Multi-Step Ubiquitin Decoding Mechanism for Proteasomal Degradation
Abstract
1. Introduction
2. Ubiquitin-Binding Proteins Related to the Proteasome: Proteasomal Ubiquitin Receptors, UBL–UBA Proteins, and p97
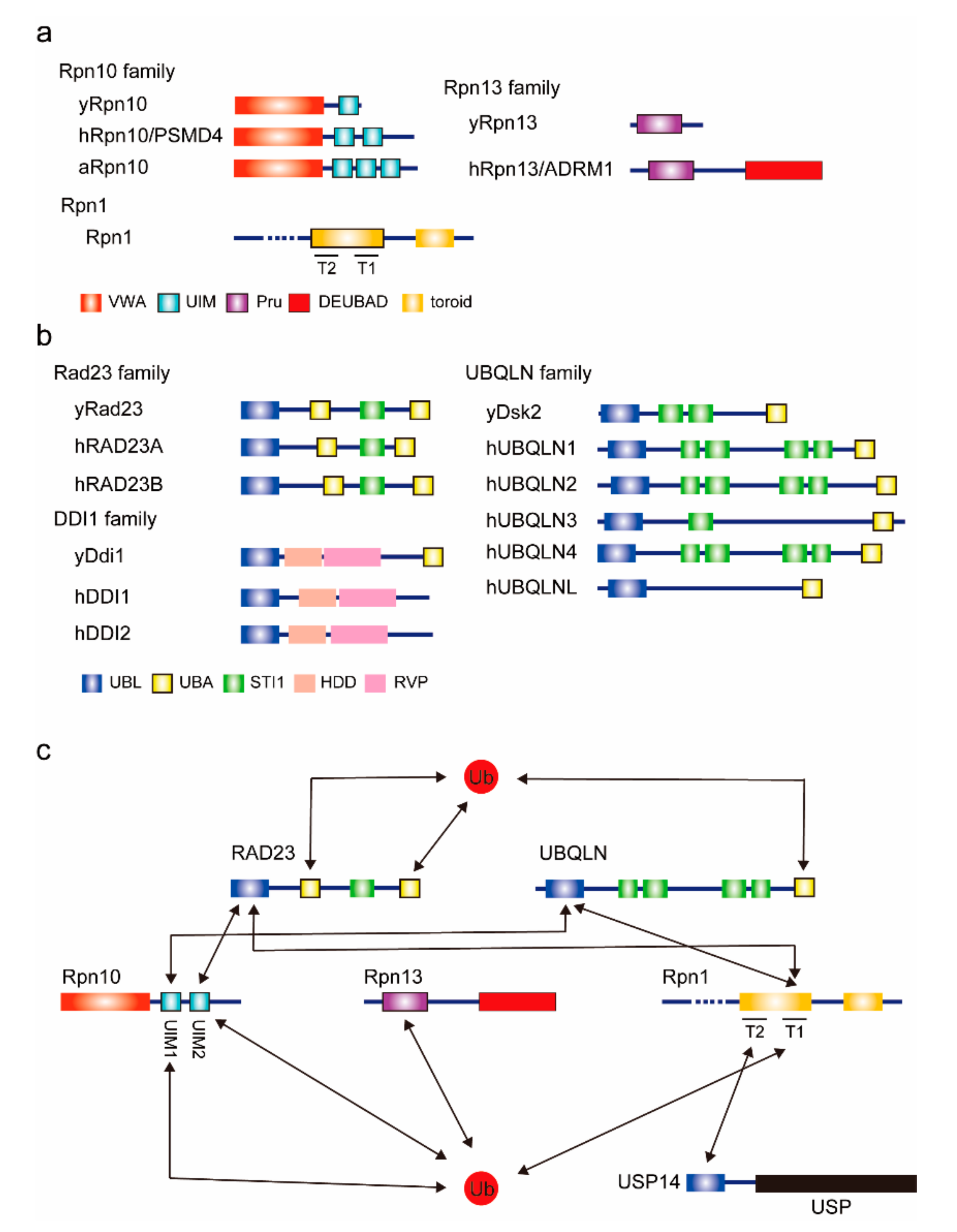
2.1. Proteasomal Ubiquitin Receptors (Rpn10, Rpn13, and Rpn1)
2.2. UBL–UBA Proteins
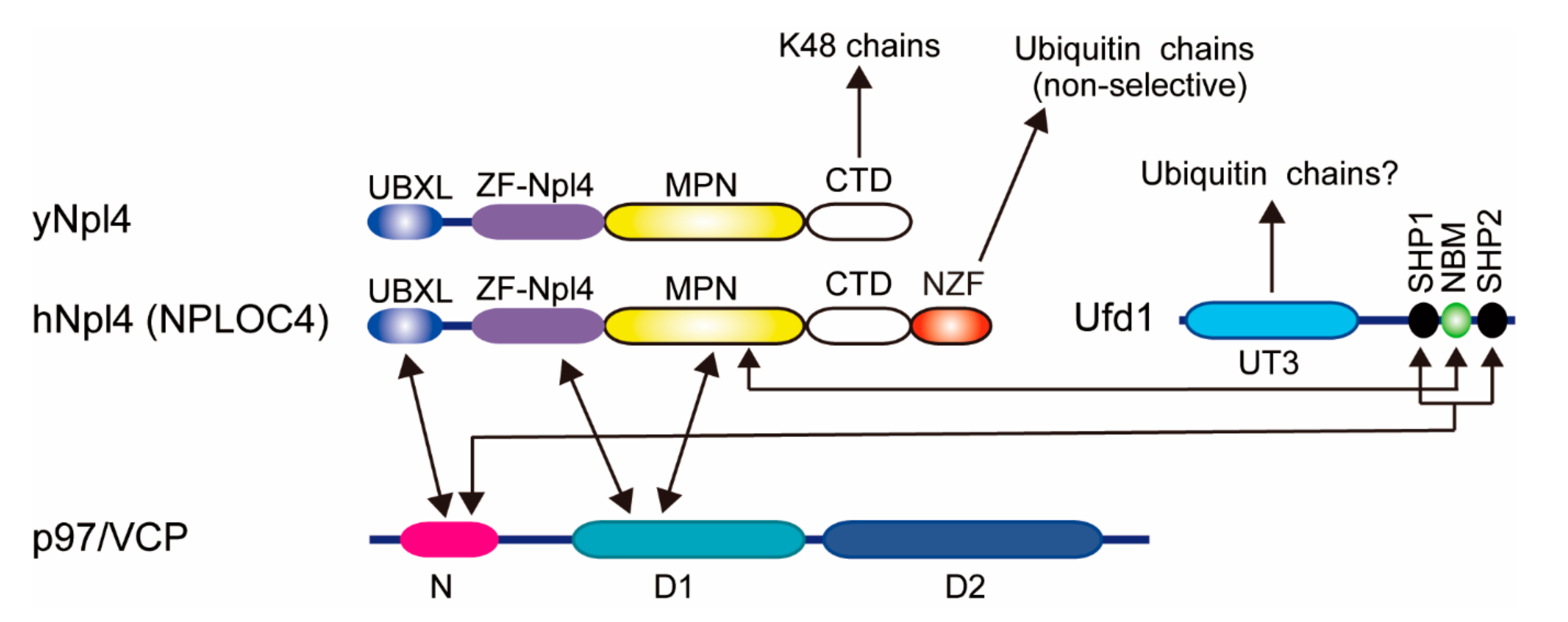
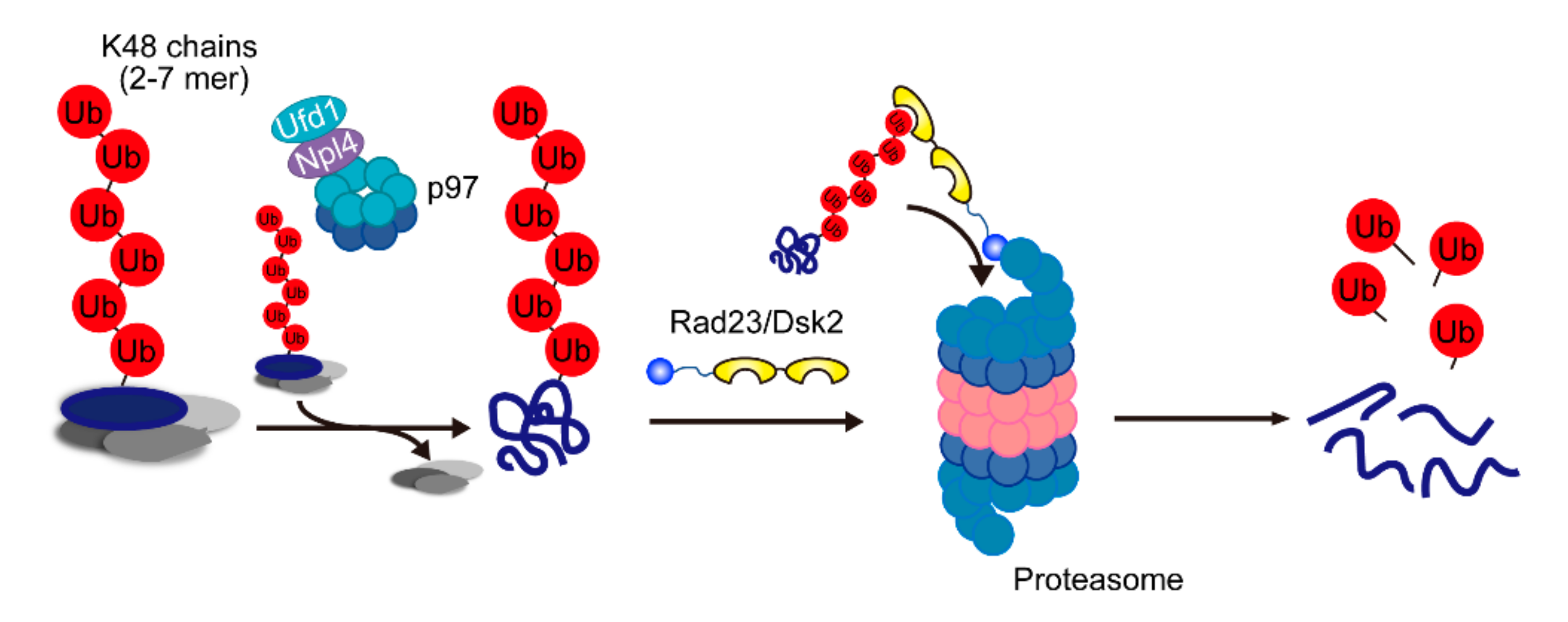
2.3. Cdc48/p97/VCP-Ufd1-Npl4 Complex
2.4. The Major Pathway for Proteasomal Degradation
3. Ubiquitin Signal for the Proteasome: Linkage Type and Length
3.1. Ubiquitin Chain Type Selectivity for Proteasomal Degradation
3.2. Ubiquitin Chain Length as a Signal for Proteasomal Degradation
4. Intracellular Dynamics of the Proteasome
4.1. Discovery of Stress-Dependent Proteasome Nuclear Foci
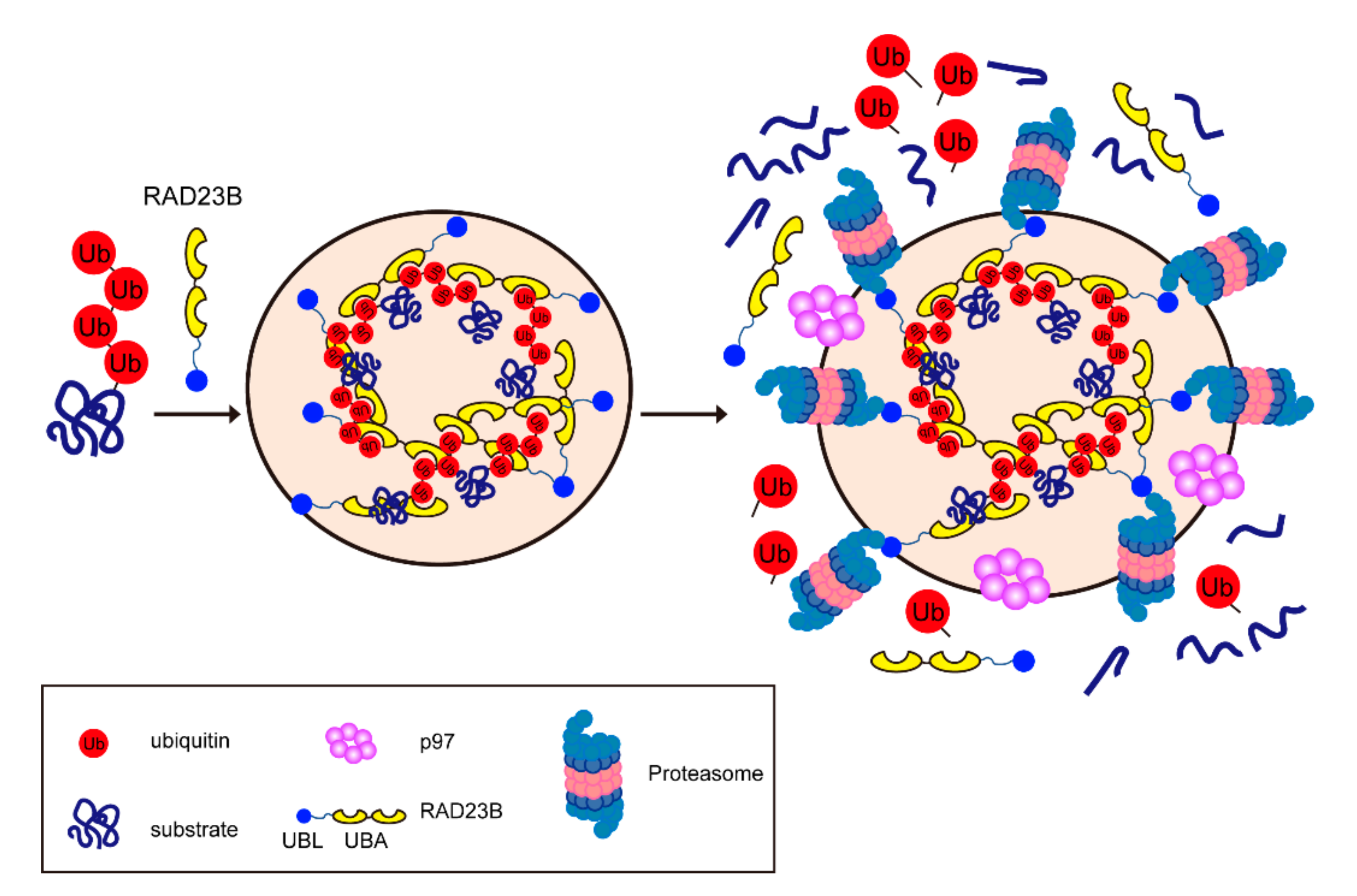
4.2. Molecular Mechanism of the Formation of Proteasome Foci
5. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Kish-Trier, E.; Hill, C.P. Structural biology of the proteasome. Annu. Rev. Biophys. 2013, 42, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Ohtake, F.; Arai, N.; Kaiho, A.; Yasuda, S.; Tanaka, K.; Saeki, Y. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol. Cell 2017, 66, 488–502. [Google Scholar] [CrossRef]
- Fatimababy, A.S.; Lin, Y.L.; Usharani, R.; Radjacommare, R.; Wang, H.T.; Tsai, H.L.; Lee, Y.; Fu, H. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 2010, 277, 796–816. [Google Scholar] [CrossRef]
- Kang, Y.; Chen, X.; Lary, J.W.; Cole, J.L.; Walters, K.J. Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J. Mol. Biol. 2007, 369, 168–176. [Google Scholar] [CrossRef]
- Chen, X.; Ebelle, D.L.; Wright, B.J.; Sridharan, V.; Hooper, E.; Walters, K.J. Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome. J. Mol. Biol. 2019, 431, 939–955. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Kim, H.T.; Goldberg, A.L. UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc. Natl. Acad. Sci. USA 2018, 115, E11642–E11650. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proc. Natl. Acad. Sci. USA 2020, 117, 4664–4674. [Google Scholar] [CrossRef]
