Abstract
Iron is an essential element required to support the health of organisms. This element is critical for regulating the activities of cellular enzymes including those involved in cellular metabolism and DNA replication. Mechanisms that underlie the tight control of iron levels are crucial in mediating the interaction between microorganisms and their host and hence, the spread of infection. Microorganisms including viruses, bacteria, and fungi have differing iron acquisition/utilization mechanisms to support their ability to acquire/use iron (e.g., from free iron and heme). These pathways of iron uptake are associated with promoting their growth and virulence and consequently, their pathogenicity. Thus, controlling microorganismal survival by limiting iron availability may prove feasible through the use of agents targeting their iron uptake pathways and/or use of iron chelators as a means to hinder development of infections. This review will serve to assimilate findings regarding iron and the pathogenicity of specific microorganisms, and furthermore, find whether treating infections mediated by such organisms via iron chelation approaches may have potential clinical benefit.
1. Introduction
1.1. Iron, an Essential Element for Survival of Both Hosts and Microorganisms
Iron is a key element needed to support fundamental cellular processes including oxygen transport, DNA replication, transcription, and metabolic processes in many living organisms [1,2]. It is also essential to support the growth, virulence, and pathogenicity of microorganisms such as viruses, microbes, and fungi [3,4], which can acquire iron from within its host environment.
Dietary iron in the host can be obtained in the form of heme from various sources including red meat, seafood, and poultry [5]. This heme iron is absorbed into cells through a mechanism that involves the Heme Carrier Protein (HCP1), a proton-coupled folate transporter (PCFT) [6]. Non-heme iron can be obtained predominantly from plant sources [6]. Non-heme iron absorption into cells can occur either as transferrin bound iron (TBI) or non-transferrin bound iron (NTBI) [7,8,9]. The absorption of iron into the bloodstream is primarily regulated by hepcidin (HAMP), a liver secreted peptide hormone [10].
In an adult human, hemoglobin from red blood cells (RBCs) contains approximately two-thirds of the total iron present in the body (~3–4 g) [5]. Not only is iron stored in liver cells and macrophages bound to ferritin, but it is also found in myoglobin of muscle cells [5]. Further, phagocytosis of RBCs by macrophages leads to the release of iron from hemoglobin and serves as a crucial source of iron [11]. Heme iron degradation involves the action of heme oxygenase 1 (HO-1) or heme oxygenase-2 (HO-2) [12], whereby the released free iron becomes part of the labile iron pool (LIP), a redox-active pool of intracellular iron [13].
The specific mechanisms that support the ability of microorganisms such as viruses, microbes, and fungi to uptake iron from sources in the host are discussed in this review, along with the effect of iron chelators which may potentially antagonize their growth and virulence.
1.2. Host Cell Iron Metabolic Pathway
It is well established that deregulated iron control can lead to detrimental effects on survival [1,2]. Since redox-active iron is a catalyst in electron transfer and free radical reactions, excessive amounts of free iron can deteriorate cell health (e.g., DNA damage, lipid peroxidation, and protein oxidation) [14]; therefore, a tightly regulated system is essential to appropriately balance intracellular iron levels [15]. This mechanism of control has been deciphered and involves a large array of mediators described below.
Transferrin, a carrier glycoprotein which binds to iron (as Fe3+-bound complex (TBI)), facilitates the transport of iron into cells via the transferrin receptor (CD71) [16]. Cellular entry of TBI occurs via an endocytic process which is followed by the release of iron from transferrin due to the reduced pH of the endosomal compartment. Subsequently, STEAP3 (Six Transmembrane Epithelial Antigen of Prostate 3) mediates reduction of the Fe3+ (ferric) to the Fe2+ (ferrous) form [17]. Once reduced, the iron is released from the endosomal compartment to the cytosol via endosomal DMT1 (Divalent Metal Transporter 1) [18].
The divalent metal transporter ZIP8 (Solute carrier family 39 member 8 (SLC39A8)) is one way through which NTBI can enter cells [19]. Another mechanism underlying NTBI uptake, specifically into small intestinal cells, involves the reduction of ferric iron via duodenal cytochrome b (DCYTB) [20] followed by its transport via cell surface localized DMT1 [21,22].
The imported iron (from either NTBI or TBI) can either (1) be stored in a complex with ferritin (FTN), (2) be added to the labile iron pool (LIP), (3) be exported extracellularly via ferroportin (FPN), or (4) be integrated within key enzymes involved in regulating cellular metabolic processes [23]. Extracellular export involving FPN is the only means of exporting iron out of cells and its levels are regulated by HAMP [1,2,23,24].
Iron-binding proteins in the host plasma (e.g., transferrin, haptoglobin, hemopexin, lactoferrin, lipocalin-1, and lipocalin-2) have the ability to sequester iron from various sources; their iron binding capacity can contribute to reducing the availability of extracellular iron that may support the growth and virulence of microorganisms within this environment [25]. Withholding of intracellular host iron (e.g., reduced iron uptake, increased storage of iron in ferritin, increased export of iron via FPN) may also contribute to reducing intracellular microorganismal growth [25]. Hence, tight regulation of the above described iron metabolic pathways is critical in mediating the balance between microorganisms and the host.
1.3. Iron Chelation and Associated Risks
In multiple diseases, iron chelation has been explored as a therapeutic regimen to reduce iron levels to promote health [26]. Efficacy of iron chelators depends on high membrane permeability and effectiveness of oral administration [27]. There are two major classes of iron chelators: (1) naturally occurring, e.g., Epigallocatechin-3-gallate (EGCG, found in green tea), phytic acid, curcuminoids, and (2) synthetic, e.g., Deferiprone (DFP or L1), Deferasirox (DFRA or DFX), 8-hydroxyquinoline derivatives such as VK-28 and M30 [28]. Some chelators are derived from bacterial sources including Desferioxamine (also known as deferoxamine (DFO), produced by Streptomyces pilosus) and Desferrithiocin ((DFT), a tridentate siderophore, produced by Streptomyces antibioticus) [28,29,30]. Moreover, phytochelators, obtained from plant components including vegetables and fruits, elicit iron-chelating activities; polyphenols are one such class with the ability to chelate iron with high affinity and promote health [31,32]. Mammalian-derived physiological iron chelators include (1) transferrin, found in blood plasma and involved in body-wide iron transport and (2) lactoferrin, enriched in neutrophils and in bodily secretions [32,33,34]. Not only do these physiological iron chelators bind iron but they also elicit anti-microbial activities to hinder the propagation of microorganisms [32,33,34].
A subset of FDA-approved iron chelators includes DFO, DFRA, and DFP [26] and these have different means of patient administration (e.g., oral versus intravenous) with divergent efficacies and blood brain barrier accessibilities [32]. Some of the above described iron chelators have shown success in combinatorial treatment strategies with antibiotics, anti-virals, and anti-fungals. However, the clinical applications of these iron chelators are noted to be associated with health risks. DFO, the first clinically applied iron chelator which is administered intravenously, is associated with side effects affecting vision, hearing, and kidney function; further, in some cases, Yersinia and Klebsiella infections may develop [35]. DFP, administered orally, is also associated with some health risks including alterations within the immune system (e.g., neutropenia, agranulocytosis, thrombocytopenia) in addition to arthropathy and adverse effects on liver function [35]. DFRA, also administered orally, is associated with adverse effects on the liver, digestive system, and skin [35]. Furthermore, the application of these iron chelators may result in anemia [35].
In this review, we summarize findings involving the application of iron chelators primarily in the context of infectious diseases (see Figure 1 for an overview of iron acquisition pathways).
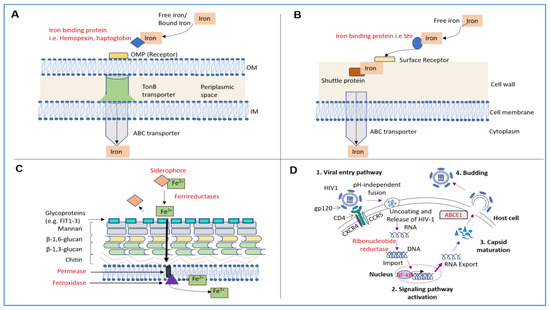
Figure 1.
Mechanisms of Iron Acquisition in Bacteria and Fungi as well as Mechanisms of Iron Utilization in Viruses. (A) In Gram-negative bacteria, the iron acquisition involves uptake of free iron from the extracellular environment. Specific proteins that bind heme or hemoglobin (e.g., hemopexin or haptoglobin) are secreted from the bacteria, to then bind to free iron or heme [36]. These complexes then interact with outer membrane protein (OMP) receptors on the surface of the outer membrane (OM) of the bacterial cell wall [37]. The iron is then moved to the TonB–dependent receptor complex [36]; after the iron reaches the ABC transporter, it finally passes the inner membrane into the bacterial cytoplasm [37]. (B) In Gram-positive bacteria, secreted specific iron/heme binding proteins interact with heme [37], following which, the iron binding protein localizes to the surface of the bacterial cell wall to bind to a specialized cell surface receptor [38]. Next, specific permeases enable the translocation of this complex across the bacterial cell wall [39]. The iron-bound molecule is subsequently transferred to a shuttle protein that then guides the iron into the ABC transporter, an integral membrane protein [39]. The ABC transporter translocates the iron-bound molecule into the bacterial cytoplasm [39]. (C) In fungi, the iron uptake system involves reductive and/or non-reductive mechanisms [40]. In the reductive iron assimilation pathway, iron acquisition is initiated via siderophores which bind to ferric iron; the iron is subsequently reduced via ferrireductases and released from the siderophores as the ferrous form [40]. The iron is then translocated to glycoproteins on the surface of the fungal cell wall and the uptake of iron is mediated by permeases followed by oxidation of ferrous to ferric iron via ferroxidase [41]. (D) HIV replication is shown herein, as an example. There exists multiple pathways for viral-host cell entry; specifically, HIV entry into a host cell requires interaction of the HIV gp120 with the CD4 receptor (associated with CCR5 or CXCR4 co-receptors) [42]; this is followed by the pH-independent fusion of the virus with the host cell membrane. Subsequently, the viral RNA is released into the cell and reverse transcribed into DNA via ribonucleotide reductase (RNR) and then imported into the nucleus [43]. Iron can increase NF-kB activation leading to upregulation of HIV gene expression. Finally, the iron-dependent ATPase transporter ABCE1 promotes HIV-1 capsid maturation [4].
2. RNA-Based Viral Infections
Overview—Viruses have adapted themselves to use host cell components to propagate. In contrast to Hepatitis C Virus (HCV) and Human Immunodeficiency Virus (HIV) which utilize SR-B1 and CD4 receptor, respectively, for host cell entry, New World Arenaviruses enter host cells by binding to transferrin receptors, a host cell receptor also responsible for iron uptake [42,44]. To support viral replication, ribonucleotide reductase (RNR), responsible for generating deoxyribonucleotides from ribonucleotides, is essential and requires iron to support its activity [45]. Host cell signaling pathways may also respond to altered iron availability (e.g., increased NF-kB activation) to support viral infections [46]. Likewise, the maturation of viral particles requires the iron-binding ATP binding cassette subfamily E member 1 (ATPase ABCE1) [4,47]. Therefore, iron appears to be a critical element to support multiple elements supporting propagation of a virus including its entry, replication, and maturation. Evidence supporting the use of iron chelators to antagonize viral propagation is presented herein.
2.1. Hepatitis C Virus (HCV)
Viral Genome and Structure—Hepatitis C Virus is a member of the Flaviviridae family and is a single-stranded positive sense virus with a diameter of 50–80 nm [48]. Its genome is comprised of 10 elements and codes for structural and non-structural components: (1) Core, capsid, (2) E1 and E2, envelope glycoproteins, (3) p7, viroporin (this protein forms pores in the host cell membrane to enable viral propagation) [49] and assembly factor, (4) NS2, autoprotease and assembly factor, (5) NS3, serine protease and helicase, assembly factor, (6) NS4A, NS3 protease co-factor, (7) NS4B, scaffold protein of the replication complex, (8) NS5A, regulator of replication and viral assembly, and (9) NS5B, DNA-dependent RNA polymerase [50]. Its entry into host cells is a complex process and involves a multitude of host cell factors such as CLDN1 (Claudin 1), OCLN (Occludin), CD81 (Cluster of Differentiation 81), and SRB1 (Scavenger receptor class B type 1) [48].
