Structure, Antioxidant and Anti-inflammatory Activities of the (4R)- and (4S)-epimers of S-Carboxymethyl-L-cysteine Sulfoxide
Abstract
1. Introduction
2. Results
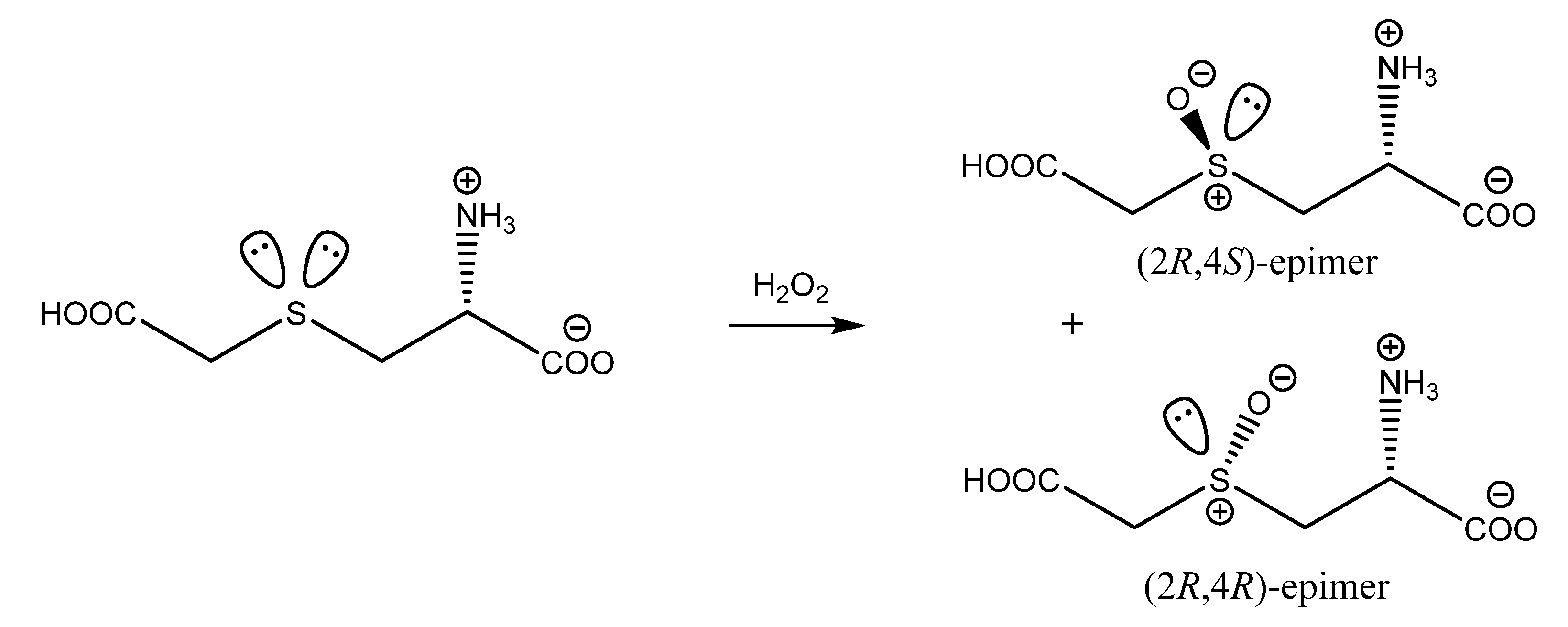
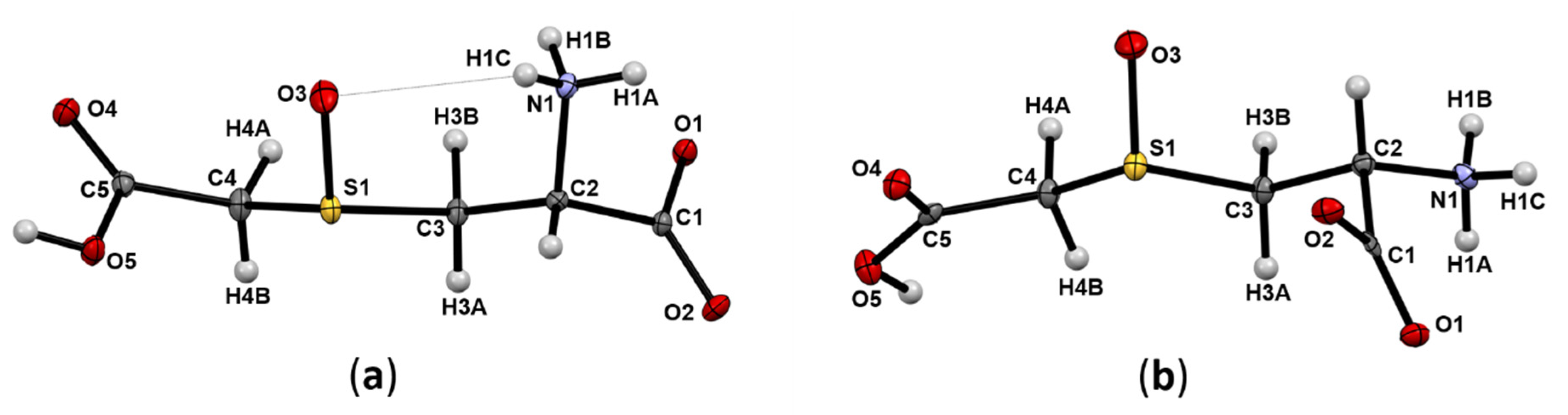
2.1. Molecular and Crystal Structures of CMCO Epimers
2.1.1. Polymorphs of the (2R,4R)-S-carboxymethylcysteine sulfoxide epimer
2.1.2. The (2R,4S)-S-carboxymethylcysteine sulfoxide epimer
2.1.3. Crystal Packing and Intermolecular Hydrogen Bonding
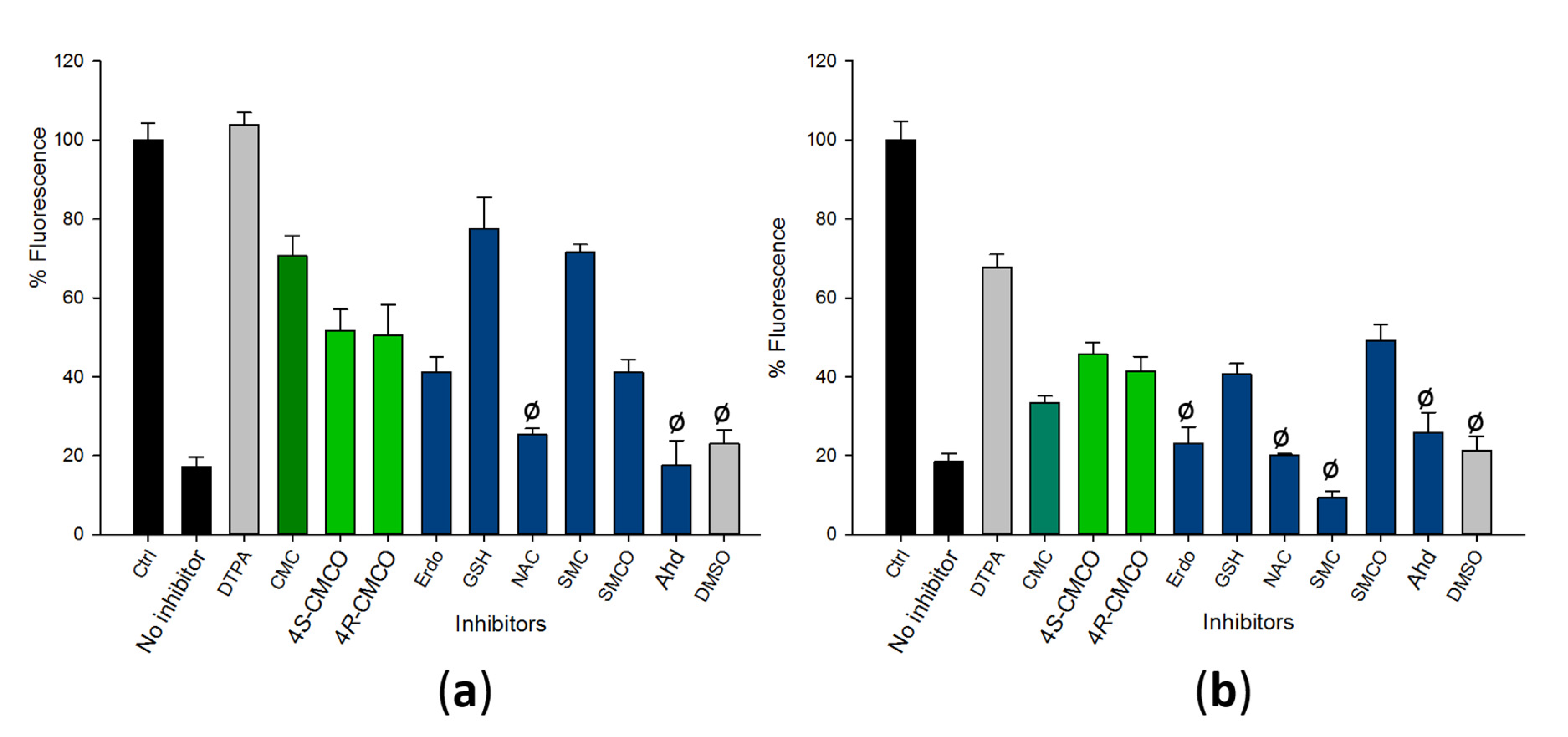
2.2. CMC and CMCO Protect DNA from Copper-Catalyzed Degradation by Hydroxyl Free Radicals
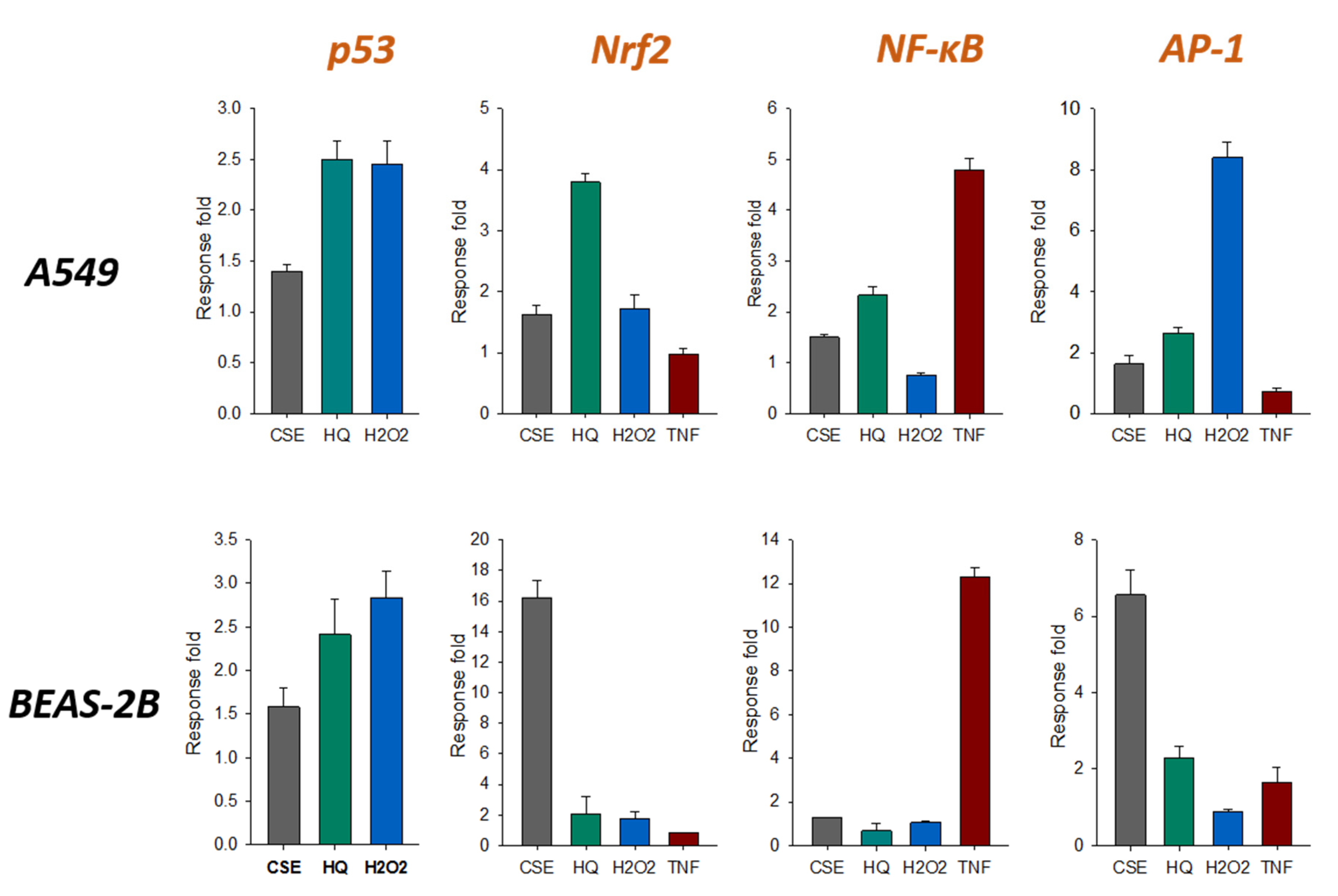
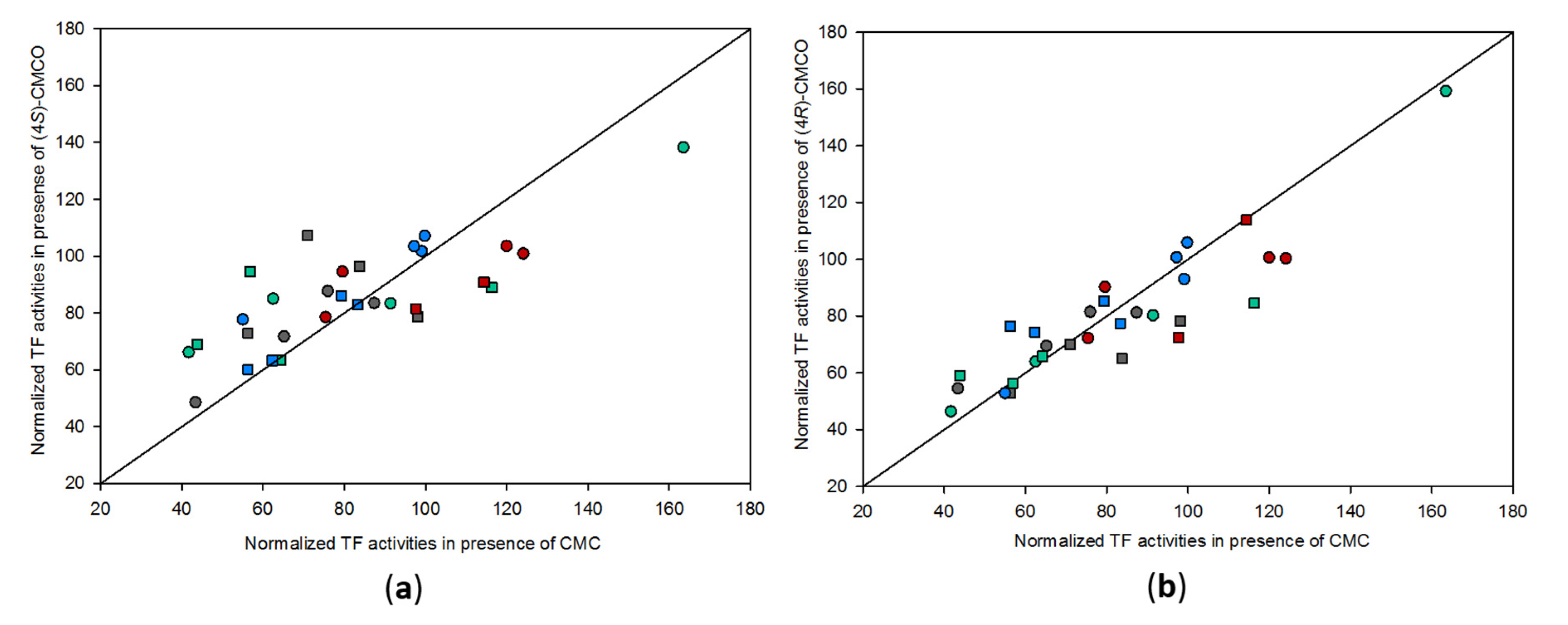
2.3. Effects of CMC and CMCO on Activation of Oxidative Stress and Pro-inflammatory Signalling Pathways in Human Bronchial Epithelial and Alveolar Cell Lines
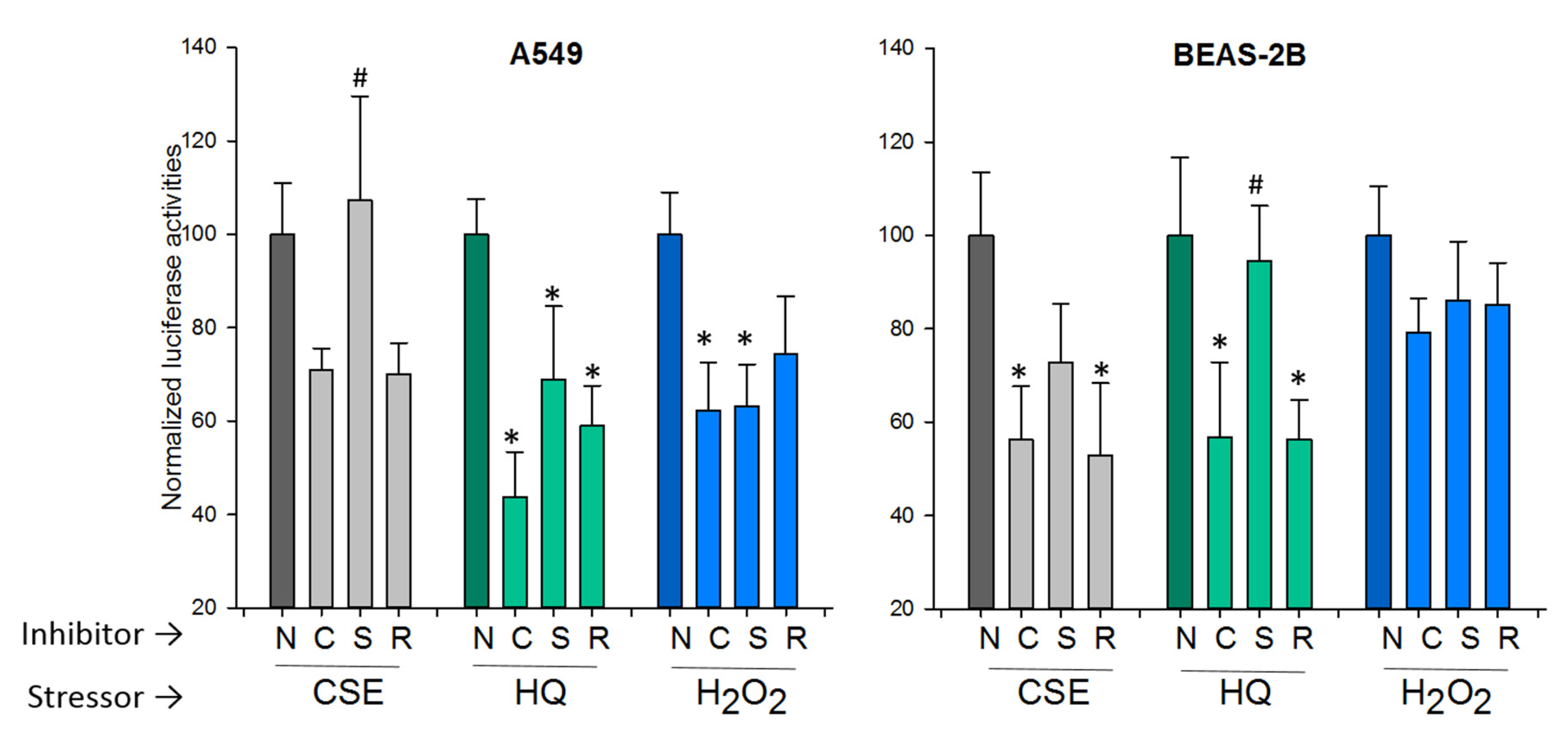
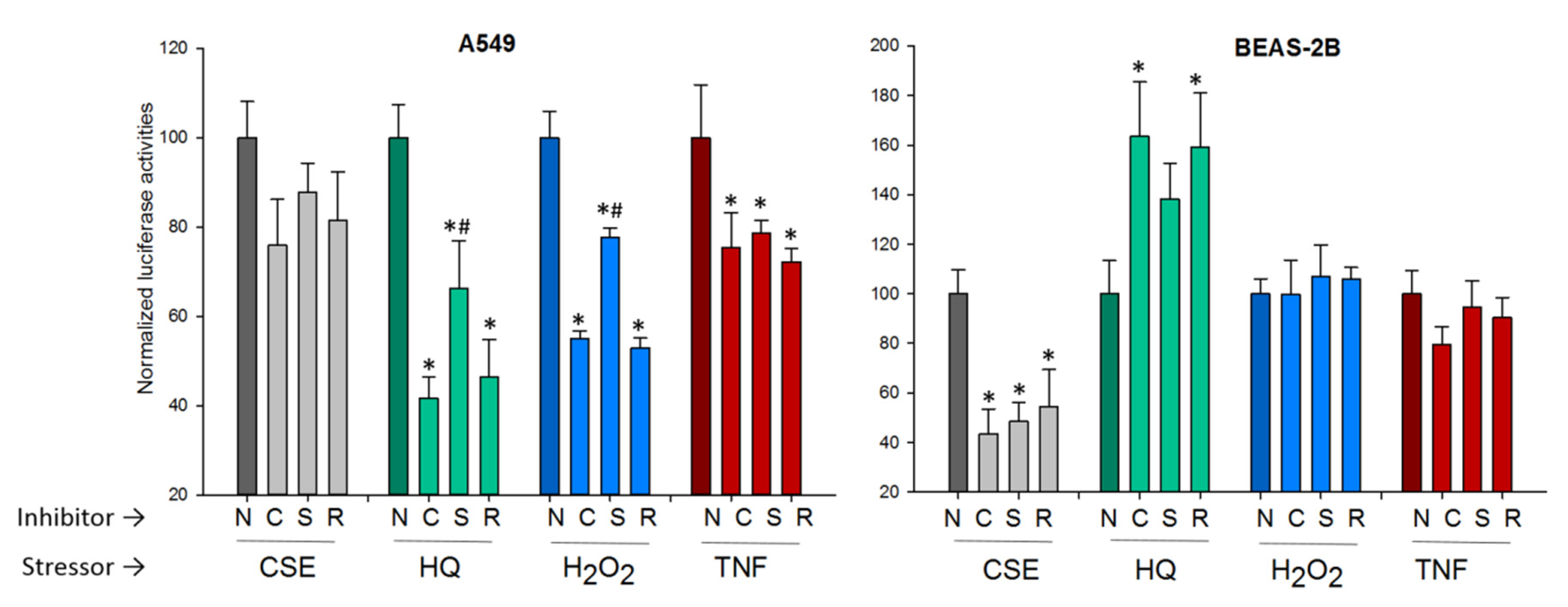
2.3.1. The p53 Pathway
2.3.2. The Nrf2 Pathway
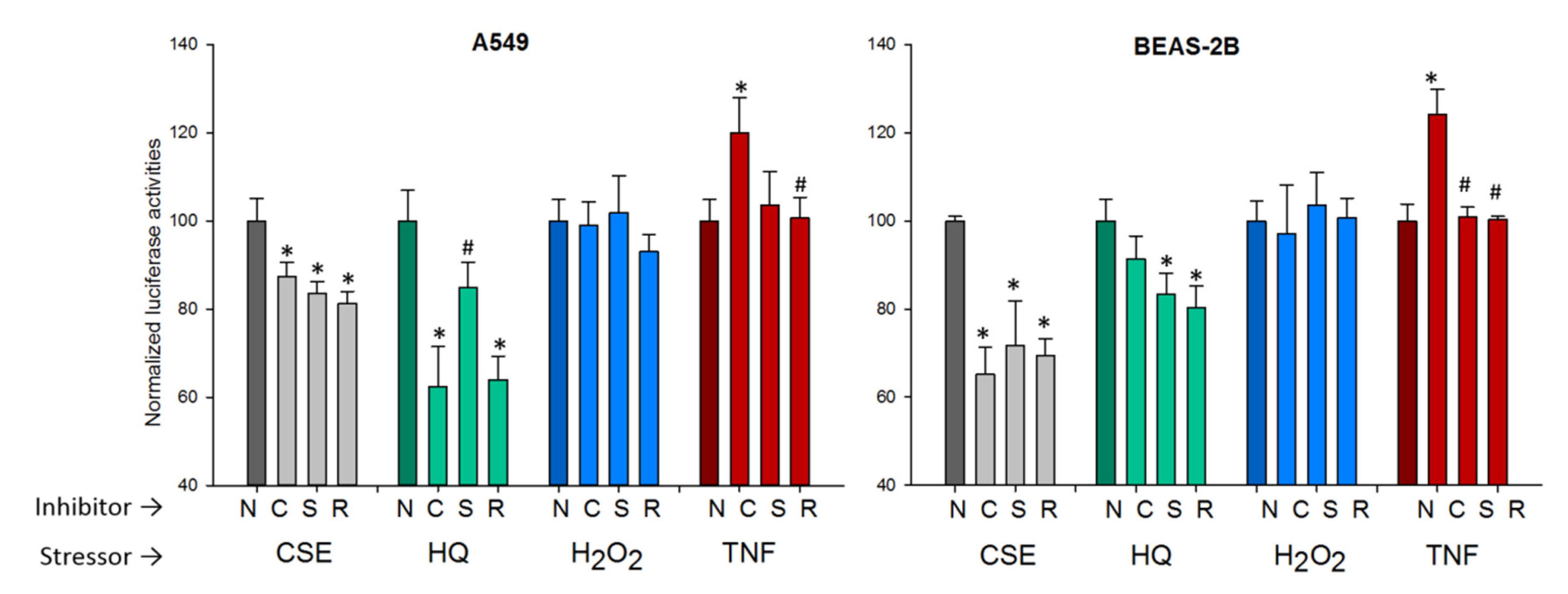
2.3.3. The NF-κB Pathway
2.3.4. The AP-1 Pathway
3. Discussion
4. Materials and Methods
4.1. Synthesis and Crystallization of S-carboxymethyl-L-cysteine sulfoxide
4.2. X-ray Diffraction Studies
4.3. In Vitro DNA Protection
4.4. Signaling Pathway Reporters
