Electrosprayed Alginate Nanoparticles as CRISPR Plasmid DNA Delivery Carrier: Preparation, Optimization, and Characterization
Abstract
1. Introduction
2. Results and Discussion
2.1. Encapsulation of CRISPR Plasmids in ALG NPs
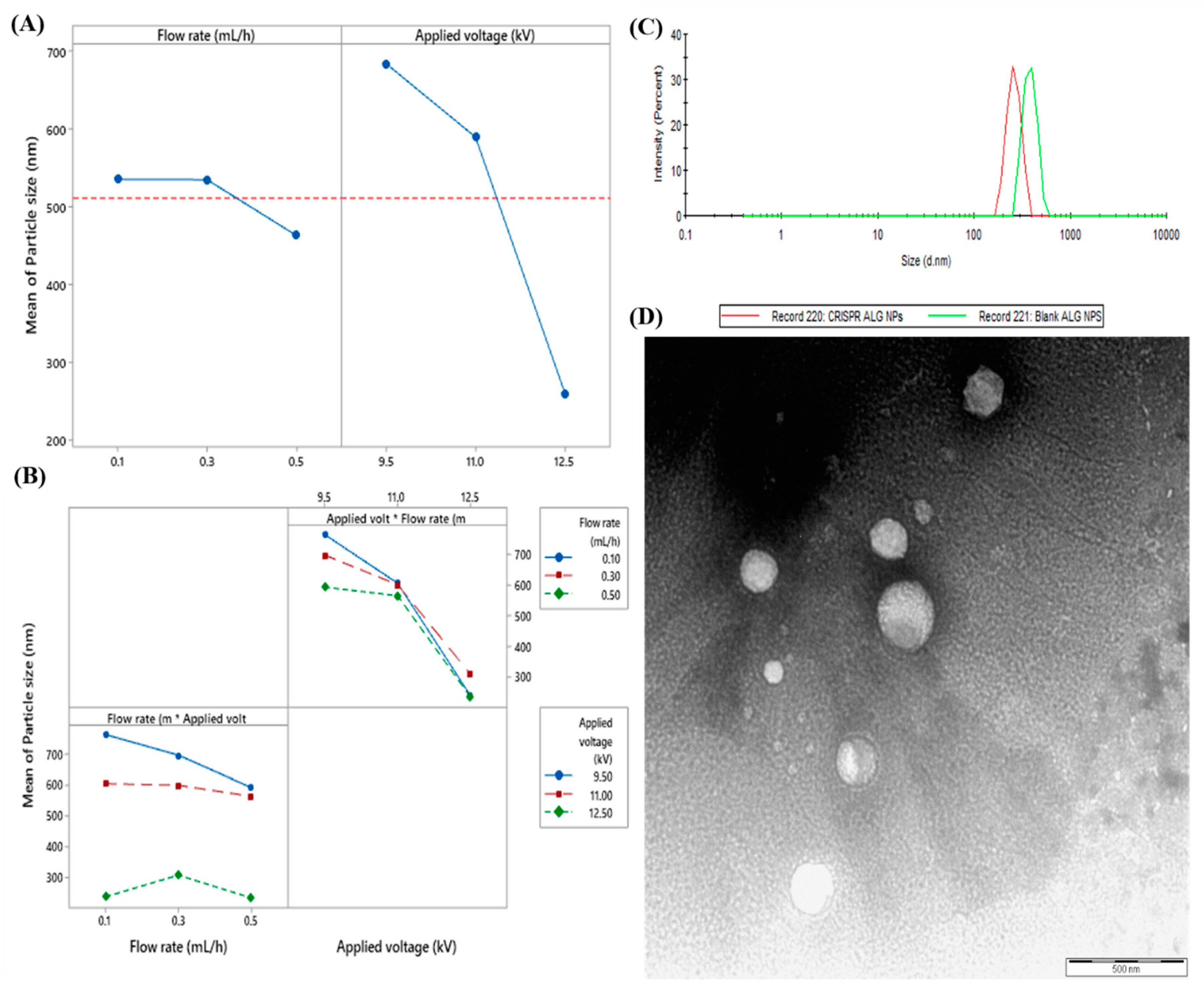
2.1.1. Optimization of Formulation Variables
2.1.2. Selection of CRISPR pDNA: ALG Ratio
2.1.3. Optimization of Process Variables
The Effect of Applied Voltage on Particles Size
The Effect of Flow Rate on Particles Size
2.2. Characterization of Optimized CRISPR ALG NPs
2.2.1. Particle Size and Zeta Potential Analysis
2.2.2. Transmission Electron Microscopy (TEM)
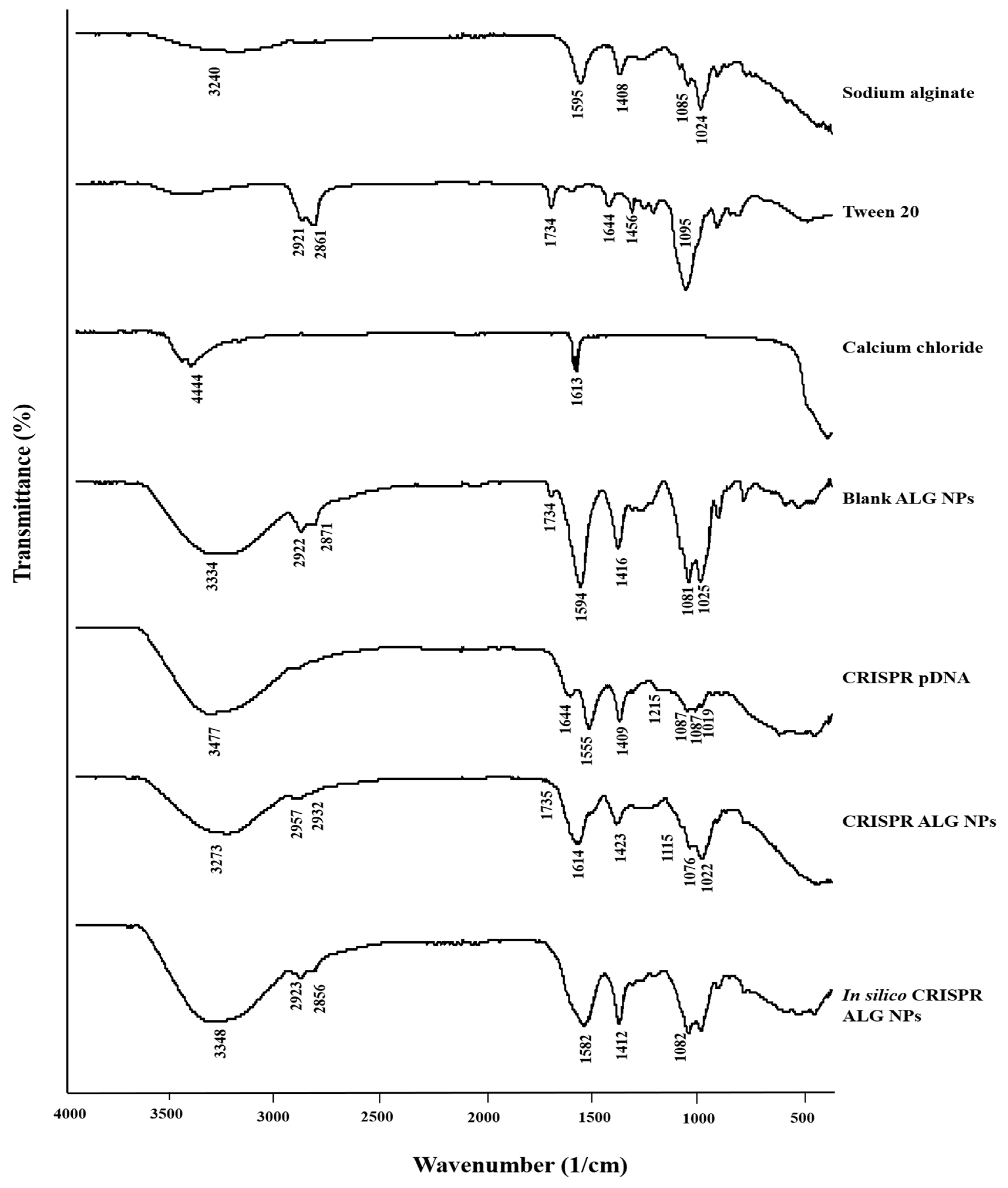
2.2.3. Attenuated Total Reflectance-Fourier Transform Infrared Spectroscopy (ATR-FTIR)
2.2.4. Encapsulation Efficiency
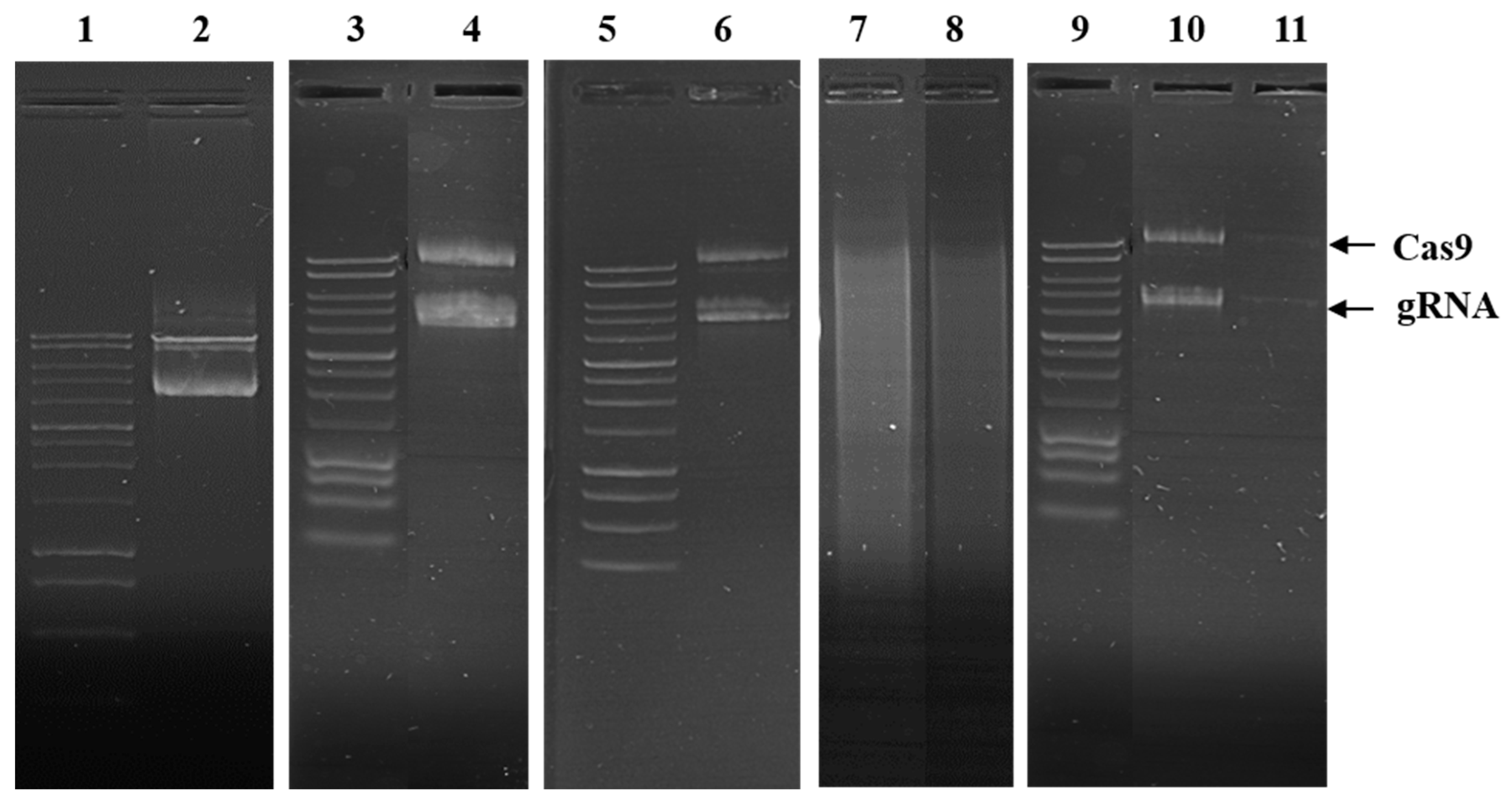
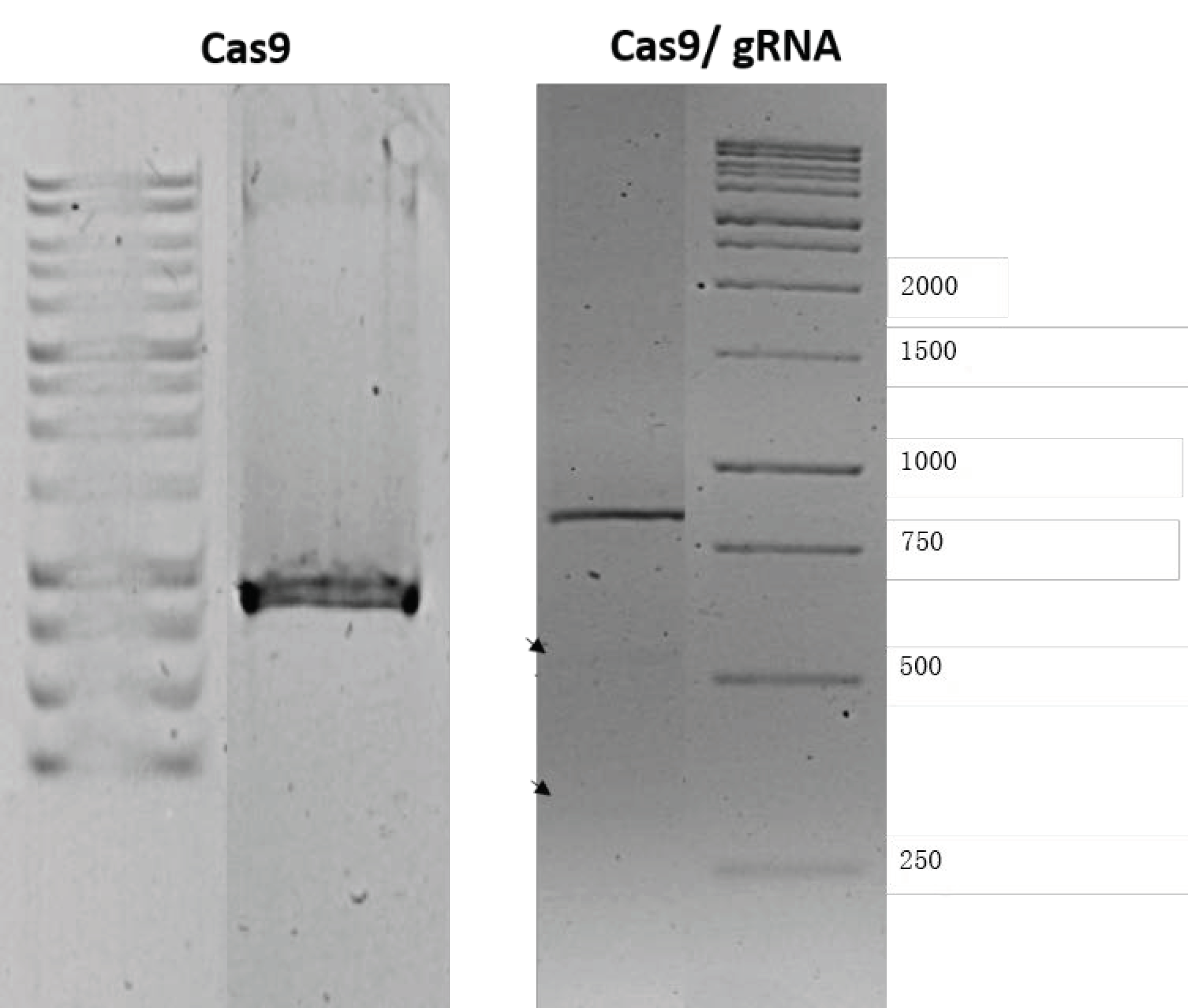
2.2.5. pDNA Integrity After Encapsulation
2.2.6. pDNA Serum Stability
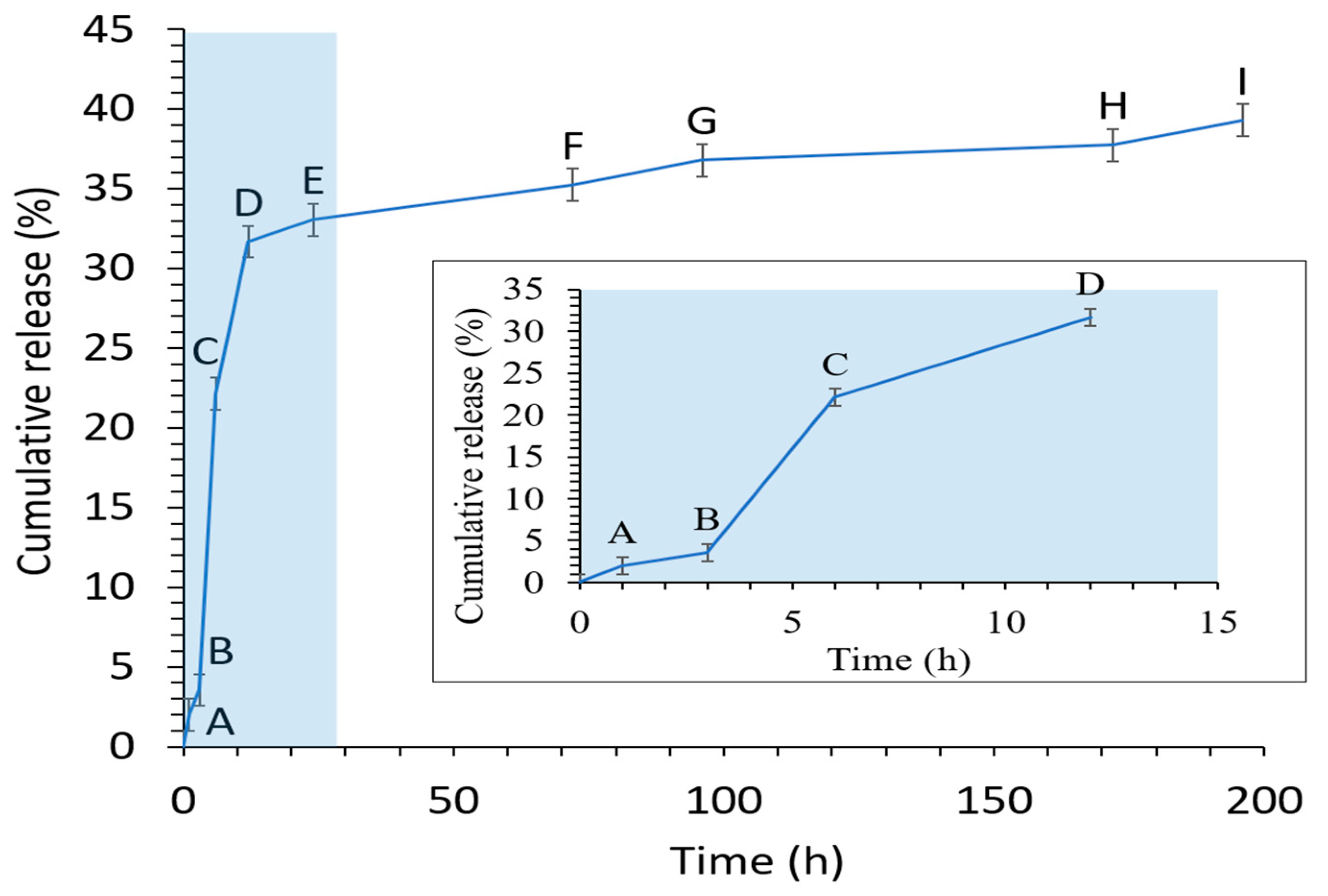
2.2.7. In Vitro Release Profile
2.2.8. NP Cytotoxicity
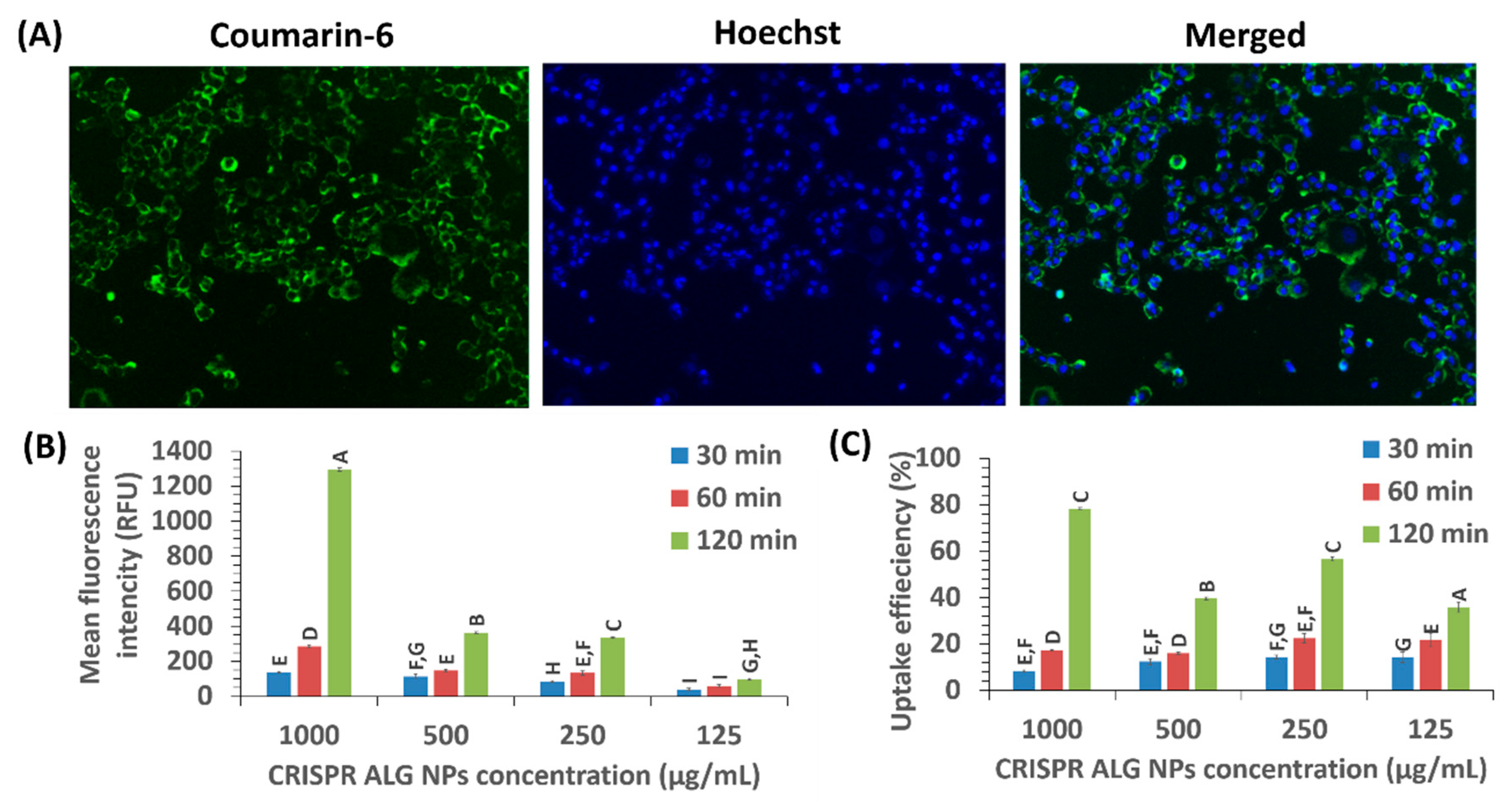
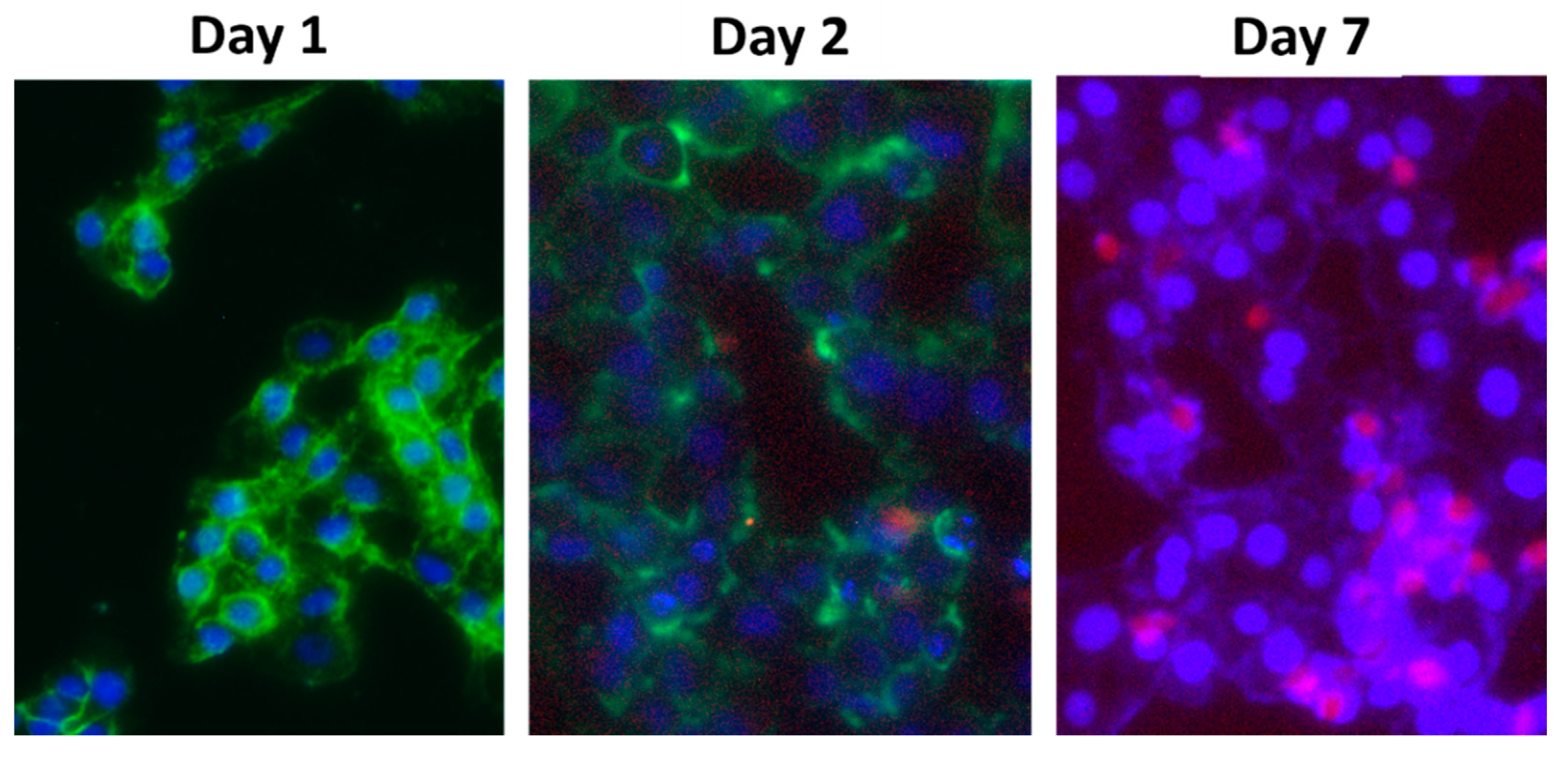
2.2.9. Cellular Uptake
2.2.10. In Vitro Transfection Efficiency
2.2.11. GFP Gene Disruption
3. Materials and Methods
3.1. Material
3.2. Encapsulation of CRISPR pDNA in ALG NPs
3.3. Optimization of CRISPR ALG NPs
3.4. Characterzation of Optimized CRISPR ALG NPs
3.4.1. Particle Size and Zeta Potential Analysis
3.4.2. TEM
3.4.3. ATR-FTIR
3.4.4. Encapsulation Efficiency
3.4.5. pDNA Integrity After Encapsulation
3.4.6. pDNA Serum Stability
3.4.7. In Vitro Release Profile
3.4.8. Cell Culture
3.4.9. Nanoparticle Cytotoxicity
3.4.10. Cellular Uptake
3.4.11. In vitro Transfection
3.4.12. Quantification of Transfection Efficiency
3.4.13. GFP Gene Disruption
3.4.14. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef] [PubMed]
- Simeonov, D.R.; Marson, A. CRISPR-Based Tools in Immunity. Annu. Rev. Immunol. 2019, 37. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.; Kim, J.-S. Therapeutic applications of CRISPR RNA-guided genome editing. Brief. Funct. Genom. 2017, 16, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Eoh, J.; Gu, L. Biomaterials as vectors for the delivery of CRISPR-Cas9. Biomater. Sci. 2019, 7, 1240–1261. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Bell, P.; McMenamin, D.; He, Z.; White, J.; Yu, H.; Xu, C.; Morizono, H.; Musunuru, K.; et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016, 34, 334–338. [Google Scholar] [CrossRef]
- Bak, R.O.; Porteus, M.H. CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep. 2017, 20, 750–756. [Google Scholar] [CrossRef]
- Huo, W.; Zhao, G.; Yin, J.; Ouyang, X.; Wang, Y.; Yang, C.; Wang, B.; Dong, P.; Wang, Z.; Watari, H.; et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J. Cancer 2017, 8, 57–64. [Google Scholar] [CrossRef]
- Wang, D.; Mou, H.; Li, S.; Li, Y.; Hough, S.; Tran, K.; Li, J.; Yin, H.; Anderson, D.G.; Sontheimer, E.J.; et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum. Gene Ther. 2015, 26, 432–442. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. In Nature Reviews Drug Discovery; Nature Publishing Group: Berlin, Germany, 2017; Volume 16, pp. 387–399. [Google Scholar] [CrossRef]
