Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer
Abstract
1. Introduction
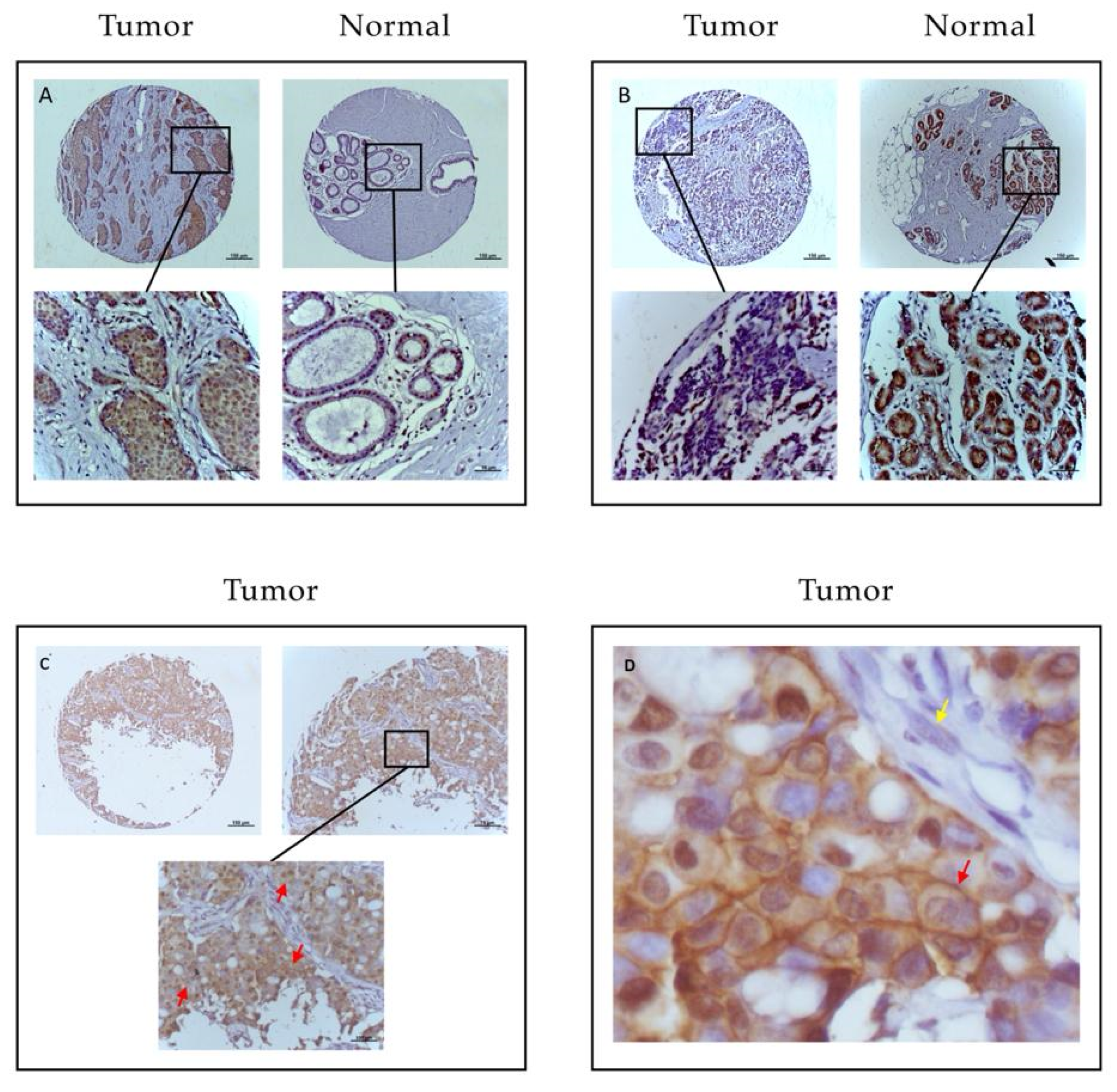
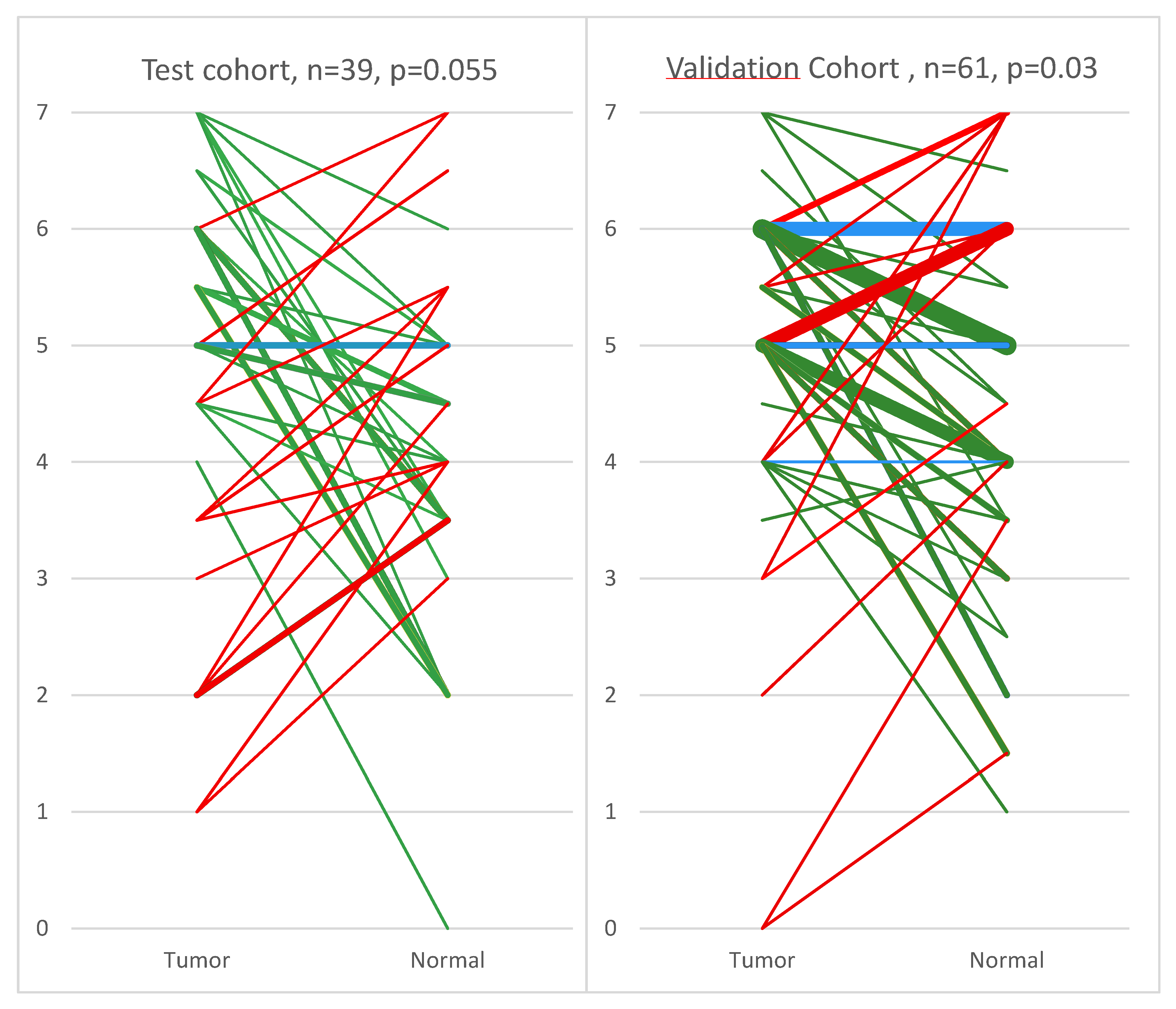
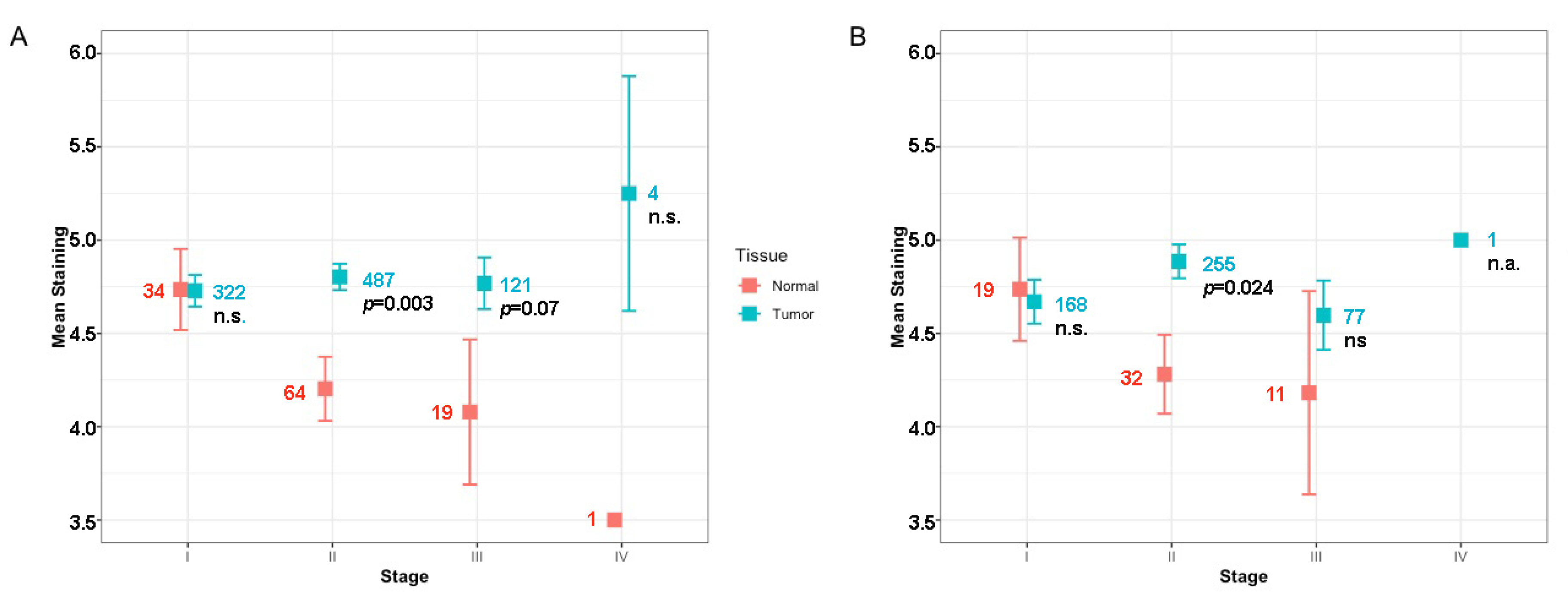
2. Results
3. Discussion
4. Materials and Methods
4.1. Patients and Tumors
4.2. Tissue Micro Array
4.3. Immunohistochemistry
4.4. Scoring Method
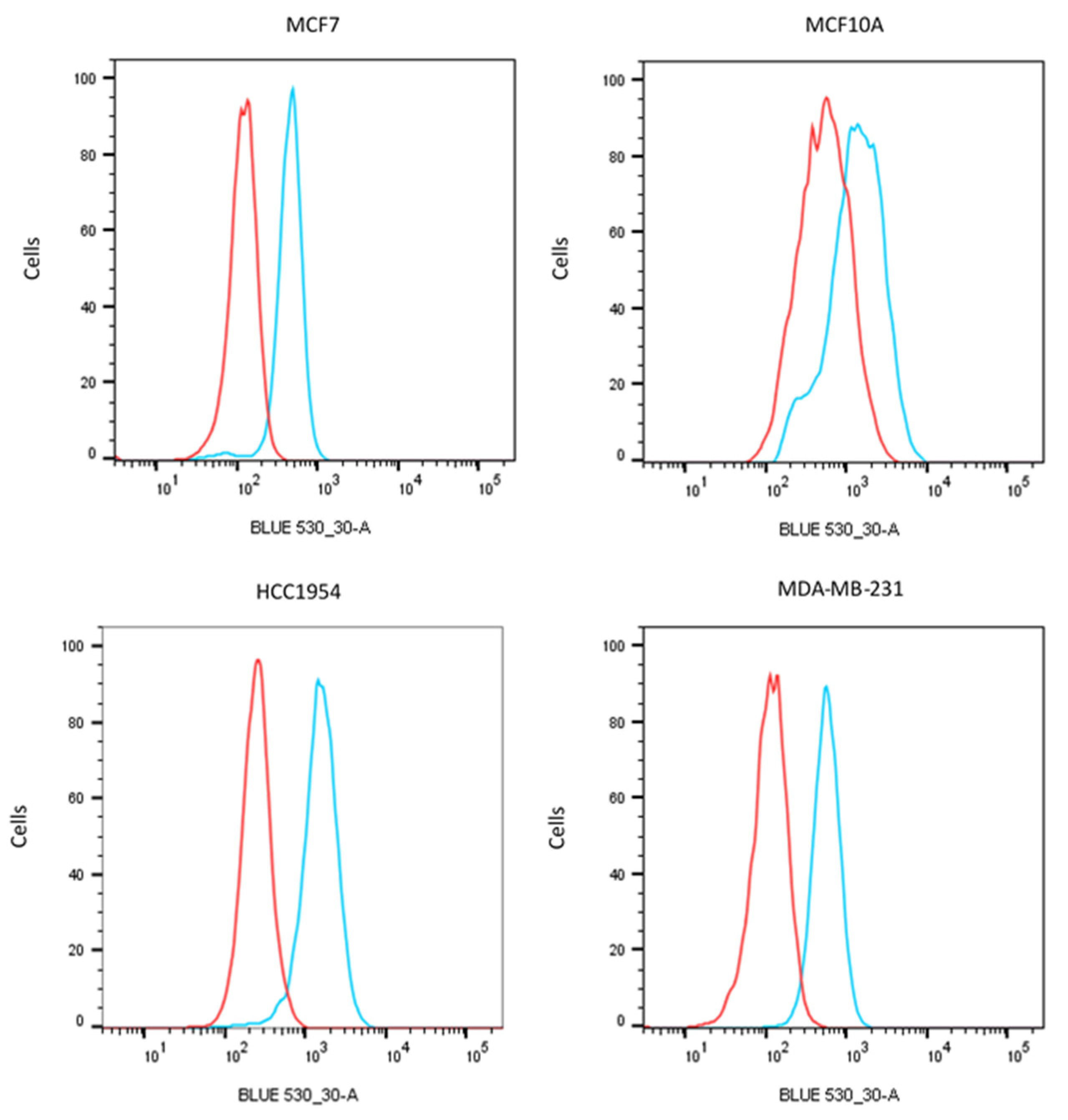
4.5. Flow Cytometry
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FIGS | Fluorescence image-guided surgery |
| PET | Positron emission tomography |
| SLM | Sentinel lymph node mapping |
| IHC | Immunohistochemistry |
| TMA | Tissue microarray |
| FFPE | Formalin-fixed paraffin-embedded |
| NIRF | Near infrared fluorescence |
| ICG | Indocyanine green |
| TBR | Tissue background ratio |
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Pleijhuis, R.G.; Graafland, M.; de Vries, J.; Bart, J.; de Jong, J.S.; van Dam, G.M. Obtaining adequate surgical margins in breast-conserving therapy for patients with early-stage breast cancer: Current modalities and future directions. Ann. Surg. Oncol. 2009, 16, 2717–2730. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, L.Z.; Brock, J.E.; Chen, Y.H.; Truong, L.; Russo, A.L.; Arvold, N.D.; Harris, J.R. Invasive lobular carcinoma of the breast: Local recurrence after breast-conserving therapy by subtype approximation and surgical margin. Breast Cancer Res. Treat. 2015, 149, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Singletary, S.E. Surgical margins in patients with early-stage breast cancer treated with breast conservation therapy. Am. J. Surg. 2002, 184, 383–393. [Google Scholar] [CrossRef]