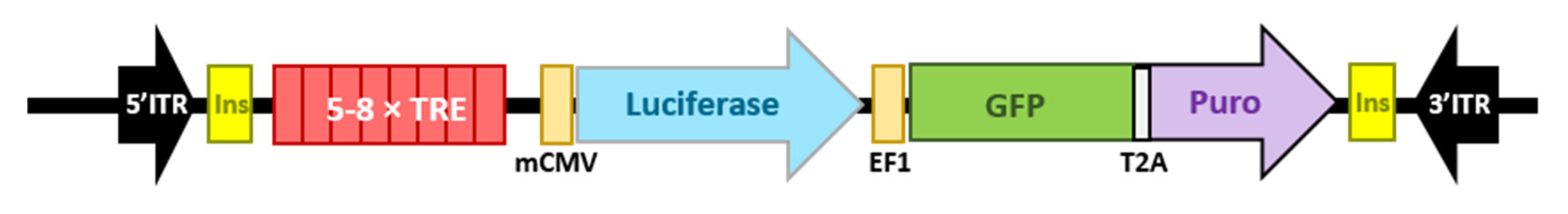
4.4.1. Reporter Vectors
4.4.2. Stable Transfections
4.4.3. Transcriptional Activity Reporter Assay
4.5. Molecular Modeling and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- López-Campos, J.L.; Tan, W.; Soriano, J.B. Global burden of COPD. Respirology 2016, 21, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Aoshiba, K.; Zhou, F.; Tsuji, T.; Nagai, A. DNA damage as a molecular link in the pathogenesis of COPD in smokers. Eur. Respir. J. 2012, 39, 1368–1376. [Google Scholar] [CrossRef] [PubMed]
- Maccio, A.; Madeddu, C.; Panzone, F.; Mantovani, G. Carbocysteine: Clinical experience and new perspectives in the treatment of chronic inflammatory diseases. Expert Opin. Pharmacother. 2009, 10, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.C.; Steventon, G.B. S-Carboxymethyl-L-cysteine. Drug Metab. Rev. 2012, 44, 129–147. [Google Scholar] [CrossRef]
- Mitchell, S.C.; Steventon, G.B. Phenylalanine 4-monooxygenase: The “sulfoxidation polymorphism”. Xenobiotica 2020, 50, 51–63. [Google Scholar] [CrossRef]
- Staffa, J.A.; Zervos, C.; Mighell, A.D.; Hubbard, C.R. S-Carboxymethyl-L-cysteine sulfoxide (configuration 2R:4R). Acta Crystallogr. 1976, B32, 3132–3135. [Google Scholar] [CrossRef]