- Donahue, R.E.; Kessler, S.W.; Bodine, D.; Mcdonagh, K.; Dunbar, C.; Goodman, S.; Agricola, B.; Byrne, E.; Raffeld, M.; Moen, R. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J. Exp. Med. 1992, 176, 1125–1135. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Chew, W.L.; Tabebordbar, M.; Cheng, J.K.; Mali, P.; Wu, E.Y.; Ng, A.H.; Zhu, K.; Wagers, A.J.; Church, G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 2016, 13, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Arai, Y.; Yamazaki, M.; Morita, S.; Kimura, M.; Itoh, M.; Abe, Y.; Hatada, I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci. Rep. 2014, 4, 4513. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lee, B.; Lee, A.Y.F.; Modzelewski, A.J.; He, L. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 2016, 291, 14457–14467. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.S.; Meyerson, M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 2014, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Qu, L.; Wang, H.; Wei, L.; Dong, Y.; Xiong, S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antivir. Res. 2015, 118, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Tu, C.-F.; Chen, C.-H. Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors. BIO-PROTOC 2017, 7. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Sakuma, T.; Sakamoto, T.; Ohmuraya, M.; Nakagata, N.; Yamamoto, T. Production of knockout mice by DNA microinjection of various CRISPR/Cas9 vectors into freeze-thawed fertilized oocytes. BMC Biotechnol. 2015, 15, 33. [Google Scholar] [CrossRef]
- Li, L.; He, Z.-Y.; Wei, X.-W.; Gao, G.-P.; Wei, Y.-Q. Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum. Gene Ther. 2015, 26, 452–462. [Google Scholar] [CrossRef]
- Wang, H.X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.W.; Bao, G.; Leong, K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef]
- Zhen, S.; Takahashi, Y.; Narita, S.; Yang, Y.-C.; Li, X. Targeted delivery of CRISPR/Cas9 to prostate cancer by modified gRNA using a flexible aptamer-cationic liposome. Oncotarget 2017, 8, 9375–9387. [Google Scholar] [CrossRef]
- Moutal, A.; Yang, X.; Li, W.; Gilbraith, K.B.; Luo, S.; Cai, S.; François-Moutal, L.; Chew, L.A.; Yeon, S.K.; Bellampalli, S.S.; et al. CRISPR/Cas9 editing of Nf1 gene identifies CRMP2 as a therapeutic target in neur ofibromatosis type 1-related pain that is reversed by (S)-Lacosamide. Pain 2017, 158, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Zuckermann, M.; Hovestadt, V.; Knobbe-Thomsen, C.B.; Zapatka, M.; Northcott, P.A.; Schramm, K.; Belic, J.; Jones, D.T.; Tschida, B.; Moriarity, B.; et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015, 6, 7391. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Hua, L.; Liu, Y.H.; Gao, L.C.; Fu, J.; Wan, D.Y.; Dong, L.H.; Song, H.F.; Gao, X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015, 22, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Kretzmann, J.A.; Ho, D.; Evans, C.W.; Plani-Lam, J.H.; Garcia-Bloj, B.; Mohamed, A.E.; O’Mara, M.L.; Ford, E.; Tan, D.E.; Lister, R.; et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem. Sci. 2017, 8, 2923–2930. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, Y.; Gong, C. Polymeric Nanocarriers for Non-Viral Gene Delivery. J. Biomed. Nanotechnol. 2015, 11, 739–770. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26349389 (accessed on 17 March 2019). [CrossRef]
