Sequence Permutation Generates Peptides with Different Antimicrobial and Antibiofilm Activities
Abstract
1. Introduction
2. Results
2.1. In Silico Properties of Sequence-Permutated Peptides
2.2. Antibacterial Activity
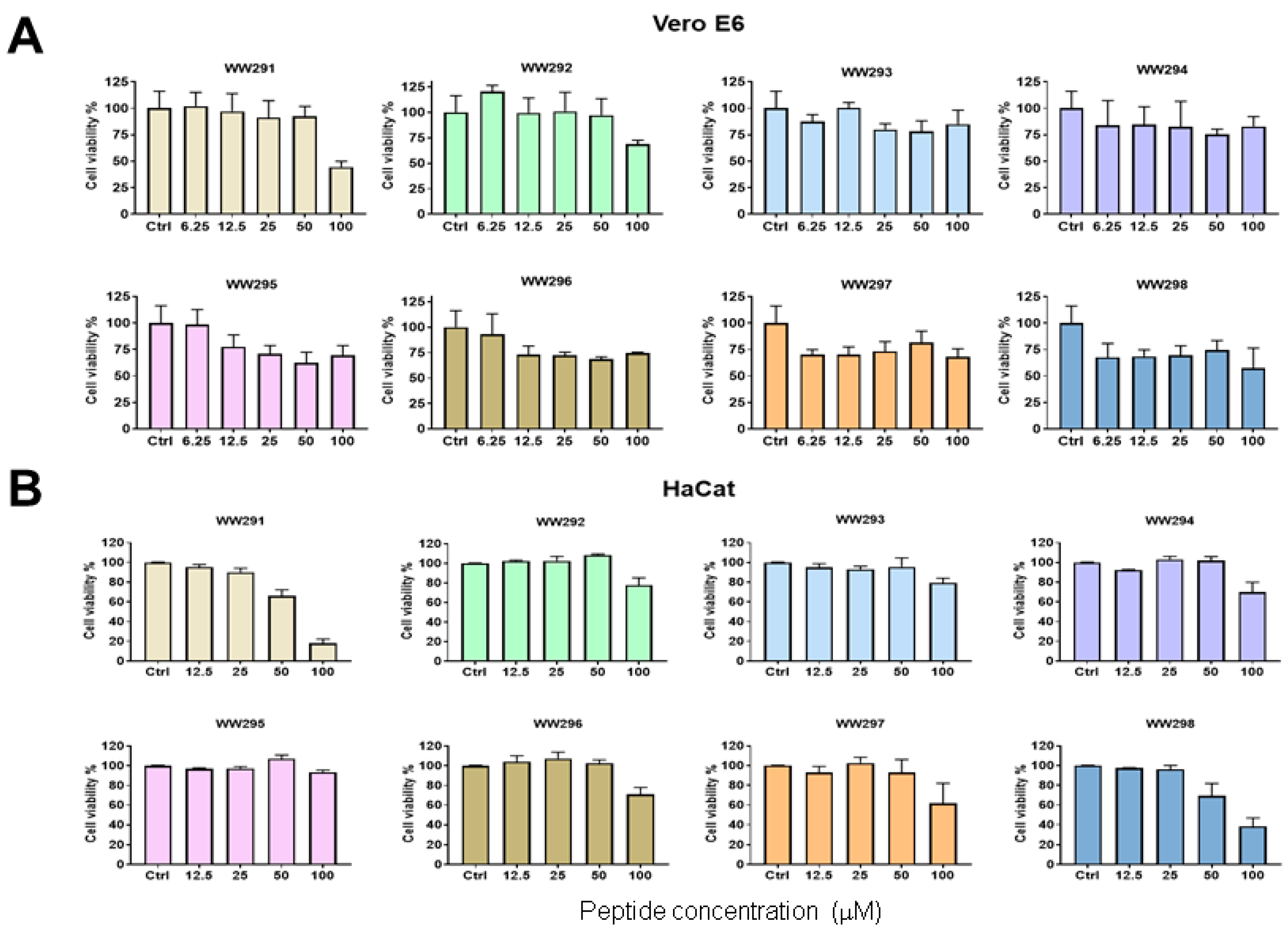
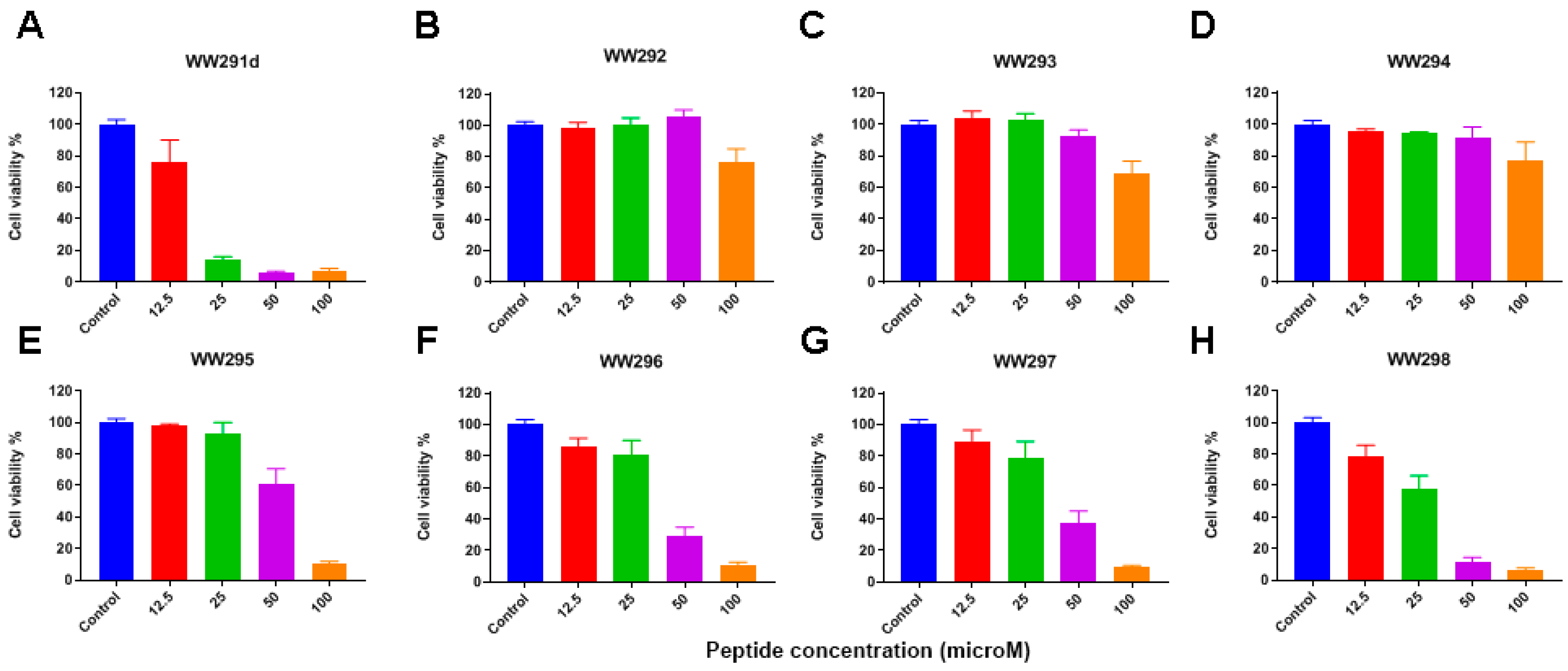