- Zysk, A.M.; Chen, K.; Gabrielson, E.; Tafra, L.; May Gonzalez, E.A.; Canner, J.K.; Schneider, E.B.; Cittadine, A.J.; Scott Carney, P.; Boppart, S.A.; et al. Intraoperative Assessment of Final Margins with a Handheld Optical Imaging Probe During Breast-Conserving Surgery May Reduce the Reoperation Rate: Results of a Multicenter Study. Ann. Surg. Oncol. 2015, 22, 3356–3362. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, M.C.; de Geus, S.W.; Prevoo, H.A.; Hawinkels, L.J.; van de Velde, C.J.; Kuppen, P.J.; Vahrmeijer, A.L.; Sier, C.F. Selecting Targets for Tumor Imaging: An Overview of Cancer-Associated Membrane Proteins. Biomark. Cancer 2016, 8, 119–133. [Google Scholar] [CrossRef]
- Stammes, M.A.; Prevoo, H.A.; Ter Horst, M.C.; Groot, S.A.; Van de Velde, C.J.; Chan, A.B.; de Geus-Oei, L.F.; Kuppen, P.J.; Vahrmeijer, A.L.; Pasquale, E.B.; et al. Evaluation of EphA2 and EphB4 as Targets for Image-Guided Colorectal Cancer Surgery. Int. J. Mol. Sci. 2017, 18, 307. [Google Scholar] [CrossRef]
- Hernot, S.; van Manen, L.; Debie, P.; Mieog, J.S.D.; Vahrmeijer, A.L. Latest developments in molecular tracers for fluorescence image-guided cancer surgery. Lancet Oncol. 2019, 20, e354–e367. [Google Scholar] [CrossRef]
- Nikolova, Z.; Djonov, V.; Zuercher, G.; Andres, A.C.; Ziemiecki, A. Cell-type specific and estrogen dependent expression of the receptor tyrosine kinase EphB4 and its ligand ephrin-B2 during mammary gland morphogenesis. J. Cell Sci. 1998, 111, 2741–2751. [Google Scholar]
- Kadife, E.; Ware, T.M.B.; Luwor, R.B.; Chan, S.T.F.; Nurgali, K.; Senior, P.V. Effects of EphB4 receptor expression on colorectal cancer cells, tumor growth, vascularization and composition. Acta Oncol. (Stockh. Swed.) 2018, 57, 1043–1056. [Google Scholar] [CrossRef]
- Lv, J.; Xia, Q.; Wang, J.; Shen, Q.; Zhang, J.; Zhou, X. EphB4 promotes the proliferation, invasion, and angiogenesis of human colorectal cancer. Exp. Mol. Pathol. 