- Liu, Y.; Zhao, G.; Xu, C.F.; Luo, Y.L.; Lu, Z.D.; Wang, J. Systemic delivery of CRISPR/Cas9 with PEG-PLGA nanoparticles for chronic myeloid leukemia targeted therapy. Biomater. Sci. 2018, 6, 1592–1603. [Google Scholar] [CrossRef]
- Sundar, S.; Kundu, J.; Kundu, S.C. Biopolymeric nanoparticles. Sci. Technol. Adv. Mater. 2010, 11, 014104. [Google Scholar] [CrossRef]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; le Breton, A.; Préat, V. PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef]
- Jo, A.; Ringel-Scaia, V.M.; McDaniel, D.K.; Thomas, C.A.; Zhang, R.; Riffle, J.S.; Allen, I.C.; Davis, R.M. Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR-Cas9 plasmid. J. Nanobiotechnol. 2020, 18, 1–14. [Google Scholar] [CrossRef]
- Peralta-Yahya, P.P.; Zhang, F.; del Cardayre, S.B.; Keasling, J.D. Microbial engineering for the production of advanced biofuels. Nature 2012, 488, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, Y.; Liu, J.; Niu, X.; Wang, Y.; Li, D. Application and performance of 3D printing in nanobiomaterials. J. Nanomater. 2013, 2013. [Google Scholar] [CrossRef]
- Field, B. Natural Polymers and their Application in Drug Delivery and biomedical field. J. PharmaSciTech 2011, 1, 16–27. [Google Scholar]
- Abedini, F.; Ebrahimi, M.; Roozbehani, A.H.; Domb, A.J.; Hosseinkhani, H. Overview on natural hydrophilic polysaccharide polymers in drug delivery. Polym. Adv. Technol. 2018, 29, 2564–2573. [Google Scholar] [CrossRef]
- Daemi, H.; Barikani, M. Synthesis and characterization of calcium alginate nanoparticles, sodium homopolymannuronate salt and its calcium nanoparticles. Sci. Iran. 2012, 19, 2023–2028. [Google Scholar] [CrossRef]
- Anirudhan, T.S.; Anila, M.M.; Franklin, S. Synthesis characterization and biological evaluation of alginate nanoparticle for the targeted delivery of curcumin. Mater. Sci. Eng. C 2017, 78, 1125–1134. [Google Scholar] [CrossRef]
- Draget, K.; Taylor, C. Chemical, physical and biological properties of alginates and their biomedical implications. Food Hydrocoll. 2011, 25, 251–256. [Google Scholar] [CrossRef]
- Goh, C.H.; Heng, P.W.S.; Chan, L.W. Alginates as a useful natural polymer for microencapsulation and therapeutic applications. In Carbohydrate Polymers; Elsevier: Amsterdam, The Netherlands, 2012; Volume 88, pp. 1–12. [Google Scholar] [CrossRef]
- Sosnik, A. Alginate Particles as Platform for Drug Delivery by the Oral Route: State-of-the-Art. ISRN Pharm. 2014, 2014, 1–17. [Google Scholar] [CrossRef]
- Ahirrao, S.P.; Gide, P.S.; Shrivastav, B.; Sharma, P. Ionotropic Gelation: A Promising Cross Linking Technique for Hydrogels. J. Pharm. Nanotechnol. 2014, 2, 1–6. [Google Scholar]
- Lopes, M.; Abrahim, B.; Veiga, F.; Seiça, R.; Cabral, L.M.; Arnaud, P.; Andrade, J.C.; Ribeiro, A.J. Preparation methods and applications behind alginate-based particles. In Expert Opinion on Drug Delivery; Taylor and Francis Ltd.: Abingdon, UK, 2017; Volume 14, pp. 769–782. [Google Scholar] [CrossRef]
- Abyadeh, M.; Zarchi, A.A.K.; Faramarzi, M.A.; Amani, A. Evaluation of factors affecting size and size distribution of chitosan-electrosprayed nanoparticles. Avicenna J. Med. Biotechnol. 2017, 9, 126–132. Available online: http://www.ncbi.nlm.nih.gov/pubmed/28706607 (accessed on 27 September 2019).