2.3. Cytotoxicity Assessment
2.4. Propidium Iodide-Based Membrane Penetration Assay
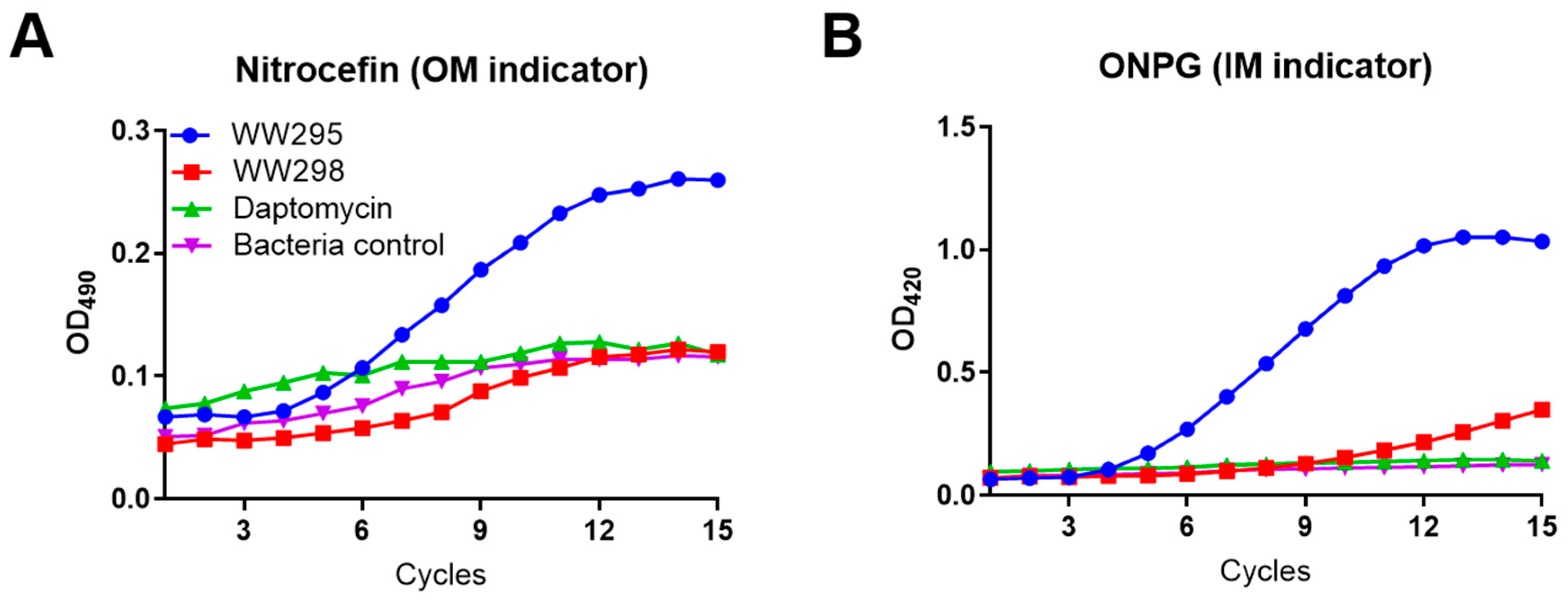