- Hirayama, N.; Shirahata, K.; Ohashi, Y.; Sasada, Y. Structure of the α form of L-glutamic acid. The α-β transition. Bull. Chem. Soc. Jpn. 1980, 53, 30–35. [Google Scholar] [CrossRef]
- Lehmann, M.S.; Nunes, A.C. A short hydrogen bond between near identical carboxyl groups in the α-modification of L-glutamic acid. Acta Crystallogr. 1980, B36, 1621–1625. [Google Scholar] [CrossRef]
- Hubbard, C.R.; Mighell, A.D.; Staffa, J.A.; Zervos, C.; Konopelski, J.P. S-Carboxymethyl-L-cysteine sulfone. Acta Crystallogr. 1976, B32, 2723–2725. [Google Scholar] [CrossRef]
- Mighell, A.D.; Hubbard, C.R.; Harris, J.; Staffa, J.A.; Zervos, C. S-Carboxymethyl-L-cysteine. Acta Crystallogr. 1979, B35, 1258–1261. [Google Scholar] [CrossRef]
- Low, J.N.; Howie, R.A.; Scrimgeour, C.M.; Watt, P.W. The structure of L-α-aminoadipic acid. Acta Crystallogr. 1988, C44, 1762–1764. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Lapenna, D.; Mezzetti, A.; de Gioia, S.; Pierdomenico, S.D.; Daniele, F.; Cuccurullo, F. Plasma copper and lipid peroxidation in cigarette smokers. Free Radic. Biol. Med. 1995, 19, 849–852. [Google Scholar] [CrossRef]
- Peng, C.; Arthur, D.; Liu, F.; Lee, J.; Xia, Q.; Lavin, M.F.; Ng, J.C. Genotoxicity of hydroquinone in A549 cells. Cell Biol. Toxicol. 2013, 29, 213–227. [Google Scholar] [CrossRef]