- Nikoo, A.M.; Kadkhodaee, R.; Ghorani, B.; Razzaq, H.; Tucker, N. Controlling the morphology and material characteristics of electrospray generated calcium alginate microhydrogels. J. Microencapsul. 2016, 33, 605–612. [Google Scholar] [CrossRef]
- Wu, Y.; Duong, A.; Lee, L.J.; Wyslouzil, B.E. Electrospray Production of Nanoparticles for Drug/Nucleic Acid Delivery. The Delivery of Nanoparticles, 1st ed.; Hashim, A.A., Ed.; InTech: Rijeka, Croatia, 2012; pp. 223–242. [Google Scholar]
- Moghaddam, M.K.; Mortazavi, S.M.; Khayamian, T. Preparation of calcium alginate microcapsules containing n-nonadecane by a melt coaxial electrospray method. J. Electrostat. 2015, 73, 56–64. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, R.; Zou, L.; Mcclements, D.J. Graphical Abstract. Food Hydrocoll. 2016. [Google Scholar] [CrossRef]
- Fasiku, V.O.; Aderibigbe, B.A.; Sadiku, E.R.; Lemmer, Y.; Owonubi, S.J.; Ray, S.S.; Mukwevho, E. Polyethylene glycol–gum acacia-based multidrug delivery system for controlled delivery of anticancer. Polym. Bull. 2019, 76, 5011–5037. [Google Scholar] [CrossRef]
- Smith, D.P.H.H. The Electrohydrodynamic Atomization of Liquids. IEEE Trans. Ind. Appl. 1986, IA-22, 527–535. [Google Scholar] [CrossRef]
- Jayasinghe, S.N.; Townsend-Nicholson, A. Stable electric-field driven cone-jetting of concentrated biosuspensions. Lab Chip 2006, 6, 1086–1090. [Google Scholar] [CrossRef]
- Drozin, V.G. The electrical dispersion of liquids as aerosols. J. Colloid Sci. 1955, 10, 158–164. [Google Scholar] [CrossRef]
- Mutoh, M.; Kaieda, S.; Kamimura, K. Convergence and disintegration of liquid jets induced by an electrostatic field. J. Appl. Phys. 1979, 50, 3174–3179. [Google Scholar] [CrossRef]
- Perez-Masia, R.; Lagaron, J.M.; Lopez-Rubio, A. Surfactant-aided electrospraying of low molecular weight carbohydrate polymers from aqueous solutions. Carbohydr. Polym. 2014, 101, 249–255. [Google Scholar] [CrossRef]
- Poncelet, D.; Neufeld, R.J.; Goosen, M.F.A.; Burgarski, B.; Babak, V. Formation of microgel beads by electric dispersion of polymer solutions. AIChE J. 1999, 45, 2018–2023. [Google Scholar] [CrossRef]
- Yaghoobi, N.; Majidi, R.F.; Faramarzi, M.a.; Baharifar, H.; Amani, A. Preparation, Optimization and Activity Evaluation of PLGA/Streptokinase Nanoparticles Using Electrospray. Adv. Pharm. Bull. 2017, 7, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Park, M.V.; Neigh, A.M.; Vermeulen, J.P.; de la Fonteyne, L.J.; Verharen, H.W.; Briedé, J.J.; van Loveren, H.; de Jong, W.H. The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles. Biomaterials 2011, 32, 9810–9817. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef]
- Jiang, W.; Kim, B.Y.S.; Rutka, J.T.; Chan, W.C.W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008, 3, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Prabha, S.; Arya, G.; Chandra, R.; Ahmed, B.; Nimesh, S. Effect of size on biological properties of nanoparticles employed in gene delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef]
- Songsurang, K.; Praphairaksit, N.; Siraleartmukul, K.; Muangsin, N. Electrospray fabrication of doxorubicin-chitosan-tripolyphosphate nanoparticles for delivery of doxorubicin. Arch. Pharm. Res. 2011, 34, 583–592. [Google Scholar] [CrossRef]