2016, 100, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nikas, I.; Ryu, H.S.; Theocharis, S. Viewing the Eph receptors with a focus on breast cancer heterogeneity. Cancer Lett. 2018, 434, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Walker-Daniels, J.; Coffman, K.; Azimi, M.; Rhim, J.S.; Bostwick, D.G.; Snyder, P.; Kerns, B.J.; Waters, D.J.; Kinch, M.S. Overexpression of the EphA2 tyrosine kinase in prostate cancer. Prostate 1999, 41, 275–280. [Google Scholar] [CrossRef]
- Yin, J.; Cui, Y.; Li, L.; Ji, J.; Jiang, W.G. Overexpression of EPHB4 Is Associated with Poor Survival of Patients with Gastric Cancer. Anticancer Res. 2017, 37, 4489–4497. [Google Scholar] [PubMed]
- Noren, N.K.; Pasquale, E.B. Paradoxes of the EphB4 receptor in cancer. Cancer Res. 2007, 67, 3994–3997. [Google Scholar] [CrossRef]
- Deken, M.M.; Bos, D.L.; Tummers, W.; March, T.L.; van de Velde, C.J.H.; Rijpkema, M.; Vahrmeijer, A.L. Multimodal image-guided surgery of HER2-positive breast cancer using [(111)In]In-DTPA-trastuzumab-IRDye800CW in an orthotopic breast tumor model. EJNMMI Res. 2019, 9, 98. [Google Scholar] [CrossRef]
- van de Ven, S.; Smit, V.T.H.B.M.; Dekker, T.J.A.; Nortier, J.W.R.; Kroep, J.R. Discordances in ER, PR and HER2 receptors after neoadjuvant chemotherapy in breast cancer. Cancer Treat. Rev. 2011, 37, 422–430. [Google Scholar] [CrossRef]
- Brantley-Sieders, D.M.; Jiang, A.; Sarma, K.; Badu-Nkansah, A.; Walter, D.L.; Shyr, Y.; Chen, J. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS ONE 2011, 6, e24426. [Google Scholar] [CrossRef]
- Vaught, D.; Brantley-Sieders, D.M.; Chen, J. Eph receptors in breast cancer: Roles in tumor promotion and tumor suppression. Breast Cancer Res. 2008, 10, 217. [Google Scholar] [CrossRef]
- Wu, Q.; Suo, Z.; Risberg, B.; Karlsson, M.G.; Villman, K.; Nesland, J.M. Expression of Ephb2 and Ephb4 in breast carcinoma. Pathol. Oncol. Res. 2004, 10, 26–33. [Google Scholar] [CrossRef]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef]