- Yang, Q.; Hergenhahn, M.; Weninger, A.; Bartsch, H. Cigarette smoke induces direct DNA damage in the human B-lymphoid cell line Raji. Carcinogenesis 1999, 20, 1769–1775. [Google Scholar] [CrossRef]
- Mossine, V.V.; Waters, J.K.; Hannink, M.; Mawhinney, T.P. piggyBac Transposon plus insulators overcome epigenetic silencing to provide for stable signaling pathway reporter cell lines. PLoS ONE 2013, 8, e85494. [Google Scholar] [CrossRef]
- Mossine, V.V.; Waters, J.K.; Chance, D.L.; Mawhinney, T.P. Transient proteotoxicity of bacterial virulence factor pyocyanin in renal tubular epithelial cells induces ER-related vacuolation and can be efficiently modulated by iron chelators. Toxicol. Sci. 2016, 154, 403–415. [Google Scholar] [CrossRef][Green Version]
- Meek, D.W. The p53 response to DNA damage. DNA Repair 2004, 3, 1049–1056. [Google Scholar] [CrossRef]
- Suzuki, T.; Yamamoto, M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824. [Google Scholar] [CrossRef]
- Sekine, T.; Hirata, T.; Mine, T.; Fukano, Y. Activation of transcription factors in human bronchial epithelial cells exposed to aqueous extracts of mainstream cigarette smoke in vitro. Toxicol. Mech. Methods 2016, 26, 22–31. [Google Scholar] [CrossRef]