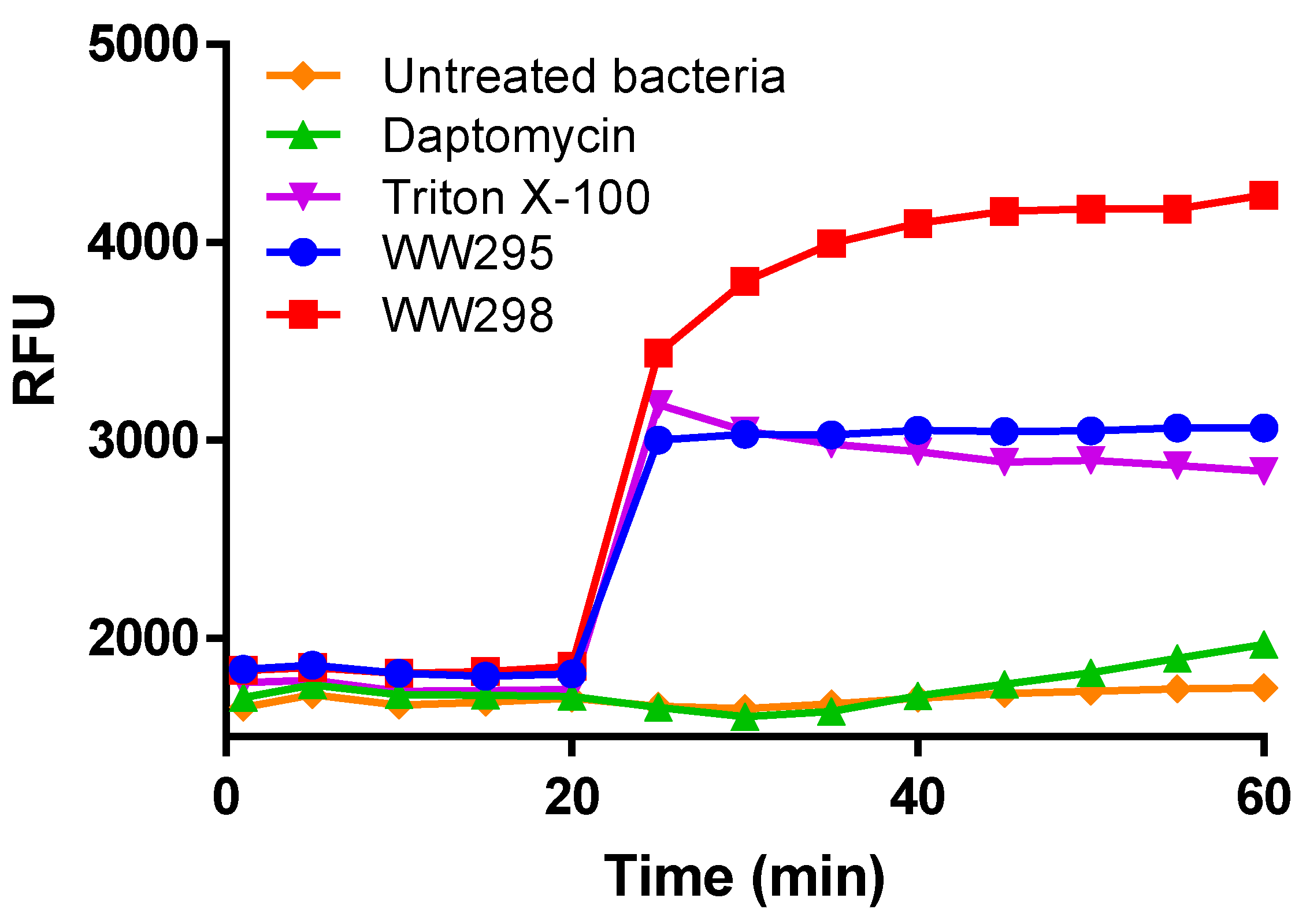
2.5. Simultaneous Detection of the Outer and Inner Membrane Permeation of E. coli
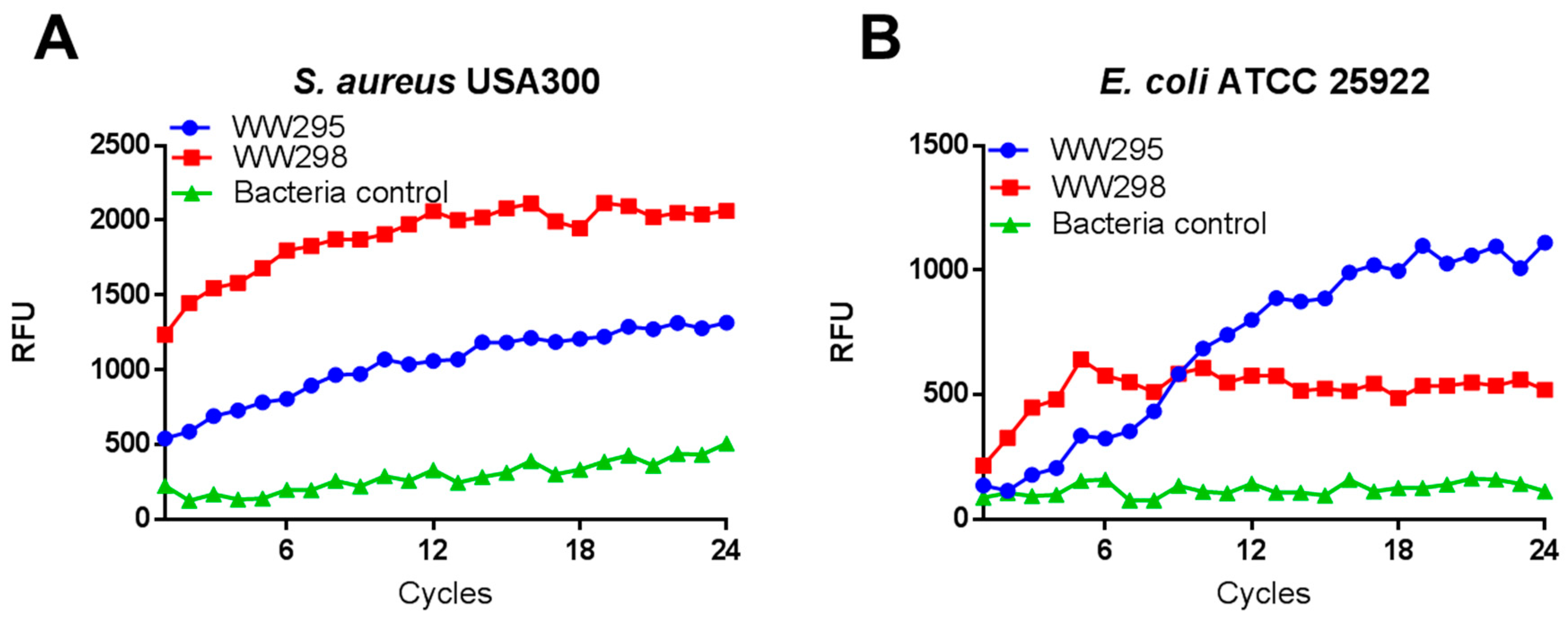