- Musaei, M.; Mokhtari, J.; Nouri, M.; Rad, Z.P. Fabrication and characterization of nanocapsules of PLGA containing BSA using electrospray technique. Nanomed. Res. J. 2017, 2, 158–164. [Google Scholar] [CrossRef]
- Abyadeh, M.; Aghajani, M.; Mahmoudabad, A.G.; Amani, A. Preparation and Optimization of Chitosan/pDNA Nanoparticles Using Electrospray. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2019, 89, 931–937. [Google Scholar] [CrossRef]
- Xu, Y.; Skotak, M.; Hanna, M. Electrospray encapsulation of water-soluble protein with polylactide. I. Effects of formulations and process on morphology and particle size. J. Microencapsul. 2006, 23, 69–78. [Google Scholar] [CrossRef]
- Khalifa, A.Z.M.; Rasool, B.K.A. Optimized Mucoadhesive Coated Niosomes as a Sustained Oral Delivery System of Famotidine. AAPS Pharmscitech 2017, 18, 3064–3075. [Google Scholar] [CrossRef] [PubMed]
- Katas, H.; Hussain, Z.; Awang, S.A. Bovine serum albumin-loaded chitosan/dextran nanoparticles: Preparation and evaluation of ex vivo colloidal stability in serum. J. Nanomater. 2013, 2013. [Google Scholar] [CrossRef]
- Enayati, M.; Ahmad, Z.; Stride, E.; Edirisinghe, M. Preparation of Polymeric Carriers for Drug Delivery with Different Shape and Size Using an Electric Jet. Curr. Pharm. Biotechnol. 2009, 10, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Nastaj, J.; Przewłocka, A.; Rajkowska-Myśliwiec, M. Biosorption of Ni(II), Pb(II) and Zn(II) on calcium alginate beads: Equilibrium, kinetic and mechanism studies. Pol. J. Chem. Technol. 2016, 18, 81–87. [Google Scholar] [CrossRef]
- Machado, L.C.; Costa, D.M.; Perotti, G.F.; Henriques, R.R.; Couto, R.A.D.; Jr, J.A.; Shiguihara, A.L. Research Article Characterization of Cellulose Ether Films Containing Tween 20 and Tween 40. J. Chem. Pharm. Res. 2016, 8, 159–166. Available online: www.jocpr.com (accessed on 12 October 2019).
- Colthup, N.B.; Daly, L.H.; Wiberley, S.E. Introduction to Infrared and Raman Spectroscopy, 3rd ed.; Academic press: London, UK, 1990; pp. 247–260. [Google Scholar]
- Mady, M.M.; Mohammed, W.A.; El-Guendy, N.M.; Elsayed, A.A. Interaction of DNA and polyethylenimine: Fourier-transform infrared (FTIR) and differential scanning calorimetry (DSC) studies. Int. J. Phys. Sci. 2011, 6, 7328–7334. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, H.T.; Shen, B.; He, W.; Liu, Y.; Lu, G.Y.; Zhu, J.J. PH-induced fabrication of DNA/chitosan/α-ZrP nanocomposite andDNA release. Nanotechnology 2010, 21, 105102. [Google Scholar] [CrossRef]
- Watson, S.M.D.; Mohamed, H.D.A.; Horrocks, B.R.; Houlton, A. Electrically conductive magnetic nanowires using an electrochemical DNA-templating route. Nanoscale 2013, 5, 5349–5359. [Google Scholar] [CrossRef]
- Phang, S.W.; Sin, L.T.; Bee, S.T.; Low, J.Y.; Tee, T.T. Release behaviour study on controlled-release phosphorous fertilizer encapsulated by starch-alginate superabsorbent composite. J. Eng. Sci. Technol. 2018, 13, 82–94. [Google Scholar]
- Hong, Y.; Li, Y.; Yin, Y.; Li, D.; Zou, G. Electrohydrodynamic atomization of quasi-monodisperse drug-loaded spherical/wrinkled microparticles. J. Aerosol Sci. 2008, 39, 525–536. [Google Scholar] [CrossRef]