- Singh, A.; Misra, V.; Thimmulappa, R.K.; Lee, H.; Ames, S.; Hoque, M.O.; Herman, J.G.; Baylin, S.B.; Sidransky, D.; Gabrielson, E.; et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006, 3, e420. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, S.R.; Lee, Y.C. Impact of oxidative stress on lung diseases. Respirology 2009, 14, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.; Calvert, J. The role for S-carboxymethylcysteine (carbocisteine) in the management of chronic obstructive pulmonary disease. Int. J. Chronic Obstr. Pulm. Dis. 2008, 3, 659–669. [Google Scholar]
- Waring, R.H.; Mitchell, S.C. The metabolism and elimination of S-carboxymethyl-L-cysteine in man. Drug Metab. Dispos. 1982, 10, 61–62. [Google Scholar]
- Nakon, R.; Beadle, E.M., Jr.; Angelici, R.J. Binding of copper(II), nickel(II), zinc(II), and cobalt(II) by 3-[(carboxymethyl)thio]-L-alanine and 3-[(-aminoethyl)thio]-L-alanine. J. Am. Chem. Soc. 1974, 96, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.; Lemke, K. Ionization constants and stability constants of copper(II) complexes of some amino acids and their sulfur-containing analogs. Hoppe-Seyler’s Z. Physiol. Chem. 1968, 349, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; Higuchi, O.; Tateshita, K. Antioxidative activity of sulfur-containing