- Rutkowski, R.; Mertens-Walker, I.; Lisle, J.E.; Herington, A.C.; Stephenson, S.A. Evidence for a dual function of EphB4 as tumor promoter and suppressor regulated by the absence or presence of the ephrin-B2 ligand. Int J. Cancer 2012, 131, E614–E624. [Google Scholar] [CrossRef] [PubMed]
- Boogerd, L.S.F.; Boonstra, M.C.; Prevoo, H.; Handgraaf, H.J.M.; Kuppen, P.J.K.; van de Velde, C.J.H.; Fish, A.; Cordfunke, R.A.; Valentijn, A.; Terwisscha van Scheltinga, A.G.; et al. Fluorescence-guided tumor detection with a novel anti-EpCAM targeted antibody fragment: Preclinical validation. Surg. Oncol. 2019, 28, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.P.; Wang, T.D. Targeted Optical Imaging Agents in Cancer: Focus on Clinical Applications. Contrast Media Mol. Imaging 2018, 2018, 2015237. [Google Scholar] [CrossRef] [PubMed]
- Tummers, W.S.; Farina-Sarasqueta, A.; Boonstra, M.C.; Prevoo, H.A.; Sier, C.F.; Mieog, J.S.; Morreau, J.; van Eijck, C.H.; Kuppen, P.J.; van de Velde, C.J.; et al. Selection of optimal molecular targets for tumor-specific imaging in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 56816–56828. [Google Scholar] [CrossRef]
- de Gouw, D.J.J.M.; Rijpkema, M.; de Bitter, T.J.J.; Baart, V.M.; Sier, C.F.M.; Hernot, S.; van Dam, G.M.; Nagtegaal, I.D.; Klarenbeek, B.R.; Rosman, C.; et al. Identifying Biomarkers in Lymph Node Metastases of Esophageal Adenocarcinoma for Tumor-Targeted Imaging. Mol. Diagn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Huisman, B.W.; Burggraaf, J.; Vahrmeijer, A.L.; Schoones, J.W.; Rissmann, R.A.; Sier, C.F.M.; van Poelgeest, M.I.E. Potential targets for tumor-specific imaging of vulvar squamous cell carcinoma: A systematic review of candidate biomarkers. Gynecol. Oncol. 2020, 156, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Baart, V.M.; Van Duijn, C.; van Egmond, S.; Dijkckmeester, W.A.; Jansen, J.C.; Vahrmeijer, A.L.; Sier, C.F.M.; Cohen, D. EGFR, uPAR and αvβ6 as promising targets for molecular imaging of cutaneous and mucosal squamous cell carcinoma of the head-and-neck region. Cancers 2020, 12, 1474. [Google Scholar] [CrossRef]