- Hamazaki, J.; Sasaki, K.; Kawahara, H.; Hisanaga, S.; Tanaka, K.; Murata, S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell Biol. 2007, 27, 6629–6638. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Song, L.; Xu, W.; DeMartino, G.N.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Cohen, R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006, 8, 994–1002. [Google Scholar] [CrossRef]
- Kajava, A.V. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J. Biol. Chem. 2002, 277, 49791–49798. [Google Scholar] [CrossRef]
- He, J.; Kulkarni, K.; da Fonseca, P.C.; Krutauz, D.; Glickman, M.H.; Barford, D.; Morris, E.P. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure 2012, 20, 513–521. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Boughton, A.J.; Krueger, S.; Fushman, D. Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1. Structure 2020, 28, 29–43.e26. [Google Scholar] [CrossRef]
- Hamazaki, J.; Hirayama, S.; Murata, S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 2015, 11, e1005401. [Google Scholar] [CrossRef]
- Lam, Y.A.; Lawson, T.G.; Velayutham, M.; Zweier, J.L.; Pickart, C.M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 416, 763–767. [Google Scholar] [CrossRef]
- Paraskevopoulos, K.; Kriegenburg, F.; Tatham, M.H.; Rosner, H.I.; Medina, B.; Larsen, I.B.; Brandstrup, R.; Hardwick, K.G.; Hay, R.T.; Kragelund, B.B.; et al. Dss1 is a 26S proteasome ubiquitin receptor. Mol. Cell 2014, 56, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R.; Seeger, M.; Hartmann-Petersen, R.; Stone, M.; Wallace, M.; Semple, C.; Gordon, C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001, 3, 939–943. [Google Scholar] [CrossRef] [PubMed]