compounds in Allium species for human LDL oxidation in vitro. BioFactors 2004, 21, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Battin, E.E.; Brumaghim, J.L. Metal specificity in DNA damage prevention by sulfur antioxidants. J. Inorg. Biochem. 2008, 102, 2036–2042. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Han, W.; Giraldo, C.; De Li, Y.; Block, E.R. Effect of cigarette smoke extract on nitric oxide synthase in pulmonary artery endothelial cells. Am. J. Respir. Cell Mol. Biol. 1998, 19, 819–825. [Google Scholar] [CrossRef]
- Meese, C.O. S-Carboxymethyl-L-cysteine (R)- and (S)-sulfoxide. Arch. Pharm. 1987, 320, 473–474. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0—New features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Mossine, V.V.; Chance, D.L.; Waters, J.K.; Mawhinney, T.P. Interaction of bacterial phenazines with colistimethate in bronchial epithelial cells. Antimicrob. Agents Chemother. 2018, 62, e02349. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]



| Molecule | Space Group | D–H ··· A | D–H | H ··· A | D ··· A | D–H ··· A | ΔC1–O 1 | ΔC5–O 2 |
|---|---|---|---|---|---|---|---|---|
| t-(4R)-CMCO | Triclinic | O5–H ··· O1 | 0.89 (2) 3 | 1.64 (3) | 2.500 (1) | 163 (2) | 0.018 | 0.083 |