- Huang, G.N.; Li, M. The role of EphB4 and IGF-IR expression in breast cancer cells. Int J. Clin. Exp. Pathol 2015, 8, 5997–6004. [Google Scholar]
- Salgia, R.; Kulkarni, P.; Gill, P.S. EphB4: A promising target for upper aerodigestive malignancies. Biochim. Biophys. Acta 2018, 1869, 128–137. [Google Scholar] [CrossRef]
- Jung, S.Y.; Han, J.H.; Park, S.J.; Lee, E.G.; Kwak, J.; Kim, S.H.; Lee, M.H.; Lee, E.S.; Kang, H.S.; Lee, K.S.; et al. The Sentinel Lymph Node Biopsy Using Indocyanine Green Fluorescence Plus Radioisotope Method Compared With the Radioisotope-Only Method for Breast Cancer Patients After Neoadjuvant Chemotherapy: A Prospective, Randomized, Open-Label, Single-Center Phase 2 Trial. Ann. Surg. Oncol. 2019, 26, 2409–2416. [Google Scholar] [PubMed]
- Tagaya, N.; Yamazaki, R.; Nakagawa, A.; Abe, A.; Hamada, K.; Kubota, K.; Oyama, T. Intraoperative identification of sentinel lymph nodes by near-infrared fluorescence imaging in patients with breast cancer. Am. J. Surg. 2008, 195, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Verbeek, F.P.; Troyan, S.L.; Mieog, J.S.; Liefers, G.J.; Moffitt, L.A.; Rosenberg, M.; Hirshfield-Bartek, J.; Gioux, S.; van de Velde, C.J.; Vahrmeijer, A.L.; et al. Near-infrared fluorescence sentinel lymph node mapping in breast cancer: A multicenter experience. Breast Cancer Res. Treat. 2014, 143, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, S.; Liu, R.; Zhou, Y.; Park, R.; Naga, K.; Krasnoperov, V.; Gill, P.S.; Li, Z.; Shan, H.; et al. EphB4-targeted imaging with antibody h131, h131-F(ab’)2 and h131-Fab. Mol. Pharm. 2013, 10, 4527–4533. [Google Scholar] [CrossRef]
- Riedl, S.J.; Pasquale, E.B. Targeting the Eph System with Peptides and Peptide Conjugates. Curr. Drug Targets 2015, 16, 1031–1047. [Google Scholar] [CrossRef]