- Bertolaet, B.L.; Clarke, D.J.; Wolff, M.; Watson, M.H.; Henze, M.; Divita, G.; Reed, S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001, 8, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Perozzi, G.; Prakash, S. RAD7 gene of Saccharomyces cerevisiae: Transcripts, nucleotide sequence analysis, and functional relationship between the RAD7 and RAD23 gene products. Mol. Cell Biol. 1986, 6, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Park, H.; Kwofie, M.A.; Lennarz, W.J. Rad23 provides a link between the Png1 deglycosylating enzyme and the 26 S proteasome in yeast. J. Biol. Chem. 2001, 276, 21601–21607. [Google Scholar] [CrossRef]
- Ng, J.M.; Vrieling, H.; Sugasawa, K.; Ooms, M.P.; Grootegoed, J.A.; Vreeburg, J.T.; Visser, P.; Beems, R.B.; Gorgels, T.G.; Hanaoka, F.; et al. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol. Cell. Biol. 2002, 22, 1233–1245. [Google Scholar] [CrossRef]
- Biggins, S.; Ivanovska, I.; Rose, M.D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1996, 133, 1331–1346. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, N.; Koepp, D.M.; Walters, K.J. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J. Mol. Biol. 2007, 365, 1093–1101. [Google Scholar] [CrossRef]
- Itakura, E.; Zavodszky, E.; Shao, S.; Wohlever, M.L.; Keenan, R.J.; Hegde, R.S. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell 2016, 63, 21–33. [Google Scholar] [CrossRef]
- Suzuki, R.; Kawahara, H. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep. 2016, 17, 842–857. [Google Scholar] [CrossRef]
- Hjerpe, R.; Bett, J.S.; Keuss, M.J.; Solovyova, A.; McWilliams, T.G.; Johnson, C.; Sahu, I.; Varghese, J.; Wood, N.; Wightman, M.; et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 2016, 166, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.-X.; Chen, W.; Hong, S.-T.; Boycott, K.M.; Gorrie, G.H.; Siddique, N.; Yang, Y.; Fecto, F.; Shi, Y.; Zhai, H.; et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011, 477, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, S.; Sugihara, M.; Morito, D.; Iemura, S.I.; Natsume, T.; Murata, S.; Nagata, K. Nuclear export of ubiquitinated proteins via the UBIN-POST system. Proc. Natl. Acad. Sci. USA 2018, 115, E4199–E4208. [Google Scholar] [CrossRef] [PubMed]