Health Implications and Current Treatments—HCV infections are associated with inflammation potentially leading to liver fibrosis, cirrhosis, and hepatocellular carcinoma [51]. Although HCV is considered curable, the health implications and risk of hepatocellular carcinoma remain a concern [51]. Interferon (IFN) has been the sole treatment regimen for HCV infected patients with only 15%–20% eliciting a sustained virological response following 11 months of treatment [52]. However, ribavirin, an immunomodulating agent described as a synthetic guanosine analogue, not only directly inhibits viral replication but also promotes efficacy of IFN, relapse response in patients infected with HCV, and a sustained virological response [53,54]. Specifically, recent data demonstrate that ribavirin promotes the JAK-STAT signaling cascade to enhance anti-viral responses against HCV [53]. Additional pathways that are activated in response to HCV include EGFR and TGF-β activation, which may contribute to disease progression and potentially offer additional targets for therapy [55]. Identification of further therapeutic regimens to combat the health complications of HCV are needed.
Iron Contribution to HCV Infections—In liver-biopsy specimens from infected HCV patients, a positive correlation between hepatic iron content and HCV was reported [56]. Efforts were also made to reduce iron levels by phlebotomy in infected HCV patients to improve outcomes. For example, in male patients in whom anti-viral therapy was ineffective, phlebotomy (administered every 1 to 3 months over a 2 year period) improved the liver histology [57]. In HCV patients characterized by elevated levels of serum alanine aminotransferase (ALT, a marker of liver damage [58]) and iron deposits in their livers, phlebotomy (performed every week or monthly over a 9 month period) improved their liver function [59]. This is supported by an independent study in which patients (resistant to IFN-α with no abnormal profile of liver iron content) responded positively to phlebotomy (over a 2 week period) with improved ALT activity [52]. However, it remains unclear whether phlebotomy can reduce HCV viral load since it was either not reported or not determined in the above described studies [52,57,59].
In in vitro studies, supplementation with FeSO4 for two days was found to increase the replicative capacity of HIV (as measured by quantification of the viral RNA) in hepatocytes [60]. Further, the iron-induced translation of HCV was mediated by factors involved in the initiation of translation including eIF3 (translation initiation factor 3) in HepG2 cells [56] and La proteins (which bind to the internal ribosome entry site (IRES) to regulate initiation of translation of HCV RNA) [61,62,63]. With respect to iron chelators, DFRA was reported to antagonize HCV-induced upregulation of these translation initiation factors in Huh-7 cells [63]; further, antisense phosphorothioate oligodeoxynucleotides targeting these initiation factors reduced iron-induced HCV translation in Huh-7 cells [63]. In contrast, iron, presented as a complex with salicylaldehyde isonicotinoyl hydrazine (lipophilic tridentate iron chelator, Fe-SIH), could mediate anti-viral effects by reducing expression of viral proteins (NS3 and Core) in Huh7.5.1 cells [64].
Although it has been suggested that therapies that reduce iron levels could be utilized as adjuvant to existing HCV therapies [60], additional studies are needed to provide support for the use of iron chelators as an adjuvant therapeutic regimen in HCV-infected patients.
2.2. Human Immunodeficiency Virus (HIV)
Viral Genome and Structure—Human immunodeficiency virus is a member of the Retroviridae family and is a single stranded RNA virus with a diameter of 100 nm [43]. HIV contains 9 elements in its genome: (1) Gag, codes for the core structural proteins (p24, p7, and p6) and the matrix (p17), (2) Pol, codes for viral replication enzymes including reverse transcriptase, integrase, and protease, (3) Vif, encodes a protein that promotes infectivity of viral progeny, (4) Vpr, codes for a protein that causes cell cycle arrest, (5) Vpu, codes for a protein involved in the release of the viral particle, (6) Tat, codes for a protein which is involved in HIV gene expression, (7) Rev, codes for a protein that allows the export of the RNA from the nucleus into the cytoplasm, (8) Env, encodes the glycoproteins in the envelope (gp120 and gp41), and (9) Nef, codes for a protein that can modulate signaling and promote viral budding [43]. The viral budding process leads to host cell lysis; however, during latency, the viral DNA lies dormant in the nucleus of specific host cells including CD4+ T Cells, which is referred to as a cellular reservoir [65,66].
Health Implications and Current Treatments—HIV can lead to acquired immunodeficiency syndrome (AIDS), a terminal stage of HIV-infections, in which patients are afflicted by opportunistic infections and cancer [67]. There is presently no cure for these patients, who are characterized by a progressive reduction of white blood cells (CD4+ T cells) as a result of the deterioration of tissues that generate lymphocytes (e.g., bone marrow and thymus) [68] and opportunistic infections (e.g., Candidiasis, Cryptococcosis, amongst others) [69] which eventually lead to mortality, if left untreated [70]. Current treatment regimens include anti-retroviral therapy (ART) which integrates three drugs including two nucleoside reverse transcriptase inhibitors, which compete with deoxynucleotides incorporated into DNA that is being replicated (e.g., emtricitabine and tenofovir), and one non-nucleoside reverse transcriptase inhibitor, integrase inhibitor, or protease inhibitor (e.g., raltegravir) [71]. Regrettably, the persistence of the virus in a cellular reservoir in latent form hinders its complete elimination even with ART treatment regimens [70]. Thus, identification of other targets to effectively deplete these reservoir holding cells are direly needed for an effective cure.
Iron Contribution to HIV Infections—In patients with HIV infections, excessive iron content in their serum [72], tissues (e.g., bone marrow [73,74], brain white matter [75,76], muscle [77]), and their cells (macrophages and microglia) [75] is suggested to contribute to the pathogenesis of the disease [78]. Further, a link between iron and HIV disease progression was identified in a male HIV patient affected by the iron overload condition, hereditary haemochromatosis (characterized by a mutation at C282Y in the HFE gene); specifically, a major reduction in viral particles was noted following an extensive 18-month phlebotomy period [79].
At a cellular level as determined from in vitro studies, the addition of exogenous iron or modulation of key modulators of host iron metabolic pathways could alter HIV-1 replication and transcription. Specifically, overexpression of FPN1, the iron export receptor, decreased transcription of HIV-1 in HEK293 cells [80]. In contrast, the addition of HAMP, which downregulates FPN1 and increases LIP, antagonized viral transcription in promonocytic cells as well as macrophages and CD4+ T cells [80,81]. Under conditions of excess ferrous sulfate heptahydrate, the survival of HIV-infected T-lymphoid CEM-syncytial sensitive cells was reduced and correlated with elevated viral replication (p24, which is a core protein of HIV encoded by gag) and reverse transcriptase activity in the cell supernatants [82]; these cellular responses were opposed by the iron chelator, DFO [82].
The mechanisms underlying these iron-associated cellular effects on HIV-1 transcription were elucidated to be mediated through NF-kB (a transcription factor that plays a role in regulating multiple cellular activities), which binds to the long-terminal repeat sequence of HIV (the control center for regulation of HIV gene expression with binding sequences for both host and viral proteins) on NF-kB response elements [83]. This pathway could be opposed by the iron chelator, DFO, in specific cell types (namely, U1 (an HIV-infected U937, a pro-monocytic myeloid leukemia cell line) and ACH-2 cells (HIV-infected acute lymphoblastic leukemia T cell line)); this was demonstrated via a gel shift assay in which DFO led to a marked reduction in NF-kB retardation complex [46].
Inhibition of the iron-dependent DNA replication enzyme, RNR, with iron chelators (DFRA and DFP) in lymphocytes could also alter HIV viral replication [84,85]. Bleomycin (BLM, an antibiotic isolated from Streptomyces verticillus which forms iron complexes generating ROS that leads to base modifications in the viral DNA) could also reduce the replicative capacity of HIV without affecting cellular viability in PBL (peripheral blood lymphocytes); whether the effects of BLM is due to iron-chelation activity is unclear [85]. Furthermore, BLM and the commonly used iron chelators DFO and DFP were capable of reducing the expression of the viral capsid core protein (p24) in macrophages and PBL [85]. Iron chelators with comparatively higher affinity for iron, CP502 and CP511 (bidentate chelators of the 3-hydroxypyridin-4-one family) were also effective in reducing viral replication (viral capsid core protein p24) by altering cellular viability (3H-thymidine) of peripheral blood lymphocytes [86].
In vitro cell studies were performed to identify changes in the expression and activities of cell cycle mediators by iron chelators in HIV-1 infected cells. Specifically, the iron chelators DFO and 311 (2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone) reduced HIV-1 viral transcription by modulating protein expression of cyclin-dependent kinases (e.g., CDK2, a mediator in cell cycle progression) in human lymphoid CEM cells [87]. Moreover, 311 and yet another iron chelator, ICL670 (4-[3,5-bis-(hydroxyphenyl)-1,2,4-triazol-1-yl]-benzoic acid), hindered HIV-1 transcription of the Tat gene by reducing CDK2 and CDK9 kinase activity on specific target proteins including cyclin T1 and the C-terminal domain of RNA polymerase II in multiple cell types including HeLa-CD4-LTR-β-gal cells, 293T cells, and CEM cells [87]. This finding is of particular importance since the Tat protein plays a key role in activating the latent virus by physically binding to CDK2 and CDK9 complexes [87]. Other novel iron chelators, including Phenyl-1-Pyridin-2yl-Ethanone-Based, inhibited CDK2 activity, reduced CDK9 levels, and increased IkBα and cytoplasmic NF-kB to mediate reduction in HIV-1 transcription of the B subtype in infected T cells [88].
Although there appears to be an abundance of data supporting the use of iron chelators in HIV-1 infected cell lines, additional research is needed to enable the use of iron chelators in clinical treatment strategies.
3. Bacterial Infections
Physical Characteristics of Gram-negative and Gram-positive Bacteria with Relevance to Iron Uptake—It is well established that iron acquisition is critical in promoting the growth and virulence of numerous pathogenic bacteria [32]. Since antibiotic resistance is of major concern in the treatment of pathogenic bacterial-induced infections, novel treatment agents are direly needed. Both Gram-negative and Gram-positive bacteria have well adapted strategies for iron uptake including specific surface chaperone proteins for acquiring heme, receptors, and ABC (ATP-binding cassette) transporters for membrane translocation, as described in detail in [89,90,91]. The iron uptake mechanisms differ between Gram-negative and Gram-positive bacteria and these differences are contributed by their physical characteristics of the outer cell membrane [92]. Specifically, Gram-positive microbes are characterized by a thick layer of peptidoglycan incorporated within the cell wall along with the extracellular exposure of teichoic acids and lipoteichoic acids as well as a diminished periplasmic space volume [90]. In contrast, Gram-negative microbes contain both an outer and inner membrane along with a larger periplasmic space volume [92]. Iron uptake mechanisms that are common across both Gram-negative and Gram-positive microbes include the involvement of ABC transporters [93].
Iron Uptake Pathway in Gram-negative Bacteria—Greater than 30 outer membrane protein (OMP) heme receptors (involved in transporting heme intracellularly) have been characterized across a wide variety of Gram-negative microbes [94]. Some specific molecules that are engaged in this process include those involved in the direct binding of heme (e.g., hemopexin) or hemoglobin (e.g., haptoglobin) to OMP receptors on the bacterial cell wall [36,37]. After delivery of the heme to the periplasmic membrane, the heme is then transferred via the ABC transporters to the cytoplasmic compartment [37]. Another acquisition mechanism involves hemophores, which interacts with free heme in the external environment and transports it to the surface of the bacterial cell membrane [37]. When the heme is transported to the bacterial cell surface, it then interacts with the TonB dependent transport pathway [37,95,96]. As a specific example, the heme acquisition system A (HasA) participates in the heme-uptake process in the pathogenic microbe, Pseudomonas aeruginosa, which we discuss in greater detail below [97]. Another example includes the heme/hemopexin utilization (HxuA) pathway which is involved in pathogenic microbes that are deficient in heme production [98]. The ferric uptake regulator (Fur) protein, a transcriptional regulator of iron uptake genes, is a key mediator of iron regulation in Gram-negative microbes as well [99].