- Zhang, R.; Xiong, C.; Huang, M.; Zhou, M.; Huang, Q.; Wen, X.; Liang, D.; Li, C. Peptide-conjugated polymeric micellar nanoparticles for Dual SPECT and optical imaging of EphB4 receptors in prostate cancer xenografts. Biomaterials 2011, 32, 5872–5879. [Google Scholar] [CrossRef][Green Version]
- Dimasi, N.; Fleming, R.; Hay, C.; Woods, R.; Xu, L.; Wu, H.; Gao, C. Development of a Trispecific Antibody Designed to Simultaneously and Efficiently Target Three Different Antigens on Tumor Cells. Mol. Pharm. 2015, 12, 3490–3501. [Google Scholar] [CrossRef]
- Liu, S.; Li, D.; Park, R.; Liu, R.; Xia, Z.; Guo, J.; Krasnoperov, V.; Gill, P.S.; Li, Z.; Shan, H.; et al. PET imaging of colorectal and breast cancer by targeting EphB4 receptor with 64Cu-labeled hAb47 and hAb131 antibodies. J. Nucl Med. 2013, 54, 1094–1100. [Google Scholar] [CrossRef]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Giordano, C.; Nuzzo, S.; Ricci-Vitiani, L.; Scognamiglio, I.; Minic, Z.; Pallini, R.; et al. Targeting Ephrin Receptor Tyrosine Kinase A2 with a Selective Aptamer for Glioblastoma Stem Cells. Mol. Ther. Nucleic Acids 2020, 20, 176–185. [Google Scholar] [CrossRef]
- Pretze, M.; Neuber, C.; Kinski, E.; Belter, B.; Köckerling, M.; Caflisch, A.; Steinbach, J.; Pietzsch, J.; Mamat, C. Synthesis, radiolabelling and initial biological characterisation of (18)F-labelled xanthine derivatives for PET imaging of Eph receptors. Org. Biomol. Chem. 2020, 18, 3104–3116. [Google Scholar] [CrossRef]
- de Kruijf, E.M.; Sajet, A.; van Nes, J.G.; Natanov, R.; Putter, H.; Smit, V.T.; Liefers, G.J.; van den Elsen, P.J.; Van de Velde, C.J.; Kuppen, P.J. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Test Cohort | Validation Cohort | ||
|---|---|---|---|---|
| n (662) | % | n (667) | % | |
| Age in years | ||||
| <45 | 124 | 18.7 | 134 | 20.1 |