- Richly, H.; Rape, M.; Braun, S.; Rumpf, S.; Hoege, C.; Jentsch, S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 2005, 120, 73–84. [Google Scholar] [CrossRef]
- Verma, R.; Oania, R.; Graumann, J.; Deshaies, R.J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 2004, 118, 99–110. [Google Scholar] [CrossRef]
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300. [Google Scholar] [CrossRef]
- Lee, D.Y.; Arnott, D.; Brown, E.J. Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 2013, 14, 373–381. [Google Scholar] [CrossRef]
- Sirkis, R.; Gerst, J.E.; Fass, D. Ddi1, a eukaryotic protein with the retroviral protease fold. J. Mol. Biol. 2006, 364, 376–387. [Google Scholar] [CrossRef]
- Krylov, D.M.; Koonin, E.V. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr. Biol. 2001, 11, R584–R587. [Google Scholar] [CrossRef]
- Kaplun, L.; Tzirkin, R.; Bakhrat, A.; Shabek, N.; Ivantsiv, Y.; Raveh, D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol. Cell Biol. 2005, 25, 5355–5362. [Google Scholar] [CrossRef]
- Nowicka, U.; Zhang, D.; Walker, O.; Krutauz, D.; Castañeda, C.A.; Chaturvedi, A.; Chen, T.Y.; Reis, N.; Glickman, M.H.; Fushman, D. DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure 2015, 23, 542–557. [Google Scholar] [CrossRef]
- Yip, M.C.J.; Bodnar, N.O.; Rapoport, T.A. Ddi1 is a ubiquitin-dependent protease. Proc. Natl. Acad. Sci. USA 2020, 117, 7776–7781. [Google Scholar] [CrossRef]
- Koizumi, S.; Irie, T.; Hirayama, S.; Sakurai, Y.; Yashiroda, H.; Naguro, I.; Ichijo, H.; Hamazaki, J.; Murata, S. The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Bug, M.; Bremer, S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012, 14, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Vembar, S.S.; Brodsky, J.L. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef] [PubMed]