Iron Uptake Pathway in Gram-positive Bacteria—In contrast to Gram-negative microbes, far fewer details have been uncovered with respect to the mechanisms underlying heme uptake in Gram-positive microbes. However, the HemT-like lipoprotein, HmuT, has been identified to participate in this process, specifically in Corynebacterium diphtheriae, a well-studied Gram-negative microbe [100]. The components of the ABC transporter pathway in Streptococcus pyogenes, another well-studied Gram-negative microbe, involves the Shr protein (which binds heme), the streptococcal heme-binding protein Shp (which relays the heme for transport across the bacterial envelope), SiaA (which is the heme-binding lipoprotein), SiaB (membrane permease), and SiaC (ATPase) [38,39]. A more well understood mechanism of the pathogenic bacterium, Staphylococcus Aureus, has been identified which uses hemolysins in its pursuit to acquire bound heme [101].
3.1. Pseudomonas Aeruginosa: A Gram-Negative Microbe Associated with Wound Infections and Cystic Fibrosis
Bacterial Features—Pseudomonas aeruginosa, a multi-drug resistant pathogen, has a genome size of 5.5–7 Mbp with the capacity to express genes underlying resistant phenotypes [102]. Together with complex metabolic processes, these features support its propagation in unfavorable environments [103]. The pathogenicity of P. aeruginosa is mediated by structural components including flagellum and pili as well as cell surface glycolipids and lectins that are involved in bacterial movement and adhesion to host cells [104]. Further, secretion of virulence factors is mediated by quorum sensing pathways leading to secretion of elastases (proteases) into the host environment [104]. In addition, P. aeruginosa is capable of injecting cytotoxins into host cells [104].
Contribution of Iron—Several iron uptake mechanisms are involved in mediating P. aeruginosa growth properties [105]. The pyoverdine (Pvd) and pyochelin (Pch) siderophores (of which pyoverdine has a higher iron affinity) are involved in the movement of extracellular iron into P. aeruginosa [106,107] using FptA and FpvA (outer membrane proteins) [108]. The addition of Pch with P. aeruginosa injected intraperitoneally in Swiss–Webster mice resulted in increased virulence [109]. Using immunosuppressed mice, mutant strains of P. aeruginosa deficient in Pvd or Pch/Pvd inoculated intranasally, elicited reduced growth in the pulmonary tissue coinciding with decreased virulence [110].
In wounds infected with P. aeruginosa, the rate of repair is diminished in multiple animal models (e.g., rabbit, murine, pig in vivo models) [104]. Specifically, in a murine wound model (in which a muscle was injured in the right rectus abdominus to which P. aeruginosa was applied), transcriptional profiling identified 7 out of 136 differentially expressed genes that were involved in pyochelin biosynthesis, including the pyochelin receptor fptA, pchH, pchG, pchE, pchD, pchB, and pchC (the biosynthesis of Pch requires the iron-regulated pchDCBA operon) [106]. In addition, the iron-sulfur cluster genes were upregulated [111]. These findings suggest that iron uptake in this bacteria contributes to its pathogenic activity within infected wounds [111]. Other pathways that are involved in iron uptake in P. aeruginosa include the citrate-mediated Fe3+ uptake pathways that engages FecA, an outer membrane ferric citrate receptor, the FeoB transporter, and the PcoA, periplasmic ferroxidase [112]. The process of iron uptake from heme involves the TonB system which involves HasA, an extracellular heme-binding protein, HasR, and PhuR encoded on the phuSTUVW operon (a gene cluster encoding an outer membrane receptor and specific ABC transporters for heme and hemoglobin uptake) [113].
In addition to wound infections, P. aeruginosa infections in lungs are frequent in patients afflicted with cystic fibrosis [114]. In an effort to determine whether iron chelation may hinder the development of such infections in cystic fibrosis, administration of aerosolized bovine lactoferrin (bLF) was performed in a mouse model of cystic fibrosis with P. aeruginosa infection. Neutrophil numbers, pro-inflammatory cytokines, and microbial numbers were reduced with bLF treatment [115]. Although lactoferrin is considered a natural anti-microbial agent present in secretions of the airways, which also has the ability to bind iron [115], it is unclear whether its iron binding potential is responsible for the observed outcomes. Nonetheless, bLF may have potential as a clinical agent to alleviate pathogenic infections and inflammation in cystic fibrosis patients.
Manuka honey, produced from the nectar of Leptospermum scoparium (manuka bush), was discovered to elicit anti-microbial activity against multiple pathogens [116]. Specifically, the honey could hinder the growth of several pathogenic microbes including P. aeruginosa, Escherichia coli, and S. aureus [116]. Although the ferrozine-based iron chelation assay was utilized to determine that the honey mediates iron chelating activity [116] and the honey simulated an environment of limiting iron availability [116], it remains unclear whether the anti-microbial effect of the Manuka honey is due to its iron chelation ability. Further investigation is needed to not only identify the potential iron chelating component in the honey but to also further investigate whether mimicking an environment with low iron content may potentially diminish the growth of P. aeruginosa, which could potentially be utilized as a strategy to overcome antibiotic resistance.
3.2. Porphyromonas Gingivalis, Prevotella Intermedia, and Fusobacterium Nucleatum: Bacteria Associated with Periodontitis
Bacterial features—The oral microbiome can be composed of up to 700 species; an imbalance of these species could lead to the development of periodontitis [117]. Specifically, Porphyromonas gingivalis and Fusobacterium nucleatum are critically important in the amalgation of late and early colonizers within the oral cavity [117]. Furthermore, P. gingivalis and Prevotella intermedia are mutualistic in terms of heme acquisition, as described in detail below. P. gingivalis (Strain W83) is a Gram-negative oral bacteria with a genome size of 2,343,479 bp. P. intermedia is described as a anaerobe that is Gram-negative with a genome size (OMA14 strain) which is represented by two circular chromosomes of 2,280,262 and 867,855 bp, respectively [118]. Genome sequencing of five subspecies of F. nucleatum, a Gram-negative anaerobic microbe, has identified a range of genome sizes, namely 1.84–2.7 Mbp [119,120]. Within the periodontal pocket, in response to an altered microbiome, an inflammatory fluid is generated, called gingival crevicular fluid [121]; this exudate contains iron containing proteins such as hemoglobin, lactoferrin, and transferrin, which may contribute to the outgrowth of pathogenic oral bacteria [122].
Contribution of iron—Since P. gingivalis is deficient in siderophores [123,124] as well as specific heme precursor enzymes [125,126,127], it acquires heme from exogenous sources [128]. This bacteria acquires iron using (1) specific outer membrane receptors, (2) proteases, and (3) lipoproteins [128]. With respect to outer membrane receptors, P. gingivalis contains proteins that physically interact with hemoglobin (e.g., haptoglobin) and heme (e.g., hemopexin) in gingival crevicular fluid [129,130,131]. For proteases, specific genes identified in this bacteria include rgpA (which codes for gingipain that cleaves arginyl peptide bonds), hagA (which codes for hemagglutinin A), and kgp (which codes for gingipain that cleaves lysyl peptide bonds) [130,132]. One mutualistic behavior between P. gingivalis and P. intermedia involves the process of heme acquisition; specifically, this involves the HmuY protein in P. gingivalis and the proteolytic activity of P. intermedia [133]. The hemolytic activity of P. intermedia, which increases free hemoglobin, is thus proposed to provide an optimal growth environment for P. gingivalis [133,134]. In addition, mutualism via InpA (proteolytically oxidizing hemoglobin in P. intermedia), supports iron (III) protoporphyrin IX generation via hmuY from P. gingivalis [133,135].
Subgingival plaque P. gingivalis could be effectively inhibited in its growth rate and adhesiveness by the iron chelator, DFO, by reducing its ability to accumulate hemin, Fe3+-protoporphyrin IX, a virulence factor [136]. The effect of iron chelating agents were also investigated on P. intermedia, another Gram-negative microbe present in periodontal lesions [137]. DFRA could effectively inhibit its growth and biofilm-forming activities [137]. A blueberry extract, containing high levels of flavonoids which has potential to elicit iron chelating activity, was able to antagonize the growth, biofilm formation, and proteolytic activity (decreased matrix metalloproteinase secretion) of F. nucleatum [138]. The microbial activity of this microbe could also be opposed by bioactive components present in green and black tea (e.g., EGCG and theaflavins) which also appear to be associated with iron chelating activities [139].
Over the past two decades, there has been a rise in antibiotic-resistant infections, which could be attributed to the overuse of antibiotics (with evidence of country dependency [140]) and antibiotic resistance gene transfer to the bacteria present within the oral cavity [141,142]. Although the above described natural agents appear to elicit anti-microbial activities which may offer some protective health benefits against periodontitis, it remains unclear whether their effects are due to their iron-chelation ability. Thus, further investigation is needed to identify the iron chelating components in the blueberry extract and the tea as well as address their potential in mediating anti-microbial activities in these pathogenic microbes.
3.3. Streptococcus Pneumoniae: A Gram-Positive Bacteria
Bacterial Features—Infections that are due to Streptococcus pneumoniae, a pathogen with a core genome size of 1,536,569 bp [143], that causes pneumonia, meningitis, and bacteremia, is associated with multiple serotypes and thus a search for candidates to target as a treatment approach remains an ongoing effort [144].
Contribution of Iron—S. pneumoniae does not express siderophores, which is unlike other microbes [144,145]. To overcome this limitation, pneumolysin is released from S. pneumoniae (as a result of autolysin (a cell wall degrading enzyme activity [146])) to elicit hemolytic activity; this activity is responsible for the lysis of erythrocytes leading to the release of heme [147]. S. pneumoniae can acquire iron from hemoglobin and heme-binding proteins; specifically, receptors present on the surface of S. pneumoniae including pit1, pit2, and ABC transporters, are involved in the uptake of released iron from heme [148]. A mutant strain of S. pneumoniae that was defective in hemin uptake was found to diminish virulence in an intraperitoneally injected mouse model [145]. S. pneumoniae contains two operons, namely piuBCDA and piaABCD (prior naming system, pit1BCDA and pit2ABCD), which code for proteins involved in iron uptake [149]. Pit1 and pit2, which are S. Pneumoniae loci which code for an ABC transporter, enables the Gram-positive bacteria to acquire iron from hemoglobin [148]. Using two different mouse models (pneumonia model in which S. pneumoniae was inoculated intranasally and a systemic model in which S. pneumoniae was inoculated via the intraperitoneal cavity), double knockouts of pit and pit2 led to a marked reduction in S. pneumoniae virulence [148]. A proteomic study, using parallel metabolic pulse labeling in S. pneumoniae, was performed in the presence of the iron chelator, 2,2′-bipyridine, to limit iron content within the environment of the pathogen [144]. Under this condition, transport and binding proteins involved in S. pneumoniae pathogenesis as well as those involved in cell division (FtsA, FtsZ, and StkP) were downregulated [144]; in contrast, molecules involved in iron uptake were increased including PiuA (the lipoprotein component of ABC transporters) [81].
Altogether, these studies suggest that targeting the iron uptake pathways and/or use of iron chelators could antagonize the growth and virulence of S. pneumoniae.
3.4. Mycobacterium Tuberculosis
Bacterial Features—Mycobacterium tuberculosis is a pathogen with a genome size (H37Rv strain) of 44.1 Mbp [150]. This bacterium is predominantly intracellular and is the causative factor in tuberculosis, a highly contagious disease [151]. However, M. tuberculosis can also be disseminated extracellularly into the blood to secondary locations (e.g., central nervous and lymphatic systems) [151]. Current treatment regimens include a cocktail containing isoniazid, rifampicin, pyrazinamide, ethambutol, and streptomycin; regrettably, these agents can lead to adverse effects including liver damage and the development of M. tuberculosis resistant strains [151].