- Almurisi, S.H.; Doolaanea, A.A.; Akkawi, M.E.; Chatterjee, B.; Sarker, M.Z.I. Taste masking of paracetamol encapsulated in chitosan-coated alginate beads. J. Drug Deliv. Sci. Technol. 2020, 56, 101520. [Google Scholar] [CrossRef]
- Chan, E.S. Preparation of Ca-alginate beads containing high oil content: Influence of process variables on encapsulation efficiency and bead properties. Carbohydr. Polym. 2011, 84, 1267–1275. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, X.; Liu, W.; Zhang, F.; Cai, Q.; Deng, X. Controlled release behaviour of protein-loaded microparticles prepared via coaxial or emulsion electrospray. J. Microencapsul. 2013, 30, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.N. Generation of biologically active nano-aerosol by an electrospray-neutralization method. J. Aerosol Sci. 2011, 42, 341–354. [Google Scholar] [CrossRef]
- Azad, A.K.; Al-Mahmood, S.M.A.; Chatterjee, B.; Sulaiman, W.M.A.W.; Elsayed, T.M.; Doolaanea, A.A. Encapsulation of black seed oil in alginate beads as a ph-sensitive carrier for intestine-targeted drug delivery: In vitro, in vivo and ex vivo study. Pharmaceutics 2020, 12, 219. [Google Scholar] [CrossRef]
- Upuy, B.; Rien, A.; Minnot, A.E. FT-IR of membranes made with alginate/polylysine complexes. Variations with the mannuronic or guluronic content of the polysaccharides. Artif. Cells Blood Substit. Biotechnol. 1994, 22, 71–82. [Google Scholar] [CrossRef]
- Samimi, M.; Validov, S. Characteristics of pDNA-loaded chitosan/alginate-dextran sulfate nanoparticles with high transfection efficiency. Rom. Biotechnol. Lett. 2018, 23, 13996. [Google Scholar] [CrossRef]
- Saeed, A.; Mirakabadi, Z.; Sarzaeem, A.; Dounighi, N.M.; Soheily, S.; Borumand, M.R. Preparation and Characterization of Sodium Alginate Nanoparticles Containing ICD-85 (Venom Derived Peptides). Int. J. Innov. Appl. Stud. 2013, 4, 534–542. Available online: http://www.ijias.issr-journals.org/abstract.php?article=IJIAS-13-218-01 (accessed on 29 September 2019).
- Amiji, M.; Jain, S. Calcium alginate microparticles as a non-condensing DNA delivery and transfection system for macrophages. Pharm. Eng. 2012, 32, 42–49. [Google Scholar]
- Wang, P.; Wu, Q.; Wang, F.; Zhang, Y.; Tong, L.; Jiang, T.; Gu, C.; Huang, S.; Wang, H.; Bu, S.; et al. Evaluating cellular uptake of gold nanoparticles in HL-7702 and HepG2 cells for plasmonic photothermal therapy. Nanomedicine 2018, 13, 2245–2259. [Google Scholar] [CrossRef]
- Panyam, J.; Labhasetwar, V. Dynamics of endocytosis and exocytosis of poly(D,L-lactide-co-glycolide) nanoparticles in vascular smooth muscle cells. Pharm. Res. 2003, 20, 212–220. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, A.; Alimohammadian, M.H.; Gazori, T.; Riazi-rad, F.; Fatemi, S.M.R.; Parizadeh, A.; Haririan, I.; Havaskary, M. Comparison of chitosan, alginate and chitosan/alginate nanoparticles with respect to their size, stability, toxicity and transfection. Asian Pac. J. Trop. Dis. 2014, 4, 372–377. [Google Scholar] [CrossRef]
- You, J.; Peng, C. Calcium-Alginate Nanoparticles for Nonviral Gene Delivery Preparation of Ca-alginate Nanoparticle. Nsti-Nanotech 2005, 1, 270–273. Available online: https://briefs.techconnect.org/papers/calcium-alginate-nanoparticles-for-nonviral-gene-delivery/ (accessed on 27 September 2019).