- Kristariyanto, Y.A.; Abdul Rehman, S.A.; Campbell, D.G.; Morrice, N.A.; Johnson, C.; Toth, R.; Kulathu, Y. K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell 2015, 58, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.H.; Wang, Y.; Warren, G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002, 21, 5645–5652. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Yoshikawa, A.; Yamashita, M.; Yamagata, A.; Fukai, S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009, 28, 3903–3909. [Google Scholar] [CrossRef]
- Twomey, E.C.; Ji, Z.; Wales, T.E.; Bodnar, N.O.; Ficarro, S.B.; Marto, J.A.; Engen, J.R.; Rapoport, T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 2019, 365. [Google Scholar] [CrossRef]
- Sato, Y.; Tsuchiya, H.; Yamagata, A.; Okatsu, K.; Tanaka, K.; Saeki, Y.; Fukai, S. Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4. Nat. Commun. 2019, 10, 5708. [Google Scholar] [CrossRef] [PubMed]
- Baek, G.H.; Kim, I.; Rao, H. The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc. Natl. Acad. Sci. USA 2011, 108, 13558–13563. [Google Scholar] [CrossRef] [PubMed]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed]
- Blythe, E.E.; Olson, K.C.; Chau, V.; Deshaies, R.J. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. USA 2017, 114, E4380–E4388. [Google Scholar] [CrossRef]
- Bodnar, N.O.; Kim, K.H.; Ji, Z.; Wales, T.E.; Svetlov, V.; Nudler, E.; Engen, J.R.; Walz, T.; Rapoport, T.A. Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Nat. Struct. Mol. Biol. 2018, 25, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Tian, L.; Ratliff, K.S.; Lehotzky, R.E.; Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004, 11, 830–837. [Google Scholar] [CrossRef]
- Tomita, T.; Huibregtse, J.M.; Matouschek, A. A masked initiation region in retinoblastoma protein regulates its proteasomal degradation. Nat. Commun. 2020, 11, 2019. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, M.M.; Williams, C.; Dong, K.C.; Martin, A. The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun. Biol. 2019, 2, 29. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Raasi, S.; Varadan, R.; Fushman, D.; Pickart, C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005, 12, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Nathan, J.A.; Kim, H.T.; Ting, L.; Gygi, S.P.; Goldberg, A.L. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013, 32, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, W.; Kirschner, M.W. Specificity of the anaphase-promoting complex: A single-molecule study. Science 2015, 348, 1248737. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lee, B.H.; King, R.W.; Finley, D.; Kirschner, M.W. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science 2015, 348, 1250834. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Ye, Y.; Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017, 8, 14274. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef]
- Samant, R.S.; Livingston, C.M.; Sontag, E.M.; Frydman, J. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 2018, 563, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Shabek, N.; Herman-Bachinsky, Y.; Buchsbaum, S.; Lewinson, O.; Haj-Yahya, M.; Hejjaoui, M.; Lashuel, H.A.; Sommer, T.; Brik, A.; Ciechanover, A. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell 2012, 48, 87–97. [Google Scholar] [CrossRef]
- Zhang, F.; Hu, M.; Tian, G.; Zhang, P.; Finley, D.; Jeffrey, P.D.; Shi, Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 2009, 34, 473–484. [Google Scholar] [CrossRef]
- Sakata, E.; Bohn, S.; Mihalache, O.; Kiss, P.; Beck, F.; Nagy, I.; Nickell, S.; Tanaka, K.; Saeki, Y.; Förster, F.; et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 1479–1484. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Burana, D.; Ohtake, F.; Arai, N.; Kaiho, A.; Komada, M.; Tanaka, K.; Saeki, Y. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat. Commun. 2018, 9, 524. [Google Scholar] [CrossRef]
- Xu, P.; Peng, J. Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal. Chem. 2008, 80, 3438–3444. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Tsuchiya, H.; Tanaka, K.; Saeki, Y. Methods to measure ubiquitin chain length and linkage. Methods Enzymol. 2019, 618, 105–133. [Google Scholar] [CrossRef]