Contribution of Iron—Infections mediated by M. tuberculosis activate a host defense pathway that limits serum iron availability causing “anemia of chronic disease” [152]. To overcome this host limitation, M. tuberculosis has evolved mechanisms to acquire intracellular iron within host macrophages and myeloid dendritic cells via a siderophore-mediated process involving mycobactin and carboxymycobactin [152,153]. An endosome/lysosome metal ion transporter, NRAMP1 or natural resistance-associated macrophage protein 1, contributes to iron uptake in the bacterium that is located in phagosomes [152]. Thus, it has been suggested that this pathway could be a potential target for drug treatment against tuberculosis, a disease associated with increased resistance to current treatment strategies. A novel pyrazolopyridinone, PZP, which elicits intracellular iron chelator activity, could hinder the growth of M. tuberculosis [153]. Using in vivo guinea pig and mouse models with M. tuberculosis strains harboring loss of iron acquisition and uptake systems (located within the ESX-3 type VII secretion system) to restrict iron accessibility, the bacterial burden was markedly reduced [153].
Further work is needed to determine clinical efficacy of targeting the above described pathway in patients infected with M. tuberculosis.
4. Fungal Infections
Fungal Cell Structure—Since the fungal cell wall is the primary barrier that is encountered in response to anti-fungals, its characteristics have been investigated in an effort to understand its role in mediating anti-fungal resistance [154]. It is important to note that the fungal cell wall is comprised of two membrane components. The inner membrane is composed of glucans and chitin and provides not only structure but is tolerant of the immense internal forces arising from the fungal cytoplasm [154]. The outer membrane is composed of glycoproteins with both N and O-linked carbohydrates [154] and its composition may be altered under variable environmental conditions altering the ability of nutrients (including iron) to pass through to the plasma membrane [40].
Fungal Iron Uptake Pathways—The virulence and growth potential of fungi depend on specific metal ions, such as copper, zinc, manganese, nickel, and of relevance to this review, iron [155]. Although the iron metabolic pathway has been well delineated in Saccharomyces cerevisiae, a model organism [156], greater attention has been placed in understanding the iron pathways in pathogenic fungi including Cryptococcus neoformans and Aspergillus fumigatus with the past decade [40]. The mechanisms of iron uptake vary in different pathogenic fungi under divergent environmental iron conditions [157].
The most commonly described mechanisms across fungal species include the (1) reductive iron assimilation (RIA) and (2) non-reductive (siderophore-mediated) iron uptake [158]. The RIA process involves reduction of ferric iron to the ferrous form via ferrireductases (e.g., FRE genes); this is then followed by its uptake into the cells by the permease FTR1 along with the oxidation of the iron via the activity of a ferroxidase (Fet3) [159]. With respect to S. cerevisiae, it primarily employs both low-affinity and high-affinity non-reductive iron transport systems in conjunction with metalloreductases (which reduce iron) and siderophore mediated iron transport mechanisms [40,41,157]. Many fungal species utilize siderophores in iron acquisition; such fungi transport iron-bound siderophore complexes via transmembrane transporters and multivesicular bodies [40,159]. These siderophores are concentrated within the fungal cell wall and within the fungal periplasmic space [40]; the presence of cell wall glycoproteins (e.g., FIT) enables the iron uptake via a siderophore-mediated process [40]. The majority of fungal siderophores are classified into two categories: (1) hydroxamates (e.g., rhodotorulic acid, coprogens, ferrichromes, and fusarinines) and (2) polycarboxylates [157]. Additional sources of fungal iron include hemin [157], heme, and hemoglobin which involve an iron uptake pathway that differs to the ones described above [159]. Specifically, a family of proteins which contain a cysteine-rich Common in Fungal Extracellular Membrane (CFEM) domain are involved (e.g., Rbt, Pga7, and Csa2) in C. albicans and S. cerevisiae and components of the endosomal sorting (ESCRT-I) complex are involved in C. neoformans [159].
Due to the vast array of iron acquisition mechanisms employed by fungi, these pathways involved in iron accumulation contributing to fungal virulence could be potentially targeted to formulate novel treatment strategies.
4.1. Cryptococcus Neoformans
Fungal Features—Cryptococcus neoformans, a basidiomycete fungus involved in meningitis, can lead to a poor outcome in patients who are immunocompromised [160]. The treatments are limited to anti-fungal agents such as amphotericin B (which binds to sterol in fungal cell membranes, leading to pore formation and ultimately fungal death [161]) and fluconazole (which disrupts the fungal membrane and ergosterol synthesis) [162]). However, these drugs elicit side effects including kidney damage [160] and thus, other agents are needed to improve treatment responses.
Contribution of Iron—It is well established that the iron permease (Cft1) and ferroxidase (Cfo1) are involved in the iron uptake pathway in C. neoformans [160]. Cft1 mutant strains of this fungi leads to reduced growth, reduced intracellular iron, and elevated susceptibility to miconazole and amphotericin B [163].
In a mouse model, intranasal inoculation of C. neoformans containing a mutation in cft1 markedly reduced fungal virulence [163]. Intranasal instillation of C. neoformans with a deficiency of Cfo1 (but not Cfo2) into mice led to a reduction in virulence but also increased the sensitivity to amphotericin B and fluconazole [164]. The mechanism underlying this finding was suggested to be due to reduction of the cofactor, heme, which is needed for enzymes involved in ergosterol biosynthesis such as Erg11, an anti-fungal drug target [164]. Further, the addition of heme or ferrioxamine (a siderophore) reduced the drug sensitivity (amphotericin B and fluconazole) in the microbe [164]. In support, decreased virulence of C. neoformans was noted with genetic deficits in these fungal iron uptake proteins coinciding with reduced resistance to the anti-fungal agents [160,163,164].
Chelation of intracellular iron with bathophenanthroline disulfonate (BPS) or DFP, in combination with fluconazole or miconazole, synergistically altered the growth capacity of C. neoformans in vitro [160]. Based on these findings, the use of iron chelators or targeting the above described iron mediators may offer alternative strategies of treatment as adjuvant drugs along with the anti-fungals [160].
4.2. Aspergillus Fumigatus
Fungal Features—Aspergillus fumigatus, an airborne saprophytic fungus that is responsible for the development of invasive pulmonary aspergillosis, is common in patients characterized by iron overload and with blood cancers [165]. First-line treatment regimens include the application of systemic antifungals such as voriconazole and isavuconazole; however, resistance to these agents has been reported [166].
Contribution of Iron—Since A. fumigatus is unable to acquire host iron directly, specifically from transferrin, ferritin, or heme, it has developed efficient iron uptake mechanisms including (1) RIA and (2) siderophore-mediated processes [167]. As described earlier, RIA involves reducing Fe3+ to Fe2+ via ferrireductases that are present within the fungal cell membrane; the imported iron is then oxidized by a ferroxidase and a permease, FetC and FtrA, respectively [168]. A. fumigatus is also capable of acquiring iron via extracellular siderophores, fusarinine C (FsC) and triacetylfusarinine C (TAFC), which are then imported into the fungus via transporters in the membrane (SIT, siderophore iron transporter) [169]. Although genetic inactivation of RIA does not alter fungal virulence, SidA, which is a critical enzyme in the biosynthesis of siderophores, was necessary for mediating virulence in a murine model involving intranasal instillation [169]. The biosynthesis of siderophores in A. fumigatus is regulated by the GATA transcriptional regulator, SreA [170]; however, A. fumigatus deficient in SreA does not differ to the wild type strain in terms of virulence [170]. Using two pulmonary invasive aspergillosis murine models (leucopenic mice that are immunosuppressed with cortisone acetate and cyclophosphamide as well as a non-leucopenic model that is immunosuppressed with cortisone acetate), the deficiency of HapX in A. fumigatus led to a reduction in the spread of the fungal infection [171]. HapX is a bZip (basic leucine zipper containing domain) transcriptional regulator involved in downregulating iron-dependent metabolic pathways and the biosynthesis of heme [171].
The combination of the iron chelator DFRA, with a liposomal preparation of Amphotericin B, was effective in reducing fungal infections in murine models of pulmonary Aspergillosis which supported murine survival [165]. Although this study, which utilizes an iron chelating treatment strategy for this disease, shows promise in an in vivo mouse model, further work is needed to determine clinical feasibility of using such iron chelators in patients infected with A. fumigatus.
4.3. Rhizopus Oryzae
Fungal Features—Rhizopus oryzae is a filamentous fungus involved in the development of mucormycosis, a common infection in (a) patients with diabetic ketoacidosis [172], (b) those who are immunocompromised as a result of cytotoxic chemotherapy [173], or (c) those undergoing organ transplantation [174,175]. Current treatments include Amphotericin B, as described above, which often leads to kidney damage [176]. Unfortunately, due to surgical disfigurement and the high mortality index, the development of improved treatment regimens remain essential [177].
Contribution of Iron—The genome sequencing project for R. oryzae has identified several genes involved in iron uptake including three ferric reductases, six copper oxidases, a high-affinity iron permease, siderophore permeases, SreA, and genes involved in the uptake of iron from heme [178,179]. The fungal receptors, FOB1 and FOB2 (ferrioxamine binding plasma membrane proteins) are involved in promoting the binding of ferrioxamine, a siderophore, [180] in order to promote iron uptake via the FTR1 permease mechanism [181]. Loss of FTR1 via genetic manipulation (via RNAi and reduction in DNA copy number) decreases iron acquisition in R. oryzae as well as fungal infections in a murine model of diabetic ketoacidosis infected with spores via the tail vein or intranasal instillation [181].
In a diabetic ketoacidosis mouse model and a DFO-treated mouse model injected with R. oryzae spores into the tail vein, FOB1 and FOB2 deficient fungi were essential for mediating virulence in the DFO-model only [180]. Furthermore, these genes were critical for mediating iron uptake into R. oryzae [180]. However, other iron chelators, such as DFP and DFRA, were successful in reducing virulence and improved survival in an in vivo diabetic ketoacidotic mouse model [182,183]. It is suggested that the acidotic condition of the diabetics may contribute to decreased binding capacity of iron to transferrin [177], increasing free iron levels to promote mucormycosis infections [172]. Treatment with DFO increased the infectivity of R. oryzae in immunocompetent guinea pigs [184] and albino guinea pigs [185].
The oral administration of DFRA, an iron chelator, resulted in clinical improvement in a 40-year old patient infected with Rhinocerebral mucormycosis (an opportunistic invasive fungal infection) in combination with liposomal amphotericin B [186]. This one clinical study warrants further clinical application of DFRA or other iron chelators in the treatment of patients infected with R. oryzae. Collectively, targeting these fungal iron pathway holds promise to improving existing therapeutic modalities for overcoming the detrimental health consequences of R. oryzae fungal infections.
5. Concluding Perspectives
Although limited applications for iron chelation therapeutic approaches are noted in clinical practice for the topics presented herein, evidence from in vitro and in vivo animal models, which cover a wide range of diseases, provides a strong positive foundation for future clinical applications. With respect to iron availability, it is essential that iron levels in the host be tightly controlled to hinder the development of microorganismal infections within the host by (1) supporting the capacity of iron-binding proteins in the host plasma to limit iron availability to hinder the growth and virulence of microorganisms and (2) diminishing the intracellular host iron by targeting TBI or NTBI uptake processes, increasing ferritin-bound iron/reducing ferritinophagic processes, and increasing FPN-mediated iron export. Although there is evidence for iron chelators in hindering microorganismal virulence, this is an area that would benefit from further research investigation. Furthermore, the identification of the iron chelating components in the natural compounds derived from plants (e.g., blueberry extract, tea, as well as honey) would also be beneficial in the identification of naturally occurring iron chelators.