| 45–55 | 163 | 24.6 | 214 | 32.1 |
| 55–65 | 149 | 22.5 | 148 | 22.2 |
| >65 | 222 | 33.5 | 171 | 25.6 |
| Missing | 4 | 0.6 | 0 | 0 |
| Stage | ||||
| I | 181 | 27.3 | 252 | 3.8 |
| II | 352 | 53.2 | 317 | 47.5 |
| III | 95 | 14.4 | 58 | 8.7 |
| IV | 0 | 0 | 4 | 0.6 |
| Unknown | 34 | 5.1 | 36 | 5.4 |
| Histological type | ||||
| Ductal | 588 | 88.8 | 536 | 80.4 |
| Lobular | 62 | 9.4 | 66 | 9.9 |
| Other | 0 | 0 | 65 | 9.7 |
| Missing | 12 | 1.8 | 0 | 0 |
| Differentiation | ||||
| Good | 109 | 16.5 | 108 | 16.2 |
| Moderate | 322 | 48.6 | 275 | 41.2 |
| Poor | 221 | 33.4 | 209 | 31.3 |
| Unknown | 10 | 1.5 | 75 | 11.2 |
| Estrogen receptor | ||||
| Positive | 368 | 55.6 | 456 | 68.4 |
| Negative | 272 | 41.1 | 140 | 21.0 |
| Missing | 22 | 3.3 | 71 | 10.6 |
| Progesterone receptor | ||||
| Positive | 327 | 49.4 | 317 | 47.5 |
| Negative | 299 | 45.2 | 260 | 39.0 |
| Missing | 36 | 5.4 | 90 | 13.5 |
| Her2/Neu status | ||||
| Positive | 52 | 7.9 | 84 | 12.6 |
| Negative | 495 | 74.8 | 247 | 37.0 |
| Missing | 115 | 17.4 | 336 | 50.4 |
| Neoadjuvant therapy | ||||
| CT | 0 | 0 | 32 | 4.8 |
| HT | 0 | 0 | 22 | 3.3 |
| CT + HT | 0 | 0 | 1 | 0.1 |
| None | 658 | 99.4 | 610 | 91.5 |
| Missing | 4 | 0.6 | 2 | 0.2 |
| Characteristic | Test Cohort | Validation Cohort | ||
|---|---|---|---|---|
| n (39) | % | n (61) | % | |
| Age | ||||
| <45 | 14 | 35.9 | 21 | 34.4 |
| 45–55 | 9 | 23.1 | 20 | 32.8 |
| 55–65 | 5 | 12.8 | 12 | 19.7 |
| >65 | 11 | 28.2 | 8 | 13.1 |
| Missing | 0 | 0 | 0 | 0 |
| Stage (%) | ||||
| I | 6 | 15.4 | 20 | 32.8 |
| II | 24 | 61.5 | 28 | 45.9 |
| III | 8 | 20.5 | 6 | 9.8 |
| IV | 0 | 0 | 1 | 1.6 |
| Unknown | 1 | 2.6 | 6 | 9.8 |
| Histological type | ||||
| Ductal | 32 | 82.1 | 53 | 86.9 |
| Lobular | 7 | 17.9 | 2 | 3.3 |
| Other | 0 | 0 | 6 | 9.8 |
| Missing | 0 | 0 | 0 | 0 |
| Differentiation | ||||
| Good | 5 | 12.8 | 10 | 16.4 |
| Moderate | 16 | 41.0 | 24 | 39.3 |