- Enenkel, C.; Lehmann, A.; Kloetzel, P.M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998, 17, 6144–6154. [Google Scholar] [CrossRef]
- Wilkinson, C.R.; Wallace, M.; Morphew, M.; Perry, P.; Allshire, R.; Javerzat, J.P.; McIntosh, J.R.; Gordon, C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998, 17, 6465–6476. [Google Scholar] [CrossRef]
- Amsterdam, A.; Pitzer, F.; Baumeister, W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. USA 1993, 90, 99–103. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Arai, N.; Tanaka, K.; Saeki, Y. Cytoplasmic proteasomes are not indispensable for cell growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2013, 436, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.K.; Porras-Yakushi, T.R.; Reitsma, J.M.; Huber, F.M.; Sweredoski, M.J.; Hoelz, A.; Hess, S.; Deshaies, R.J. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife 2016, 5. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Prado, M.A.; Schmidt, P.J.; Sendamarai, A.K.; Wilson-Grady, J.T.; Min, M.; Campagna, D.R.; Tian, G.; Shi, Y.; Dederer, V.; et al. UBE2O remodels the proteome during terminal erythroid differentiation. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Yanagitani, K.; Juszkiewicz, S.; Hegde, R.S. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 2017, 357, 472–475. [Google Scholar] [CrossRef]
- Jacobson, A.D.; MacFadden, A.; Wu, Z.; Peng, J.; Liu, C.W. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell 2014, 25, 1824–1835. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Ali, R.; Jiou, J.; Fung, H.Y.J.; Burke, K.A.; Kim, S.J.; Lin, Y.; Peeples, W.B.; Saltzberg, D.; Soniat, M.; et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173, 693–705.e622. [Google Scholar] [CrossRef] [PubMed]
- Conicella, A.E.; Dignon, G.L.; Zerze, G.H.; Schmidt, H.B.; D’Ordine, A.M.; Kim, Y.C.; Rohatgi, R.; Ayala, Y.M.; Mittal, J.; Fawzi, N.L. TDP-43 alpha-helical structure tunes liquid-liquid phase separation and function. Proc. Natl. Acad. Sci. USA 2020, 117, 5883–5894. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef]