Author Contributions
R.C., N.S., R.S., A.C., and A.S. performed PubMed literature searches and co-wrote the first draft of this review. M.N., R.C. and A.C. generated Figure 1. M.N. conceived the overall content, developed the independent topics for this review, performed PubMed Literature searches, wrote draft, modified figure, and revised final draft. All authors critically reviewed and approved the manuscript for final submission.
Funding
We gratefully acknowledge funding to support this work provided by NCI R21 CA178468-01A1 awarded to M.N.
Acknowledgments
We regret not having cited all articles relevant to this topic within this review due to space limitations.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABC | ATP-binding cassette |
| AIDS | acquired immune deficiency syndrome |
| ALT | alanine aminotransferase |
| ART | anti-retroviral therapy |
| ATPase ABCE1 | ATP binding cassette subfamily E member 1 |
| bLF | bovine lactoferrin |
| NRAMP1 | natural resistance-associated macrophage protein 1 |
| BLM | bleomycin |
| BPS | bathophenanthroline disulfonate |
| bZIP | basic leucine zipper containing domain |
| CD81 | Cluster of Differentiation 81 |
| CDK | cyclin dependent kinases |
| CFEM | Common in Fungal Extracellular Membrane |
| CLDN1 | Claudin 1 |
| DCYTB | duodenal cytochrome b |
| DFO | Desferioxamine or deferoxamine |
| DFP or L1 | Deferiprone |
| DFRA or DFX | Deferasirox |
| DFT | Desferrithiocin |
| DMT1 | Divalent Metal Transporter 1 |
| DNA | deoxyribonucleic acid |
| EGCG | Epigallocatechin-3-gallate |
| EGFR | epidermal growth factor receptor |
| eIF3 | Eukaryotic initiation factor 3 |
| FDA | Food and Drug Administration |
| Fe-SIH | iron-salicylaldehyde isonicotinoyl hydrazine |
| FeSO4 | iron (II) sulfate |
| FPN1 | ferroportin |
| FsC | fusarinine C |
| FTN | ferritin |
| Fur | ferric uptake regulator |
| GATA | Globin Transcription Factor |
| HAMP | hepcidin |
| HasA | heme acquisition system A |
| HCP1 | Heme Carrier Protein |
| HCV | Hepatitis C Virus |
| HIV | Human Immunodeficiency Virus |
| HO-1 | heme oxygenase 1 |
| HO-2 | heme oxygenase 2 |
| IFN | Interferon |
| IRES | internal ribosome entry site |
| JAK | Janus Kinase |
| LIP | labile iron pool |
| NF-kB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NS3 | non-structural protein 3 |
| NTBI | non-transferring bound iron |
| OCLN | Occludin |
| OMP | outer membrane protein |
| PBL | peripheral blood lymphocytes |
| PCFT | proton-coupled folate transporter |
| Pch | pyochelin |
| Pvd | pyoverdine |
| PZP | pyrazolopyridinone |
| RBC | red blood cells |
| RIA | reductive iron assimilation |
| RNA | ribonucleic acid |
| RNR | ribonucleotide reductase |
| ROS | reactive oxygen species |
| SIT | siderophore iron transporter |
| SLC39A8 | Solute carrier family 39 member 8 |
| SRB1 | Scavenger receptor class B type 1 |
| STAT | Signal Transducer and Activator of Transcription |
| STEAP3 | Six Transmembrane Epithelial Antigen of Prostate 3 |
| TAFC | triacetylfusarinine C |
| TBI | transferrin-bound iron |
| TGF-β | transforming growth factor beta |
References
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164–174. [Google Scholar]
- Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Regulators of iron homeostasis: New players in metabolism, cell death, and disease. Trends Biochem. Sci. 2016, 41, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Cassat, J.E.; Skaar, E.P. Iron in infection and immunity. Cell Host Microbe 2013, 13, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Drakesmith, H.; Prentice, A. Viral infection and iron metabolism. Nat. Rev. Microbiol. 2008, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Systemic iron homeostasis. Physiol. Rev. 2013, 93, 1721–1741. [Google Scholar] [CrossRef]
- Shayeghi, M.; Latunde-Dada, G.O.; Oakhill, J.S.; Laftah, A.H.; Takeuchi, K.; Halliday, N.; Khan, Y.; Warley, A.; McCann, F.E.; Hider, R.C.; et al. Identification of an intestinal heme transporter. Cell 2005, 122, 789–801. [Google Scholar] [CrossRef]
- Ji, C.; Kosman, D.J. Molecular mechanisms of non-transferrin-bound and transferring-bound iron uptake in primary hippocampal neurons. J. Neurochem. 2015, 133, 668–683. [Google Scholar] [CrossRef]
- Silva, B.; Faustino, P. An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim. Biophys. Acta 2015, 1852, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Knutson, M.D. Iron transport proteins: Gateways of cellular and systemic iron homeostasis. J. Biol. Chem. 2017, 292, 12735–12743. [Google Scholar] [CrossRef]
- Sangkhae, V.; Nemeth, E. Regulation of the iron homeostatic hormone hepcidin. Adv. Nutr. 2017, 8, 126–136. [Google Scholar] [CrossRef]
- Dev, S.; Babitt, J.L. Overview of iron metabolism in health and disease. Hemodial. Int. 2017, 21 (Suppl. 1), S6–S20. [Google Scholar]
- Yoshida, T.; Noguchi, M.; Kikuchi, G. Oxygenated form of heme. Heme oxygenase complex and requirement for second electron to initiate heme degradation from the oxygenated complex. J. Biol. Chem. 1980, 255, 4418–4420. [Google Scholar] [PubMed]
- Kakhlon, O.; Cabantchik, Z.I. The labile iron pool: Characterization, measurement, and participation in cellular processes (1). Free Radic. Biol. Med. 2002, 33, 1037–1046. [Google Scholar] [PubMed]
- MacKenzie, E.L.; Iwasaki, K.; Tsuji, Y. Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 997–1030. [Google Scholar] [CrossRef]
- Coffey, R.; Ganz, T. Iron homeostasis: An anthropocentric perspective. J. Biol. Chem. 2017, 292, 12727–12734. [Google Scholar]
- Kawabata, H. Transferrin and transferrin receptors update. Free Radic. Biol. Med. 2019, 133, 46–54. [Google Scholar] [CrossRef]
- Sendamarai, A.K.; Ohgami, R.S.; Fleming, M.D.; Lawrence, C.M. Structure of the membrane proximal oxidoreductase domain of human steap3, the dominant ferrireductase of the erythroid transferrin cycle. Proc. Natl. Acad. Sci. USA 2008, 105, 7410–7415. [Google Scholar] [CrossRef]
- Lane, D.J.; Bae, D.H.; Merlot, A.M.; Sahni, S.; Richardson, D.R. Duodenal cytochrome b (dcytb) in iron metabolism: An update on function and regulation. Nutrients 2015, 7, 2274–2296. [Google Scholar]
- van Raaij, S.E.G.; Srai, S.K.S.; Swinkels, D.W.; van Swelm, R.P.L. Iron uptake by zip8 and zip14 in human proximal tubular epithelial cells. Biometals 2019, 32, 211–226. [Google Scholar]
- McKie, A.T.; Barrow, D.; Latunde-Dada, G.O.; Rolfs, A.; Sager, G.; Mudaly, E.; Mudaly, M.; Richardson, C.; Barlow, D.; Bomford, A.; et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 2001, 291, 1755–1759. [Google Scholar] [CrossRef]
- Mackenzie, B.; Hediger, M.A. Slc11 family of h+-coupled metal-ion transporters nramp1 and dmt1. Pflug. Arch. Eur. J. Physiol. 2004, 447, 571–579. [Google Scholar] [CrossRef]
- Gunshin, H.; Mackenzie, B.; Berger, U.V.; Gunshin, Y.; Romero, M.F.; Boron, W.F.; Nussberger, S.; Gollan, J.L.; Hediger, M.A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 1997, 388, 482–488. [Google Scholar] [CrossRef]
- Wang, J.; Pantopoulos, K. Regulation of cellular iron metabolism. Biochem. J. 2011, 434, 365–381. [Google Scholar] [CrossRef]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef]
- Nairz, M.; Schroll, A.; Sonnweber, T.; Weiss, G. The struggle for iron-a metal at the host-pathogen interface. Cell. Microbiol. 2010, 12, 1691–1702. [Google Scholar] [CrossRef]
- Mobarra, N.; Shanaki, M.; Ehteram, H.; Nasiri, H.; Sahmani, M.; Saeidi, M.; Goudarzi, M.; Pourkarim, H.; Azad, M. A review on iron chelators in treatment of iron overload syndromes. Int. J. Hematol. Oncol. Stem Cell Res. 2016, 10, 239–247. [Google Scholar]
- Nunez, M.T.; Chana-Cuevas, P. New perspectives in iron chelation therapy for the treatment of neurodegenerative diseases. Pharmaceuticals 2018, 11, 109. [Google Scholar] [CrossRef]
- Hatcher, H.C.; Singh, R.N.; Torti, F.M.; Torti, S.V. Synthetic and natural iron chelators: Therapeutic potential and clinical use. Future Med. Chem. 2009, 1, 1643–1670. [Google Scholar] [CrossRef]
- Chiani, M.; Akbarzadeh, A.; Farhangi, A.; Mazinani, M.; Saffari, Z.; Emadzadeh, K.; Mehrabi, M.R. Optimization of culture medium to increase the production of desferrioxamine b (desferal) in streptomyces pilosus. Pak. J. Biol. Sci. 2010, 13, 546–550. [Google Scholar] [CrossRef]
- Bergeron, R.J.; Wiegand, J.; McManis, J.S.; Bharti, N. Desferrithiocin: A search for clinically effective iron chelators. J. Med. Chem. 2014, 57, 9259–9291. [Google Scholar] [CrossRef]
- Kontoghiorghe, C.N.; Kolnagou, A.; Kontoghiorghes, G.J. Phytochelators intended for clinical use in iron overload, other diseases of iron imbalance and free radical pathology. Molecules 2015, 20, 20841–20872. [Google Scholar] [CrossRef]
- Kontoghiorghes, G.J.; Kontoghiorghe, C.N. Iron and chelation in biochemistry and medicine: New approaches to controlling iron metabolism and treating related diseases. Cells 2020, 9, 1456. [Google Scholar] [CrossRef]
- Rosa, L.; Cutone, A.; Lepanto, M.S.; Paesano, R.; Valenti, P. Lactoferrin: A natural glycoprotein involved in iron and inflammatory homeostasis. Int. J. Mol. Sci. 2017, 18, 1985. [Google Scholar] [CrossRef]
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019, 59, 580–596. [Google Scholar] [CrossRef]
- Poggiali, E.; Cassinerio, E.; Zanaboni, L.; Cappellini, M.D. An update on iron chelation therapy. Blood Transfus. 2012, 10, 411–422. [Google Scholar]
- Stojiljkovic, I.; Hantke, K. Hemin uptake system of yersinia enterocolitica: Similarities with other tonb-dependent systems in gram-negative bacteria. EMBO J. 1992, 11, 4359–4367. [Google Scholar] [CrossRef]
- Tong, Y.; Guo, M. Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 2009, 481, 1–15. [Google Scholar] [CrossRef]
- Sook, B.R.; Block, D.R.; Sumithran, S.; Montanez, G.E.; Rodgers, K.R.; Dawson, J.H.; Eichenbaum, Z.; Dixon, D.W. Characterization of siaa, a streptococcal heme-binding protein associated with a heme abc transport system. Biochemistry 2008, 47, 2678–2688. [Google Scholar] [CrossRef]
- Zhu, H.; Liu, M.; Lei, B. The surface protein shr of streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein shp. BMC Microbiol. 2008, 8, 15. [Google Scholar] [CrossRef]
- Philpott, C.C. Iron uptake in fungi: A system for every source. Biochim. Biophys. Acta 2006, 1763, 636–645. [Google Scholar] [CrossRef]