- Yin, W.; Xiang, P.; Li, Q. Investigations of the effect of DNA size in transient transfection assay using dual luciferase system. Anal. Biochem. 2005, 346, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Capito, R.M.; Spector, M. Plasmid size influences chitosan nanoparticle mediated gene transfer to chondrocytes. J. Biomed. Mater. Res. Part A 2008, 84, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Frank, L.A.; Onzi, G.R.; Morawski, A.S.; Pohlmann, A.R.; Guterres, S.S.; Contri, R.V. Chitosan as a coating material for nanoparticles intended for biomedical applications. React. Funct. Polym. 2020, 147, 104459. [Google Scholar] [CrossRef]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Doolaanea, A.A.; Mansor, N.I.; Nor, N.H.M.; Mohamed, F. Co-encapsulation of Nigella sativa oil and plasmid DNA for enhanced gene therapy of Alzheimers disease. J. Microencapsul. 2016, 33, 114–126. [Google Scholar] [CrossRef]
- Doolaanea, A.A.; Mansor, N.I.; Nor, N.H.M.; Mohamed, F. Cellular uptake of Nigella sativa oil-PLGA microparticle by PC-12 cell line. J. Microencapsul. 2014, 31, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Chea, K.; Pyda, R.; Chua, M.; Dominguez, I. Comparative analysis of non-viral transfection methods in mouse embryonic fibroblast cells. J. Biomol. Tech. 2017, 28, 67–74. [Google Scholar] [CrossRef] [PubMed]
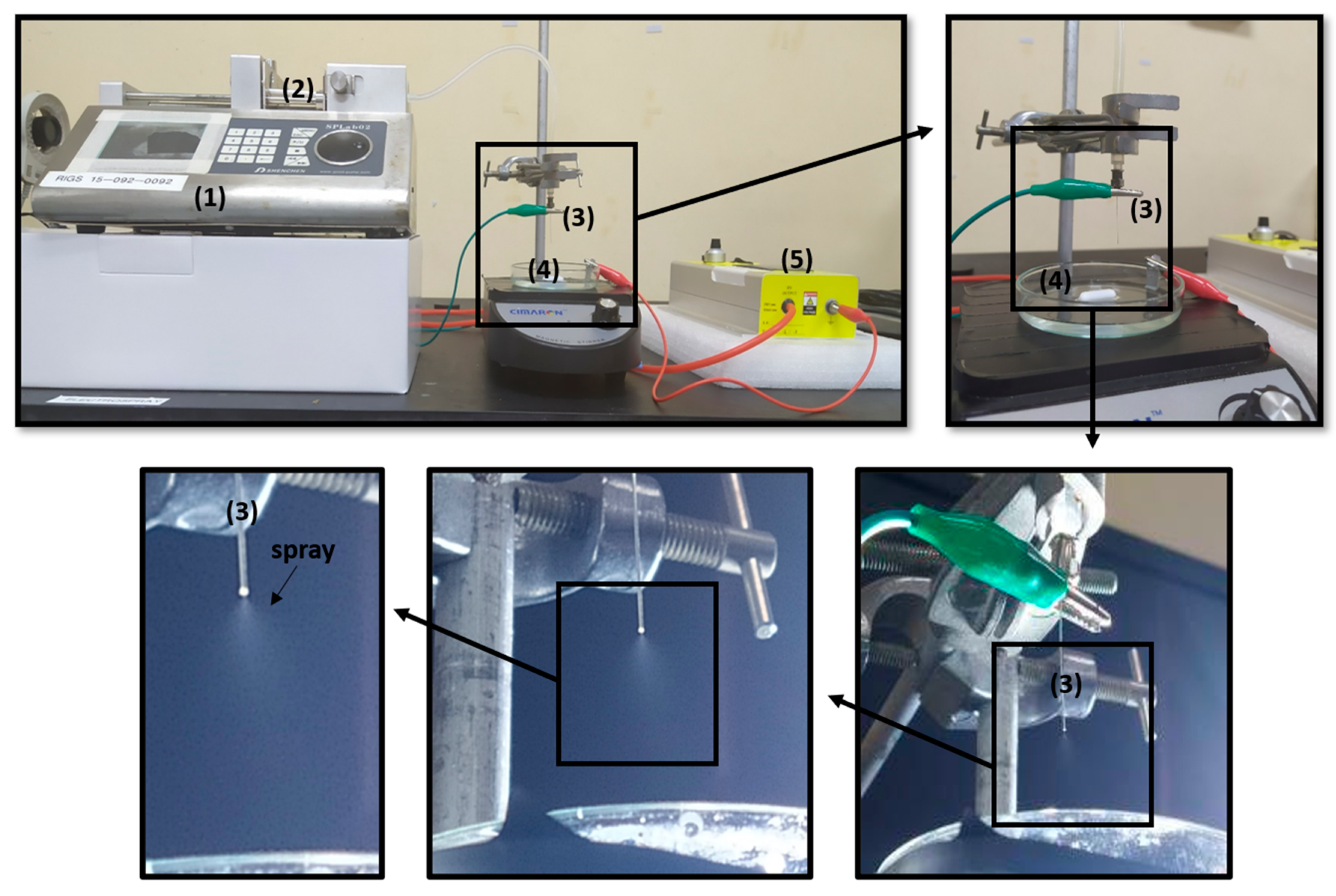
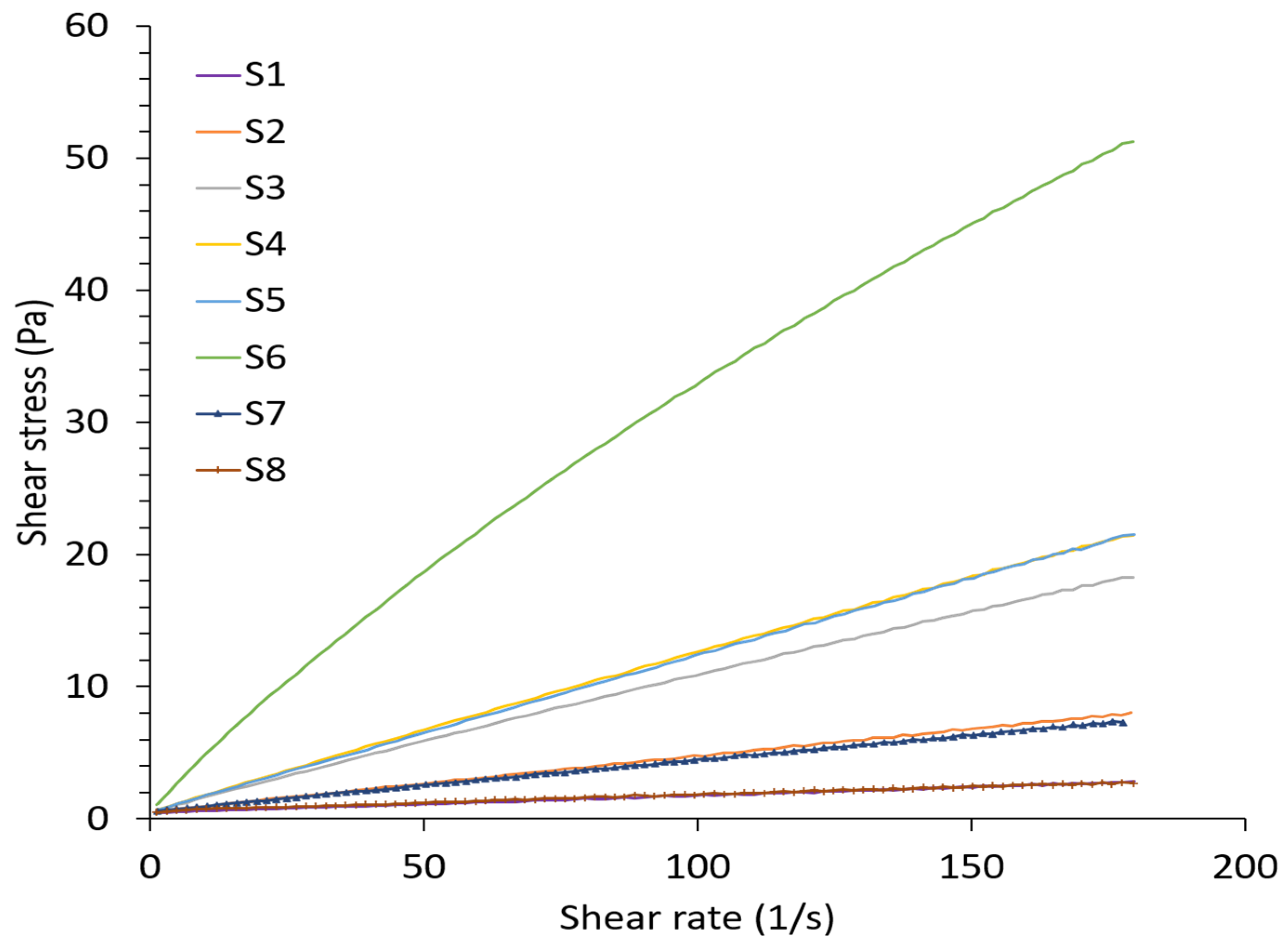
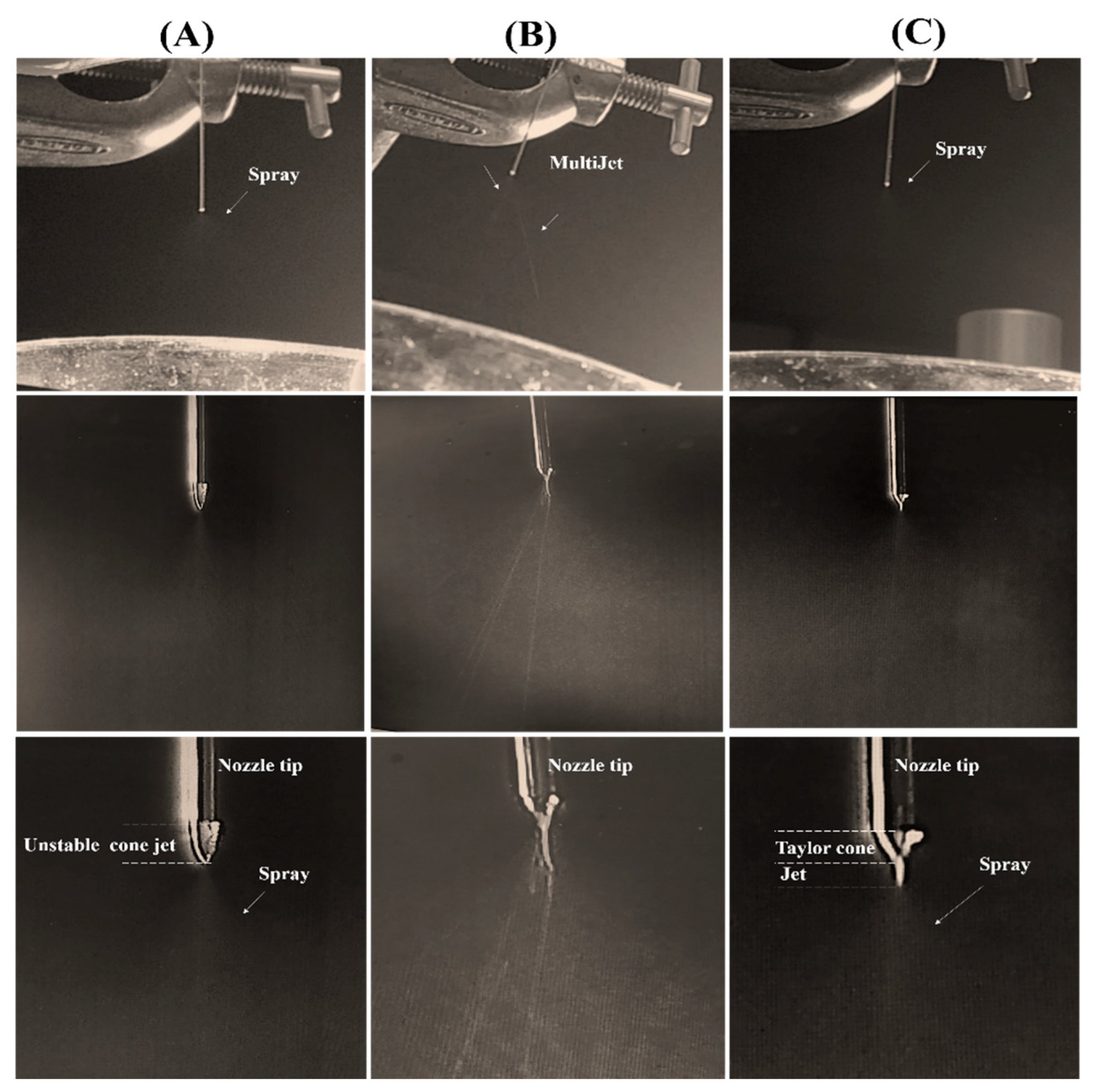


| Solution | ALG Concentration % w/v | Tween 20 % v/v | Electrical Conductivity (mS/cm) | Viscosity (mPa S) | Surface Tension (mN/m) | Density (kg/m3) | pH | Spray | |
|---|---|---|---|---|---|---|---|---|---|
| Stability | Mode | ||||||||
| S1 | 0.5 | - | 1.92 ± 0.09 A | 24.45 ± 0.04 D | 62.90 ± 0.27 A | 1003 ± 0.00 C | 6.57 ± 0.010 C | unstable | cone jet |
| S2 | 1 | - | 2.64 ± 0.06 B | 59.03 ± 0.05 C | 65.02 ± 0.01 B | 1004 ± 0.01 C | 6.57 ± 0.017 C | unstable | cone jet |