- Raasi, S.; Orlov, I.; Fleming, K.G.; Pickart, C.M. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J. Mol. Biol. 2004, 341, 1367–1379. [Google Scholar] [CrossRef]
- Dao, T.P.; Kolaitis, R.M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castañeda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.P.; Martyniak, B.; Canning, A.J.; Lei, Y.; Colicino, E.G.; Cosgrove, M.S.; Hehnly, H.; Castañeda, C.A. ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Structure 2019, 27, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018, 28, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Salin, B.; Daignan-Fornier, B.; Sagot, I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008, 181, 737–745. [Google Scholar] [CrossRef]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef]
- Janer, A.; Martin, E.; Muriel, M.P.; Latouche, M.; Fujigasaki, H.; Ruberg, M.; Brice, A.; Trottier, Y.; Sittler, A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J. Cell Biol. 2006, 174, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Turakhiya, A.; Meyer, S.R.; Marincola, G.; Böhm, S.; Vanselow, J.T.; Schlosser, A.; Hofmann, K.; Buchberger, A. ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol. Cell 2018, 70, 906–919.e907. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchiya, H.; Endo, A.; Saeki, Y. Multi-Step Ubiquitin Decoding Mechanism for Proteasomal Degradation. Pharmaceuticals 2020, 13, 128. https://doi.org/10.3390/ph13060128
Tsuchiya H, Endo A, Saeki Y. Multi-Step Ubiquitin Decoding Mechanism for Proteasomal Degradation. Pharmaceuticals. 2020; 13(6):128. https://doi.org/10.3390/ph13060128
Chicago/Turabian StyleTsuchiya, Hikaru, Akinori Endo, and Yasushi Saeki. 2020. "Multi-Step Ubiquitin Decoding Mechanism for Proteasomal Degradation" Pharmaceuticals 13, no. 6: 128. https://doi.org/10.3390/ph13060128
APA StyleTsuchiya, H., Endo, A., & Saeki, Y. (2020). Multi-Step Ubiquitin Decoding Mechanism for Proteasomal Degradation. Pharmaceuticals, 13(6), 128. https://doi.org/10.3390/ph13060128