| o-(4R)-CMCO | Orthorhombic | O5–H ··· O1 | 1.15 (3) | 2.44 (3) | 3.247 (2) | 126 (2) | 0.039 | 0.055 |
| O5–H ··· O2 | 1.15 (3) | 1.32 (3) | 2.450 (2) | 168 (3) | ||||
| m-(4S)-CMCO | Monoclinic | O5–H ··· O2 | 0.82 (4) | 1.67 (4) | 2.489 (3) | 176 (4) | 0.010 | 0.083 |
| CMC [10] | Monoclinic | O5–H ··· O2 | 0.98 (3) | 1.58 (3) | 2.548 (2) | 169 (3) | 0.008 | 0.102 |
| (4R)-CMCO [6] | Orthorhombic | O2–H ··· O5 | 1.04 (4) | 1.41 (4) | 2.449 (3) | 176 (4) | 0.032 | 0.058 |
| CMCO2 [9] | Orthorhombic | O5–H ··· O2 | 0.96 (5) | 1.54 (5) | 2.504 (5) | 175 (5) | 0.048 | 0.098 |
| Molecule | H ··· O | H ··· H | H ··· S | H ··· C | O ··· O | O ··· S | O ··· C | C ··· C | S ··· C |
|---|---|---|---|---|---|---|---|---|---|
| CMC [10] | 58.5 | 22.2 | 9.1 | 2.2 | 2.5 | 2.9 | 0.6 | 0.6 | 1.5 |
| t-(4R)-CMCO | 68.8 | 18.1 | 4.6 | 0.6 | 2.4 | 2.6 | 2.9 | 0 | 0 |
| o-(4R)-CMCO | 77.6 | 10.3 | 4.9 | 2.9 | 2.0 | 0.7 | 1.6 | 0 | 0 |
| m-(4S)-CMCO | 68.4 | 18.2 | 4.9 | 0.4 | 2.8 | 2.0 | 2.4 | 0 | 0.9 |
| CMCO2 [9] | 78.4 | 11.2 | 0 | 1.4 | 5.8 | 0 | 3.2 | 0 | 0 |
| Structure | E Electrostatic | E Polar | E Dispersion | E Repulsion | E Total |
|---|---|---|---|---|---|
| CMC [10] | −302.7 | −124.6 | −103.4 | 277.3 | −331.1 |
| t-(4R)-CMCO | −309.6 | −130.2 | −118.8 | 319.2 | −329.9 |
