ± 0.67 | 30.51 ± 0.26 |
| Norfloxacin | 11.60 ± 0.31 | 12.02 ± 0.73 |
| Novobiocin | 27.38 ± 1.24 | 28.39 ± 1.56 |
| Trimethoprim | 22.18 ± 1.05 | 20.39 ± 1.20 |
| Cephalexin | 14.45 ± 0.72 | 13.39 ± 1.23 |
| Genome Position | Gene | Locus Tag a (Old Locus Tag) | Mutation b | Change | Information |
|---|---|---|---|---|---|
| 776,659 | - | RS03770 (0702) | SNV: A993T | Glu331Asp | Allophanate hydrolase |
| 1,118,342 | - | RS05495 (1021) | SNV: T26G | Ile9Ser | Hypothetical protein (DUF2129 domain containing protein) |
| 1,526,963 | hepT | RS07410 (1359) | SNV: C272T | Thr91Ile | heptaprenyl diphosphate synthase subunit II |
| 1,808,243 | accA | RS08985 (1646) | SNV: C481T | Pro161Ser | Acetyl-CoA carboxylase carboxyl transferase subunit alpha |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berdejo, D.; Pagán, E.; Merino, N.; Pagán, R.; García-Gonzalo, D. Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance. Pharmaceuticals 2020, 13, 239. https://doi.org/10.3390/ph13090239
Berdejo D, Pagán E, Merino N, Pagán R, García-Gonzalo D. Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance. Pharmaceuticals. 2020; 13(9):239. https://doi.org/10.3390/ph13090239
Chicago/Turabian StyleBerdejo, Daniel, Elisa Pagán, Natalia Merino, Rafael Pagán, and Diego García-Gonzalo. 2020. "Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance" Pharmaceuticals 13, no. 9: 239. https://doi.org/10.3390/ph13090239
APA StyleBerdejo, D., Pagán, E., Merino, N., Pagán, R., & García-Gonzalo, D. (2020). Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance. Pharmaceuticals, 13(9), 239. https://doi.org/10.3390/ph13090239