- Lesuisse, E.; Labbe, P. Reductive and non-reductive mechanisms of iron assimilation by the yeast saccharomyces cerevisiae. J. Gen. Microbiol. 1989, 135, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Marsh, M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011, 195, 1071–1082. [Google Scholar] [PubMed]
- Fanales-Belasio, E.; Raimondo, M.; Suligoi, B.; Butto, S. Hiv virology and pathogenetic mechanisms of infection: A brief overview. Annali Dell’istituto Superiore di Sanita 2010, 46, 5–14. [Google Scholar] [CrossRef]
- Choe, H.; Jemielity, S.; Abraham, J.; Radoshitzky, S.R.; Farzan, M. Transferrin receptor 1 in the zoonosis and pathogenesis of new world hemorrhagic fever arenaviruses. Curr. Opin. Microbiol. 2011, 14, 476–482. [Google Scholar] [CrossRef]
- Romeo, A.M.; Christen, L.; Niles, E.G.; Kosman, D.J. Intracellular chelation of iron by bipyridyl inhibits DNA virus replication: Ribonucleotide reductase maturation as a probe of intracellular iron pools. J. Biol. Chem. 2001, 276, 24301–24308. [Google Scholar] [CrossRef]
- Sappey, C.; Boelaert, J.R.; Legrand-Poels, S.; Forceille, C.; Favier, A.; Piette, J. Iron chelation decreases nf-kappa b and hiv type 1 activation due to oxidative stress. AIDS Res. Hum. Retrovir. 1995, 11, 1049–1061. [Google Scholar]
- Dooher, J.E.; Schneider, B.L.; Reed, J.C.; Lingappa, J.R. Host abce1 is at plasma membrane hiv assembly sites and its dissociation from gag is linked to subsequent events of virus production. Traffic 2007, 8, 195–211. [Google Scholar]
- Dubuisson, J.; Cosset, F.L. Virology and cell biology of the hepatitis c virus life cycle: An update. J. Hepatol. 2014, 61, S3–S13. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar]
- Chevaliez, S.; Pawlotsky, J.M. Hcv genome and life cycle. In Hepatitis C Viruses: Genomes and Molecular Biology; Tan, S.L., Ed.; Horizon Bioscience: Norfolk, UK, 2006. [Google Scholar]
- Wong, R.J.; Gish, R.G. Metabolic manifestations and complications associated with chronic hepatitis c virus infection. Gastroenterol. Hepatol. 2016, 12, 293–299. [Google Scholar]
- Herrera, J.L. Iron depletion is not effective in inducing a virologic response in patients with chronic hepatitis c who failed to respond to interferon therapy. Am. J. Gastroenterol. 1999, 94, 3571–3575. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, N.J.; Murphy, A.G.; Bourke, N.M.; Keogh, C.A.; Hegarty, J.E.; O’Farrelly, C. Ribavirin enhances ifn-alpha signalling and mxa expression: A novel immune modulation mechanism during treatment of hcv. PLoS ONE 2011, 6, e27866. [Google Scholar]
- McHutchison, J.G.; Gordon, S.C.; Schiff, E.R.; Shiffman, M.L.; Lee, W.M.; Rustgi, V.K.; Goodman, Z.D.; Ling, M.H.; Cort, S.; Albrecht, J.K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis c. Hepatitis interventional therapy group. N. Engl. J. Med. 1998, 339, 1485–1492. [Google Scholar]
- Goto, K.; Roca Suarez, A.A.; Wrensch, F.; Baumert, T.F.; Lupberger, J. Hepatitis c virus and hepatocellular carcinoma: When the host loses its grip. Int. J. Mol. Sci. 2020, 21, 3057. [Google Scholar] [CrossRef] [PubMed]
- Theurl, I.; Zoller, H.; Obrist, P.; Datz, C.; Bachmann, F.; Elliott, R.M.; Weiss, G. Iron regulates hepatitis c virus translation via stimulation of expression of translation initiation factor 3. J. Infect. Dis. 2004, 190, 819–825. [Google Scholar] [CrossRef]
- Sartori, M.; Andorno, S.; Rossini, A.; Boldorini, R.; Bozzola, C.; Carmagnola, S.; Del Piano, M.; Albano, E. Phlebotomy improves histology in chronic hepatitis c males with mild iron overload. World J. Gastroenterol. 2010, 16, 596–602. [Google Scholar] [CrossRef]
- Akkaya, O.; Kiyici, M.; Yilmaz, Y.; Ulukaya, E.; Yerci, O. Clinical significance of activity of alt enzyme in patients with hepatitis c virus. World J. Gastroenterol. 2007, 13, 5481–5485. [Google Scholar] [CrossRef]
- Hayashi, H.; Takikawa, T.; Nishimura, N.; Yano, M.; Isomura, T.; Sakamoto, N. Improvement of serum aminotransferase levels after phlebotomy in patients with chronic active hepatitis c and excess hepatic iron. Am. J. Gastroenterol. 1994, 89, 986–988. [Google Scholar]
- Kakizaki, S.; Takagi, H.; Horiguchi, N.; Toyoda, M.; Takayama, H.; Nagamine, T.; Mori, M.; Kato, N. Iron enhances hepatitis c virus replication in cultured human hepatocytes. Liver 2000, 20, 125–128. [Google Scholar] [CrossRef]
- Ali, N.; Siddiqui, A. The la antigen binds 5’ noncoding region of the hepatitis c virus rna in the context of the initiator aug codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 1997, 94, 2249–2254. [Google Scholar] [CrossRef]
- Wolin, S.L.; Cedervall, T. The la protein. Annu. Rev. Biochem. 2002, 71, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, Y.; An, D.; Diao, H.; Xu, W.; He, X.; Sun, R.; Wei, L.; Li, L. Regulation of hepatitis c virus translation initiation by iron: Role of eif3 and la protein. Virus Res. 2012, 167, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Fillebeen, C.; Pantopoulos, K. Iron inhibits replication of infectious hepatitis c virus in permissive huh7.5.1 cells. J. Hepatol. 2010, 53, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Alexaki, A.; Liu, Y.; Wigdahl, B. Cellular reservoirs of hiv-1 and their role in viral persistence. Curr. HIV Res. 2008, 6, 388–400. [Google Scholar] [CrossRef]
- Siliciano, R.F.; Greene, W.C. Hiv latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. [Google Scholar] [CrossRef]
- Justiz Vaillant, A.A.; Gulick, P.G. HIV Disease; Statpearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Ford, E.S.; Puronen, C.E.; Sereti, I. Immunopathogenesis of asymptomatic chronic hiv infection: The calm before the storm. Curr. Opin. HIV AIDS 2009, 4, 206–214. [Google Scholar] [CrossRef]
- Kappe, R.; Levitz, S.; Harrison, T.S.; Ruhnke, M.; Ampel, N.M.; Just-Nubling, G. Recent advances in cryptococcosis, candidiasis and coccidioidomycosis complicating hiv infection. Med. Mycol. 1998, 36 (Suppl. 1), 207–215. [Google Scholar]
- Sadowski, I.; Hashemi, F.B. Strategies to eradicate hiv from infected patients: Elimination of latent provirus reservoirs. Cell. Mol. Life Sci. 2019, 76, 3583–3600. [Google Scholar] [CrossRef]
- Kemnic, T.R.; Gulick, P.G. HIV Antiretroviral Therapy; Statpearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Riera, A.; Gimferrer, E.; Cadafalch, J.; Remacha, A.; Martin, S. Prevalence of high serum and red cell ferritin levels in hiv-infected patients. Haematologica 1994, 79, 165–167. [Google Scholar]
- Diebold, J.; Tabbara, W.; Marche, C.; Audouin, J.; Le Tourneau, A. Bone marrow changes at several stages of hiv infection, studied on bone marrow biopsies in 85 patients. Arch. Anat. Cytol. Pathol. 1991, 39, 137–146. [Google Scholar]
- Geremia, G.K.; McCluney, K.W.; Adler, S.S.; Charletta, D.A.; Hoile, R.D.; Huckman, M.S.; Ramsey, R.G. The magnetic resonance hypointense spine of aids. J. Comput. Assist. Tomogr. 1990, 14, 785–789. [Google Scholar] [CrossRef]
- Gelman, B.B.; Rodriguez-Wolf, M.G.; Wen, J.; Kumar, S.; Campbell, G.R.; Herzog, N. Siderotic cerebral macrophages in the acquired immunodeficiency syndrome. Arch. Pathol. Lab. Med. 1992, 116, 509–516. [Google Scholar] [PubMed]
- Smith, T.W.; DeGirolami, U.; Henin, D.; Bolgert, F.; Hauw, J.J. Human immunodeficiency virus (hiv) leukoencephalopathy and the microcirculation. J. Neuropathol. Exp. Neurol. 1990, 49, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, R.K.; Mhiri, C.; Baudrimont, M.; Roullet, E.; Berry, J.P.; Poirier, J. Iron pigment deposits, small vessel vasculitis, and erythrophagocytosis in the muscle of human immunodeficiency virus-infected patients. Hum. Pathol. 1991, 22, 1187–1194. [Google Scholar] [CrossRef]
- Savarino, A.; Pescarmona, G.P.; Boelaert, J.R. Iron metabolism and hiv infection: Reciprocal interactions with potentially harmful consequences? Cell Biochem. Funct. 1999, 17, 279–287. [Google Scholar] [CrossRef]
- Greaves, D.E.; Griffiths, W.J.; Lever, A.M. Does venesection reduce hiv viral load in patients with hereditary haemochromatosis? Antivir. Ther. 2013, 18, 135–138. [Google Scholar] [CrossRef]
- Xu, M.; Kashanchi, F.; Foster, A.; Rotimi, J.; Turner, W.; Gordeuk, V.R.; Nekhai, S. Hepcidin induces hiv-1 transcription inhibited by ferroportin. Retrovirology 2010, 7, 104. [Google Scholar] [CrossRef]
- Whalan, R.H.; Funnell, S.G.; Bowler, L.D.; Hudson, M.J.; Robinson, A.; Dowson, C.G. Distribution and genetic diversity of the abc transporter lipoproteins piua and piaa within streptococcus pneumoniae and related streptococci. J. Bacteriol. 2006, 188, 1031–1038. [Google Scholar] [CrossRef]
- Traore, H.N.; Meyer, D. The effect of iron overload on in vitro hiv-1 infection. J. Clin. Virol. 2004, 31 (Suppl. 1), S92–S98. [Google Scholar] [CrossRef]
- Nabel, G.; Baltimore, D. An inducible transcription factor activates expression of human immunodeficiency virus in t cells. Nature 1987, 326, 711–713. [Google Scholar] [CrossRef]
- Hoffbrand, A.V.; Ganeshaguru, K.; Hooton, J.W.; Tattersall, M.H. Effect of iron deficiency and desferrioxamine on DNA synthesis in human cells. Br. J. Haematol. 1976, 33, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, N.A.; van der Bruggen, T.; Oudshoorn, M.; Nottet, H.S.; Marx, J.J.; van Asbeck, B.S. Inhibition of human immunodeficiency virus type 1 replication in human mononuclear blood cells by the iron chelators deferoxamine, deferiprone, and bleomycin. J. Infect. Dis. 2000, 181, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, N.A.; van der Bruggen, T.; Oudshoorn, M.; Hider, R.C.; Marx, J.J.; van Asbeck, B.S. Human immunodeficiency virus type 1 replication inhibition by the bidentate iron chelators cp502 and cp511 is caused by proliferation inhibition and the onset of apoptosis. Eur. J. Clin. Investig. 2002, 32 (Suppl. 1), 91–96. [Google Scholar] [CrossRef] [PubMed]
- Debebe, Z.; Ammosova, T.; Jerebtsova, M.; Kurantsin-Mills, J.; Niu, X.; Charles, S.; Richardson, D.R.; Ray, P.E.; Gordeuk, V.R.; Nekhai, S. Iron chelators icl670 and 311 inhibit hiv-1 transcription. Virology 2007, 367, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Iordanskiy, S.; Kovalskyy, D.; Breuer, D.; Niu, X.; Lin, X.; Xu, M.; Gavrilenko, K.; Kashanchi, F.; Dhawan, S.; et al. Phenyl-1-pyridin-2yl-ethanone-based iron chelators increase ikappab-alpha expression, modulate cdk2 and cdk9 activities, and inhibit hiv-1 transcription. Antimicrob. Agents Chemother. 2014, 58, 6558–6571. [Google Scholar] [CrossRef] [PubMed]
- Caza, M.; Kronstad, J.W. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 3, 80. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef]