| o-(4R)-CMCO | −336.8 | −148.7 | −117.3 | 350.2 | −351.8 |
| m-(4S)-CMCO | −323.4 | −157.7 | −118.7 | 318.4 | −365.6 |
| CMCO2 [9] | −335.7 | −142.4 | −126 | 319.7 | −372.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, J.K.; Kelley, S.P.; Mossine, V.V.; Mawhinney, T.P. Structure, Antioxidant and Anti-inflammatory Activities of the (4R)- and (4S)-epimers of S-Carboxymethyl-L-cysteine Sulfoxide. Pharmaceuticals 2020, 13, 270. https://doi.org/10.3390/ph13100270
Waters JK, Kelley SP, Mossine VV, Mawhinney TP. Structure, Antioxidant and Anti-inflammatory Activities of the (4R)- and (4S)-epimers of S-Carboxymethyl-L-cysteine Sulfoxide. Pharmaceuticals. 2020; 13(10):270. https://doi.org/10.3390/ph13100270
Chicago/Turabian StyleWaters, James K., Steven P. Kelley, Valeri V. Mossine, and Thomas P. Mawhinney. 2020. "Structure, Antioxidant and Anti-inflammatory Activities of the (4R)- and (4S)-epimers of S-Carboxymethyl-L-cysteine Sulfoxide" Pharmaceuticals 13, no. 10: 270. https://doi.org/10.3390/ph13100270
APA StyleWaters, J. K., Kelley, S. P., Mossine, V. V., & Mawhinney, T. P. (2020). Structure, Antioxidant and Anti-inflammatory Activities of the (4R)- and (4S)-epimers of S-Carboxymethyl-L-cysteine Sulfoxide. Pharmaceuticals, 13(10), 270. https://doi.org/10.3390/ph13100270