| S3 | 1.5 | - | 3.76 ± 0.05 C | 121.26 ± 0.05 B | 66.46 ± 0.30 C | 1070 ± 0.01 B,C | 6.66 ± 0.025 B | unstable | multijet |
| S4 | 2 | - | 4.67 ± 0.02 D | 134.85 ± 0.04 B | 68.13 ± 0.74 D | 1070 ± 0.03 B,C | 6.64 ± 0.040 B | unstable | multijet |
| S5 | 2.5 | - | 6.76 ± 0.02 E | 138.04 ± 0.05 B | 69.99 ± 0.12 E | 1090 ± 0.04 B | 6.68 ± 0.012 A,B | unstable | multijet |
| S6 | 3 | - | 8.59 ± 0.05 F | 356.10 ± 0.08 A | 71.30 ± 0.50 F | 1170 ± 0.04 A | 6.73 ± 0.012 A | unstable | multijet |
| S7 | 0.5 | 1 | 1.73 ± 0.03 G | 30.36 ± 0.04 D | 34.86 ±0.16 G | 1009 ± 0.00 C | 5.47 ± 0.040 D | stable | cone jet |
| S8 | 1 | 1 | 2.88 ± 0.121 H | 59.03 ± 0.05 C | 36.29 ±0.16 H | 1006 ± 0.00 C | 5.47 ± 0.020 D | stable | cone jet |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alallam, B.; Altahhan, S.; Taher, M.; Mohd Nasir, M.H.; Doolaanea, A.A. Electrosprayed Alginate Nanoparticles as CRISPR Plasmid DNA Delivery Carrier: Preparation, Optimization, and Characterization. Pharmaceuticals 2020, 13, 158. https://doi.org/10.3390/ph13080158
Alallam B, Altahhan S, Taher M, Mohd Nasir MH, Doolaanea AA. Electrosprayed Alginate Nanoparticles as CRISPR Plasmid DNA Delivery Carrier: Preparation, Optimization, and Characterization. Pharmaceuticals. 2020; 13(8):158. https://doi.org/10.3390/ph13080158
Chicago/Turabian StyleAlallam, Batoul, Sara Altahhan, Muhammad Taher, Mohd Hamzah Mohd Nasir, and Abd Almonem Doolaanea. 2020. "Electrosprayed Alginate Nanoparticles as CRISPR Plasmid DNA Delivery Carrier: Preparation, Optimization, and Characterization" Pharmaceuticals 13, no. 8: 158. https://doi.org/10.3390/ph13080158
APA StyleAlallam, B., Altahhan, S., Taher, M., Mohd Nasir, M. H., & Doolaanea, A. A. (2020). Electrosprayed Alginate Nanoparticles as CRISPR Plasmid DNA Delivery Carrier: Preparation, Optimization, and Characterization. Pharmaceuticals, 13(8), 158. https://doi.org/10.3390/ph13080158