| Poor | 18 | 46.2 | 23 | 37.7 |
| Unknown | 0 | 0 | 4 | 6.6 |
| ER | ||||
| Positive | 21 | 53.8 | 41 | 67.2 |
| Negative | 18 | 46.2 | 14 | 23.0 |
| Missing | 0 | 0 | 6 | 9.8 |
| PG | ||||
| Positive | 21 | 53.8 | 29 | 47.5 |
| Negative | 18 | 46.2 | 21 | 34.4 |
| Missing | 0 | 0 | 11 | 18 |
| HER2 | ||||
| Positive | 3 | 7.7 | 10 | 16.4 |
| Negative | 32 | 82.1 | 17 | 27.9 |
| Missing | 4 | 10.3 | 34 | 55.7 |
| Neoadj therapy | ||||
| CT | 0 | 0 | 3 | 4.9 |
| HT | 0 | 0 | 2 | 3.3 |
| CT + HT | 0 | 0 | 0 | 0 |
| None | 39 | 100 | 56 | 91.8 |
| Missing | 0 | 0 | 0 | 0.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Muijnck, C.; van Gorkom, Y.; van Duijvenvoorde, M.; Eghtesadi, M.; Dekker-Ensink, G.; Bhairosingh, S.S.; Affinito, A.; Kuppen, P.J.K.; Vahrmeijer, A.L.; Sier, C.F.M. Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer. Pharmaceuticals 2020, 13, 172. https://doi.org/10.3390/ph13080172
de Muijnck C, van Gorkom Y, van Duijvenvoorde M, Eghtesadi M, Dekker-Ensink G, Bhairosingh SS, Affinito A, Kuppen PJK, Vahrmeijer AL, Sier CFM. Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer. Pharmaceuticals. 2020; 13(8):172. https://doi.org/10.3390/ph13080172
Chicago/Turabian Stylede Muijnck, Cansu, Yoren van Gorkom, Maurice van Duijvenvoorde, Mina Eghtesadi, Geeske Dekker-Ensink, Shadhvi S. Bhairosingh, Alessandra Affinito, Peter J. K. Kuppen, Alexander L. Vahrmeijer, and Cornelis F. M. Sier. 2020. "Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer" Pharmaceuticals 13, no. 8: 172. https://doi.org/10.3390/ph13080172
APA Stylede Muijnck, C., van Gorkom, Y., van Duijvenvoorde, M., Eghtesadi, M., Dekker-Ensink, G., Bhairosingh, S. S., Affinito, A., Kuppen, P. J. K., Vahrmeijer, A. L., & Sier, C. F. M. (2020). Evaluation of EphB4 as Target for Image-Guided Surgery of Breast Cancer. Pharmaceuticals, 13(8), 172. https://doi.org/10.3390/ph13080172