- Garmory, H.S.; Titball, R.W. Atp-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004, 72, 6757–6763. [Google Scholar] [CrossRef]
- Radkov, A.D.; Hsu, Y.P.; Booher, G.; VanNieuwenhze, M.S. Imaging bacterial cell wall biosynthesis. Annu. Rev. Biochem. 2018, 87, 991–1014. [Google Scholar] [CrossRef]
- Delepelaire, P. Bacterial abc transporters of iron containing compounds. Res. Microbiol. 2019, 170, 345–357. [Google Scholar] [CrossRef]
- Runyen-Janecky, L.J. Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 2013, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Yue, W.W.; Buchanan, S.K. Recognition of iron-free siderophores by tonb-dependent iron transporters. Mol. Microbiol. 2004, 54, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.A.; Foster-Hartnett, D.; McIntosh, M.A.; Postle, K. Regions of escherichia coli tonb and fepa proteins essential for in vivo physical interactions. J. Bacteriol. 1997, 179, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Letoffe, S.; Redeker, V.; Wandersman, C. Isolation and characterization of an extracellular haem-binding protein from pseudomonas aeruginosa that shares function and sequence similarities with the serratia marcescens hasa haemophore. Mol. Microbiol. 1998, 28, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Cope, L.D.; Thomas, S.E.; Hrkal, Z.; Hansen, E.J. Binding of heme-hemopexin complexes by soluble hxua protein allows utilization of this complexed heme by haemophilus influenzae. Infect. Immun. 1998, 66, 4511–4516. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Vasil, M.L. Gene repression by the ferric uptake regulator in pseudomonas aeruginosa: Cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 1996, 93, 4409–4414. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.P.; Drazek, E.S. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic corynebacterium species. J. Bacteriol. 2001, 183, 1476–1481. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Skaar, E.P.; Gaspar, A.H.; Humayun, M.; Gornicki, P.; Jelenska, J.; Joachmiak, A.; Missiakas, D.M.; Schneewind, O. Passage of heme-iron across the envelope of staphylococcus aureus. Science 2003, 299, 906–909. [Google Scholar] [CrossRef]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.F.; Tummler, B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Ruffin, M.; Brochiero, E. Repair process impairment by pseudomonas aeruginosa in epithelial tissues: Major features and potential therapeutic avenues. Front. Cell. Infect. Microbiol. 2019, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of iron uptake systems in pseudomonas aeruginosa virulence and airway infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef] [PubMed]
- Brandel, J.; Humbert, N.; Elhabiri, M.; Schalk, I.J.; Mislin, G.L.; Albrecht-Gary, A.M. Pyochelin, a siderophore of pseudomonas aeruginosa: Physicochemical characterization of the iron(iii), copper(ii) and zinc(ii) complexes. Dalton Trans. 2012, 41, 2820–2834. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Graham, R. Isolation of an iron-binding compound from pseudomonas aeruginosa. J. Bacteriol. 1979, 137, 357–364. [Google Scholar] [CrossRef]
- Mislin, G.L.; Hoegy, F.; Cobessi, D.; Poole, K.; Rognan, D.; Schalk, I.J. Binding properties of pyochelin and structurally related molecules to fpta of pseudomonas aeruginosa. J. Mol. Biol. 2006, 357, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D. Effect of pyochelin on the virulence of pseudomonas aeruginosa. Infect. Immun. 1982, 36, 17–23. [Google Scholar] [CrossRef]
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of siderophore production on pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 2000, 68, 1834–1839. [Google Scholar] [CrossRef]
- Kim, M.; Christley, S.; Khodarev, N.N.; Fleming, I.; Huang, Y.; Chang, E.; Zaborina, O.; Alverdy, J.C. Pseudomonas aeruginosa wound infection involves activation of its iron acquisition system in response to fascial contact. J. Trauma Acute Care Surg. 2015, 78, 823–829. [Google Scholar] [CrossRef]
- Marshall, B.; Stintzi, A.; Gilmour, C.; Meyer, J.M.; Poole, K. Citrate-mediated iron uptake in pseudomonas aeruginosa: Involvement of the citrate-inducible feca receptor and the feob ferrous iron transporter. Microbiology 2009, 155, 305–315. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Johnson, Z.; Vasil, M.L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in pseudomonas aeruginosa. Microbiology 2000, 146 Pt 1, 185–198. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Aali, M.; Caldwell, A.; House, K.; Zhou, J.; Chappe, V.; Lehmann, C. Iron chelation as novel treatment for lung inflammation in cystic fibrosis. Med. Hypotheses 2017, 104, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Ankley, L.M.; Monteiro, M.P.; Camp, K.M.; O’Quinn, R.; Castillo, A.R. Manuka honey chelates iron and impacts iron regulation in key bacterial pathogens. J. Appl. Microbiol. 2020, 128, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Valm, A.M. The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J. Mol. Biol. 2019, 431, 2957–2969. [Google Scholar] [CrossRef]
- Naito, M.; Ogura, Y.; Itoh, T.; Shoji, M.; Okamoto, M.; Hayashi, T.; Nakayama, K. The complete genome sequencing of prevotella intermedia strain oma14 and a subsequent fine-scale, intra-species genomic comparison reveal an unusual amplification of conjugative and mobile transposons and identify a novel prevotella-lineage-specific repeat. DNA Res. 2016, 23, 11–19. [Google Scholar]
- Ang, M.Y.; Dutta, A.; Wee, W.Y.; Dymock, D.; Paterson, I.C.; Choo, S.W. Comparative genome analysis of fusobacterium nucleatum. Genome Biol. Evol. 2016, 8, 2928–2938. [Google Scholar] [CrossRef]
- Kabwe, M.; Brown, T.L.; Dashper, S.; Speirs, L.; Ku, H.; Petrovski, S.; Chan, H.T.; Lock, P.; Tucci, J. Genomic, morphological and functional characterisation of novel bacteriophage fnu1 capable of disrupting fusobacterium nucleatum biofilms. Sci. Rep. 2019, 9, 9107. [Google Scholar] [CrossRef]
- Subbarao, K.C.; Nattuthurai, G.S.; Sundararajan, S.K.; Sujith, I.; Joseph, J.; Syedshah, Y.P. Gingival crevicular fluid: An overview. J. Pharm. Bioallied Sci. 2019, 11, S135–S139. [Google Scholar] [CrossRef]
- Duchesne, P.; Grenier, D.; Mayrand, D. Binding and utilization of human transferrin by prevotella nigrescens. Infect. Immun. 1999, 67, 576–580. [Google Scholar] [CrossRef]
- Nelson, K.E.; Fleischmann, R.D.; DeBoy, R.T.; Paulsen, I.T.; Fouts, D.E.; Eisen, J.A.; Daugherty, S.C.; Dodson, R.J.; Durkin, A.S.; Gwinn, M.; et al. Complete genome sequence of the oral pathogenic bacterium porphyromonas gingivalis strain w83. J. Bacteriol. 2003, 185, 5591–5601. [Google Scholar] [CrossRef]
- Bramanti, T.E.; Holt, S.C. Roles of porphyrins and host iron transport proteins in regulation of growth of porphyromonas gingivalis w50. J. Bacteriol. 1991, 173, 7330–7339. [Google Scholar] [CrossRef] [PubMed]
- Wyss, C. Growth of porphyromonas gingivalis, treponema denticola, t. Pectinovorum, t. Socranskii, and t. Vincentii in a chemically defined medium. J. Clin. Microbiol. 1992, 30, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Shibata, Y.; Shiroza, T.; Yukitake, H.; Peng, B.; Chen, Y.Y.; Sato, K.; Naito, M.; Abiko, Y.; Reynolds, E.C.; et al. Characterization of hemin-binding protein 35 (hbp35) in porphyromonas gingivalis: Its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Schifferle, R.E.; Shostad, S.A.; Bayers-Thering, M.T.; Dyer, D.W.; Neiders, M.E. Effect of protoporphyrin ix limitation on porphyromonas gingivalis. J. Endod. 1996, 22, 352–355. [Google Scholar] [CrossRef]
- Olczak, T.; Simpson, W.; Liu, X.; Genco, C.A. Iron and heme utilization in porphyromonas gingivalis. FEMS Microbiol. Rev. 2005, 29, 119–144. [Google Scholar] [CrossRef]
- Bramanti, T.E.; Holt, S.C. Hemin uptake in porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J. Bacteriol. 1993, 175, 7413–7420. [Google Scholar] [CrossRef][Green Version]
- Sroka, A.; Sztukowska, M.; Potempa, J.; Travis, J.; Genco, C.A. Degradation of host heme proteins by lysine-and arginine-specific cysteine proteinases (gingipains) of porphyromonas gingivalis. J. Bacteriol. 2001, 183, 5609–5616. [Google Scholar] [CrossRef]
- Lewis, J.P. Metal uptake in host-pathogen interactions: Role of iron in porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 2010, 52, 94–116. [Google Scholar] [CrossRef]
- Shi, Y.; Ratnayake, D.B.; Okamoto, K.; Abe, N.; Yamamoto, K.; Nakayama, K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of porphyromonas gingivalis. Construction of mutants with a combination of rgpa, rgpb, kgp, and haga. J. Biol. Chem. 1999, 274, 17955–17960. [Google Scholar] [CrossRef]
- Byrne, D.P.; Potempa, J.; Olczak, T.; Smalley, J.W. Evidence of mutualism between two periodontal pathogens: Co-operative haem acquisition by the hmuy haemophore of porphyromonas gingivalis and the cysteine protease interpain a (inpa) of prevotella intermedia. Mol. Oral Microbiol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Okamoto, M.; Maeda, N.; Kondo, K.; Leung, K.P. Hemolytic and hemagglutinating activities of prevotella intermedia and prevotella nigrescens. FEMS Microbiol. Lett. 1999, 178, 299–304. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Byrne, D.P.; Wawrzonek, K.; Jaworska, A.; Birss, A.J.; Potempa, J.; Smalley, J.W. Role of the cysteine protease interpain a of prevotella intermedia in breakdown and release of haem from haemoglobin. Biochem. J. 2009, 425, 257–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moon, J.H.; Herr, Y.; Kim, S.W.; Lee, J.Y. In vitro activity of deferoxamine against porphyromonas gingivalis. FEMS Microbiol. Lett. 2011, 323, 61–67. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moon, J.H.; Kim, C.; Lee, H.S.; Kim, S.W.; Lee, J.Y. Antibacterial and antibiofilm effects of iron chelators against prevotella intermedia. J. Med. Microbiol. 2013, 62, 1307–1316. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Dudonne, S.; Desjardins, Y.; Grenier, D. Wild blueberry (vaccinium angustifolium ait.) polyphenols target fusobacterium nucleatum and the host inflammatory response: Potential innovative molecules for treating periodontal diseases. J. Agric. Food Chem. 2015, 63, 6999–7008. [Google Scholar] [CrossRef]
- Ben Lagha, A.; Haas, B.; Grenier, D. Tea polyphenols inhibit the growth and virulence properties of fusobacterium nucleatum. Sci. Rep. 2017, 7, 44815. [Google Scholar] [CrossRef]
- Ardila, C.M.; Granada, M.I.; Guzman, I.C. Antibiotic resistance of subgingival species in chronic periodontitis patients. J. Periodontal Res. 2010, 45, 557–563. [Google Scholar] [CrossRef]
- Walker, C.B.; Bueno, L.C. Antibiotic resistance in an oral isolate of prevotella intermedia. Clin. Infect. Dis. 1997, 25 (Suppl. 2), S281–S283. [Google Scholar] [CrossRef]
- Bostanci, N.; Belibasakis, G.N. Porphyromonas gingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 2012, 333, 1–9. [Google Scholar] [CrossRef]
- Donati, C.; Hiller, N.L.; Tettelin, H.; Muzzi, A.; Croucher, N.J.; Angiuoli, S.V.; Oggioni, M.; Dunning Hotopp, J.C.; Hu, F.Z.; Riley, D.R.; et al. Structure and dynamics of the pan-genome of streptococcus pneumoniae and closely related species. Genome Biol. 2010, 11, R107. [Google Scholar] [CrossRef]
- Hoyer, J.; Bartel, J.; Gomez-Mejia, A.; Rohde, M.; Hirschfeld, C.; Hess, N.; Sura, T.; Maass, S.; Hammerschmidt, S.; Becher, D. Proteomic response of streptococcus pneumoniae to iron limitation. Int. J. Med. Microbiol. 2018, 308, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Tai, S.S.; Lee, C.J.; Winter, R.E. Hemin utilization is related to virulence of streptococcus pneumoniae. Infect. Immun. 1993, 61, 5401–5405. [Google Scholar] [CrossRef] [PubMed]
- Whatmore, A.M.; Dowson, C.G. The autolysin-encoding gene (lyta) of streptococcus pneumoniae displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect. Immun. 1999, 67, 4551–4556. [Google Scholar] [CrossRef] [PubMed]
- Steinfort, C.; Wilson, R.; Mitchell, T.; Feldman, C.; Rutman, A.; Todd, H.; Sykes, D.; Walker, J.; Saunders, K.; Andrew, P.W.; et al. Effect of streptococcus pneumoniae on human respiratory epithelium in vitro. Infect. Immun. 1989, 57, 2006–2013. [Google Scholar] [CrossRef]
- Brown, J.S.; Gilliland, S.M.; Holden, D.W. A streptococcus pneumoniae pathogenicity island encoding an abc transporter involved in iron uptake and virulence. Mol. Microbiol. 2001, 40, 572–585. [Google Scholar] [CrossRef]
- Brown, J.S.; Gilliland, S.M.; Ruiz-Albert, J.; Holden, D.W. Characterization of pit, a streptococcus pneumoniae iron uptake abc transporter. Infect. Immun. 2002, 70, 4389–4398. [Google Scholar] [CrossRef] [PubMed]
- Ehrt, S.; Schnappinger, D.; Rhee, K.Y. Metabolic principles of persistence and pathogenicity in mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 496–507. [Google Scholar] [CrossRef]
- Chao, A.; Sieminski, P.J.; Owens, C.P.; Goulding, C.W. Iron acquisition in mycobacterium tuberculosis. Chem. Rev. 2019, 119, 1193–1220. [Google Scholar] [CrossRef]
- Olakanmi, O.; Kesavalu, B.; Abdalla, M.Y.; Britigan, B.E. Iron acquisition by mycobacterium tuberculosis residing within myeloid dendritic cells. Microb. Pathog. 2013, 65, 21–28. [Google Scholar] [CrossRef]
- Dragset, M.S.; Poce, G.; Alfonso, S.; Padilla-Benavides, T.; Ioerger, T.R.; Kaneko, T.; Sacchettini, J.C.; Biava, M.; Parish, T.; Arguello, J.M.; et al. A novel antimycobacterial compound acts as an intracellular iron chelator. Antimicrob. Agents Chemother. 2015, 59, 2256–2264. [Google Scholar] [CrossRef]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal cell wall: Emerging antifungals and drug resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42. [Google Scholar] [CrossRef] [PubMed]
- Outten, C.E.; Albetel, A.N. Iron sensing and regulation in saccharomyces cerevisiae: Ironing out the mechanistic details. Curr. Opin. Microbiol. 2013, 16, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.H. Acquisition, transport, and storage of iron by pathogenic fungi. Clin. Microbiol. Rev. 1999, 12, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.H. Iron gathering by zoopathogenic fungi. FEMS Immunol. Med. Microbiol. 2004, 40, 95–100. [Google Scholar] [CrossRef][Green Version]
- Bairwa, G.; Hee Jung, W.; Kronstad, J.W. Iron acquisition in fungal pathogens of humans. Met. Integr. Biometal Sci. 2017, 9, 215–227. [Google Scholar] [CrossRef]
- Kim, J.; Cho, Y.J.; Do, E.; Choi, J.; Hu, G.; Cadieux, B.; Chun, J.; Lee, Y.; Kronstad, J.W.; Jung, W.H. A defect in iron uptake enhances the susceptibility of cryptococcus neoformans to azole antifungal drugs. Fungal Genet. Biol. 2012, 49, 955–966. [Google Scholar] [CrossRef]
- Holz, R.W. The effects of the polyene antibiotics nystatin and amphotericin b on thin lipid membranes. Ann. N. Y. Acad. Sci. 1974, 235, 469–479. [Google Scholar] [CrossRef]
- Thamban Chandrika, N.; Shrestha, S.K.; Ngo, H.X.; Howard, K.C.; Garneau-Tsodikova, S. Novel fluconazole derivatives with promising antifungal activity. Bioorgan. Med. Chem. 2018, 26, 573–580. [Google Scholar] [CrossRef]
- Jung, W.H.; Sham, A.; Lian, T.; Singh, A.; Kosman, D.J.; Kronstad, J.W. Iron source preference and regulation of iron uptake in cryptococcus neoformans. PLoS Pathog. 2008, 4, e45. [Google Scholar] [CrossRef]
- Jung, W.H.; Hu, G.; Kuo, W.; Kronstad, J.W. Role of ferroxidases in iron uptake and virulence of cryptococcus neoformans. Eukaryot. Cell 2009, 8, 1511–1520. [Google Scholar] [PubMed]
- Ibrahim, A.S.; Gebremariam, T.; French, S.W.; Edwards, J.E., Jr.; Spellberg, B. The iron chelator deferasirox enhances liposomal amphotericin b efficacy in treating murine invasive pulmonary aspergillosis. J. Antimicrob. Chemother. 2010, 65, 289–292. [Google Scholar] [PubMed]
- Jenks, J.D.; Hoenigl, M. Treatment of aspergillosis. J. Fungi 2018, 4, 98. [Google Scholar]
- Haas, H. Fungal siderophore metabolism with a focus on aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276. [Google Scholar]
- Blatzer, M.; Binder, U.; Haas, H. The metalloreductase freb is involved in adaptation of aspergillus fumigatus to iron starvation. Fungal Genet. Biol. 2011, 48, 1027–1033. [Google Scholar]
- Schrettl, M.; Bignell, E.; Kragl, C.; Joechl, C.; Rogers, T.; Arst, H.N., Jr.; Haynes, K.; Haas, H. Siderophore biosynthesis but not reductive iron assimilation is essential for aspergillus fumigatus virulence. J. Exp. Med. 2004, 200, 1213–1219. [Google Scholar]
- Schrettl, M.; Kim, H.S.; Eisendle, M.; Kragl, C.; Nierman, W.C.; Heinekamp, T.; Werner, E.R.; Jacobsen, I.; Illmer, P.; Yi, H.; et al. Srea-mediated iron regulation in aspergillus fumigatus. Mol. Microbiol. 2008, 70, 27–43. [Google Scholar]
- Schrettl, M.; Beckmann, N.; Varga, J.; Heinekamp, T.; Jacobsen, I.D.; Jochl, C.; Moussa, T.A.; Wang, S.; Gsaller, F.; Blatzer, M.; et al. Hapx-mediated adaption to iron starvation is crucial for virulence of aspergillus fumigatus. PLoS Pathog. 2010, 6, e1001124. [Google Scholar]
- Artis, W.M.; Fountain, J.A.; Delcher, H.K.; Jones, H.E. A mechanism of susceptibility to mucormycosis in diabetic ketoacidosis: Transferrin and iron availability. Diabetes 1982, 31, 1109–1114. [Google Scholar]
- Yamauchi, T.; Misaki, H.; Arai, H.; Iwasaki, H.; Naiki, H.; Ueda, T. An autopsy case of disseminated mucormycosis in a neutropenic patient receiving chemotherapy for the underlying solid malignancy. J. Infect. Chemother. 2002, 8, 103–105. [Google Scholar]
- Singh, N.; Aguado, J.M.; Bonatti, H.; Forrest, G.; Gupta, K.L.; Safdar, N.; John, G.T.; Pursell, K.J.; Munoz, P.; Patel, R.; et al. Zygomycosis in solid organ transplant recipients: A prospective, matched case-control study to assess risks for disease and outcome. J. Infect. Dis. 2009, 200, 1002–1011. [Google Scholar] [CrossRef]
- Almyroudis, N.G.; Sutton, D.A.; Linden, P.; Rinaldi, M.G.; Fung, J.; Kusne, S. Zygomycosis in solid organ transplant recipients in a tertiary transplant center and review of the literature. Am. J. Transpl. 2006, 6, 2365–2374. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, B.P.; Weihprecht, H.; Campbell, W.R.; Lorenz, J.N.; Webb, R.C.; Briggs, J.P.; Schnermann, J. Direct vasoconstriction as a possible cause for amphotericin b-induced nephrotoxicity in rats. J. Clin. Investig. 1991, 87, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.S.; Spellberg, B.; Edwards, J., Jr. Iron acquisition: A novel perspective on mucormycosis pathogenesis and treatment. Curr. Opin. Infect. Dis. 2008, 21, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.J.; Ibrahim, A.S.; Skory, C.; Grabherr, M.G.; Burger, G.; Butler, M.; Elias, M.; Idnurm, A.; Lang, B.F.; Sone, T.; et al. Genomic analysis of the basal lineage fungus rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009, 5, e1000549. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.S.; Spellberg, B.; Walsh, T.J.; Kontoyiannis, D.P. Pathogenesis of mucormycosis. Clin. Infect. Dis. 2012, 54 (Suppl. 1), S16–S22. [Google Scholar] [CrossRef]
- Liu, M.; Lin, L.; Gebremariam, T.; Luo, G.; Skory, C.D.; French, S.W.; Chou, T.F.; Edwards, J.E., Jr.; Ibrahim, A.S. Fob1 and fob2 proteins are virulence determinants of rhizopus oryzae via facilitating iron uptake from ferrioxamine. PLoS Pathog. 2015, 11, e1004842. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Gebremariam, T.; Lin, L.; Luo, G.; Husseiny, M.I.; Skory, C.D.; Fu, Y.; French, S.W.; Edwards, J.E., Jr.; Spellberg, B. The high affinity iron permease is a key virulence factor required for rhizopus oryzae pathogenesis. Mol. Microbiol. 2010, 77, 587–604. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Gebermariam, T.; Fu, Y.; Lin, L.; Husseiny, M.I.; French, S.W.; Schwartz, J.; Skory, C.D.; Edwards, J.E., Jr.; Spellberg, B.J. The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J. Clin. Investig. 2007, 117, 2649–2657. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Edwards, J.E., Jr.; Fu, Y.; Spellberg, B. Deferiprone iron chelation as a novel therapy for experimental mucormycosis. J. Antimicrob. Chemother. 2006, 58, 1070–1073. [Google Scholar] [CrossRef]
- Boelaert, J.R.; de Locht, M.; Van Cutsem, J.; Kerrels, V.; Cantinieaux, B.; Verdonck, A.; Van Landuyt, H.W.; Schneider, Y.J. Mucormycosis during deferoxamine therapy is a siderophore-mediated infection. In vitro and in vivo animal studies. J. Clin. Investig. 1993, 91, 1979–1986. [Google Scholar] [CrossRef] [PubMed]
- Boelaert, J.R.; Van Cutsem, J.; de Locht, M.; Schneider, Y.J.; Crichton, R.R. Deferoxamine augments growth and pathogenicity of rhizopus, while hydroxypyridinone chelators have no effect. Kidney Int. 1994, 45, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.; Ibrahim, A.; Edwards, J.E., Jr.; Walot, I.; Spellberg, B. Deferasirox, an iron-chelating agent, as salvage therapy for rhinocerebral mucormycosis. Antimicrob. Agents Chemother. 2006, 50, 3968–3969. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).