2.6. Evidence of Membrane Depolarization of S. aureus USA300
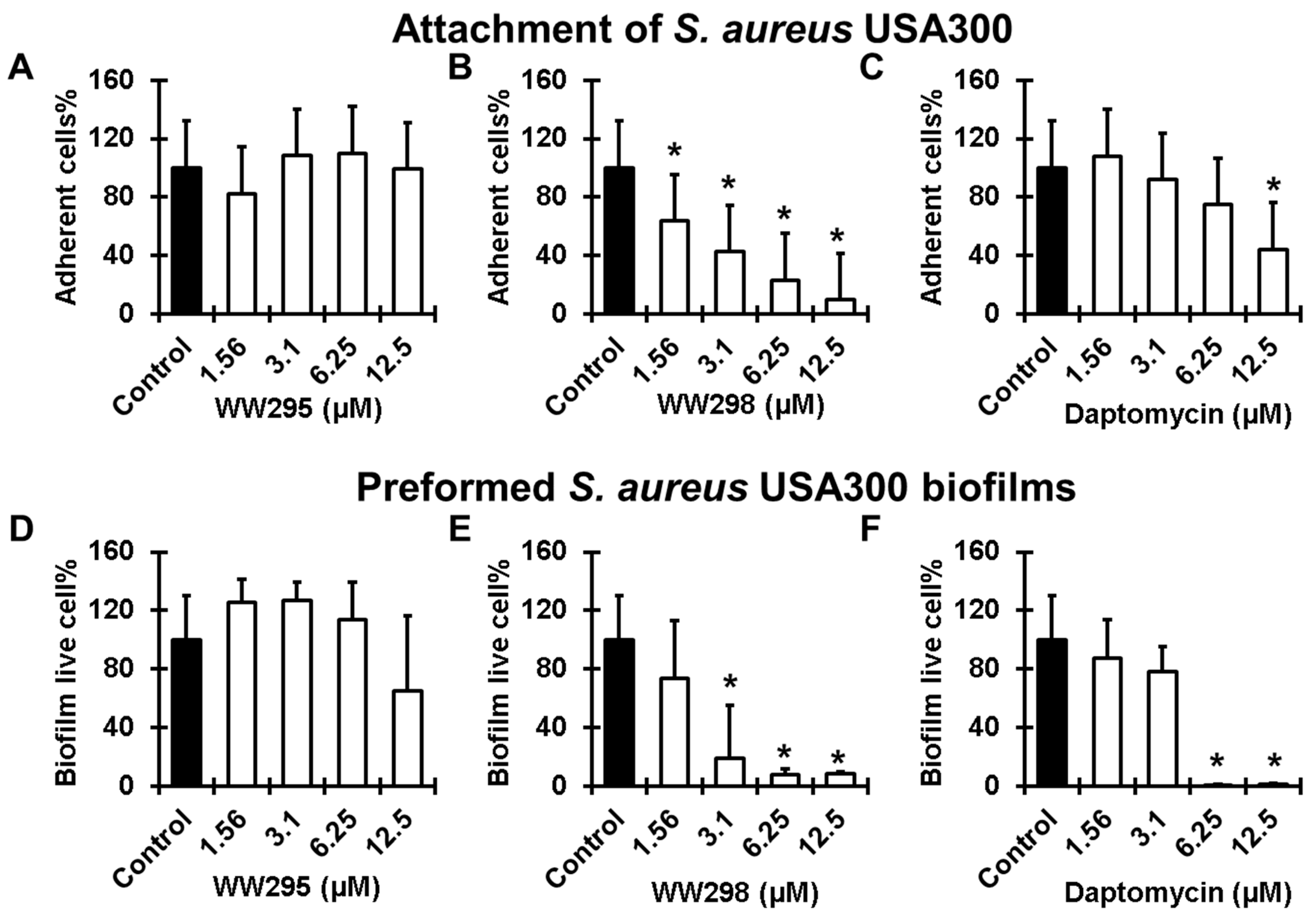
2.7. Antibiofilm Effects on MRSA
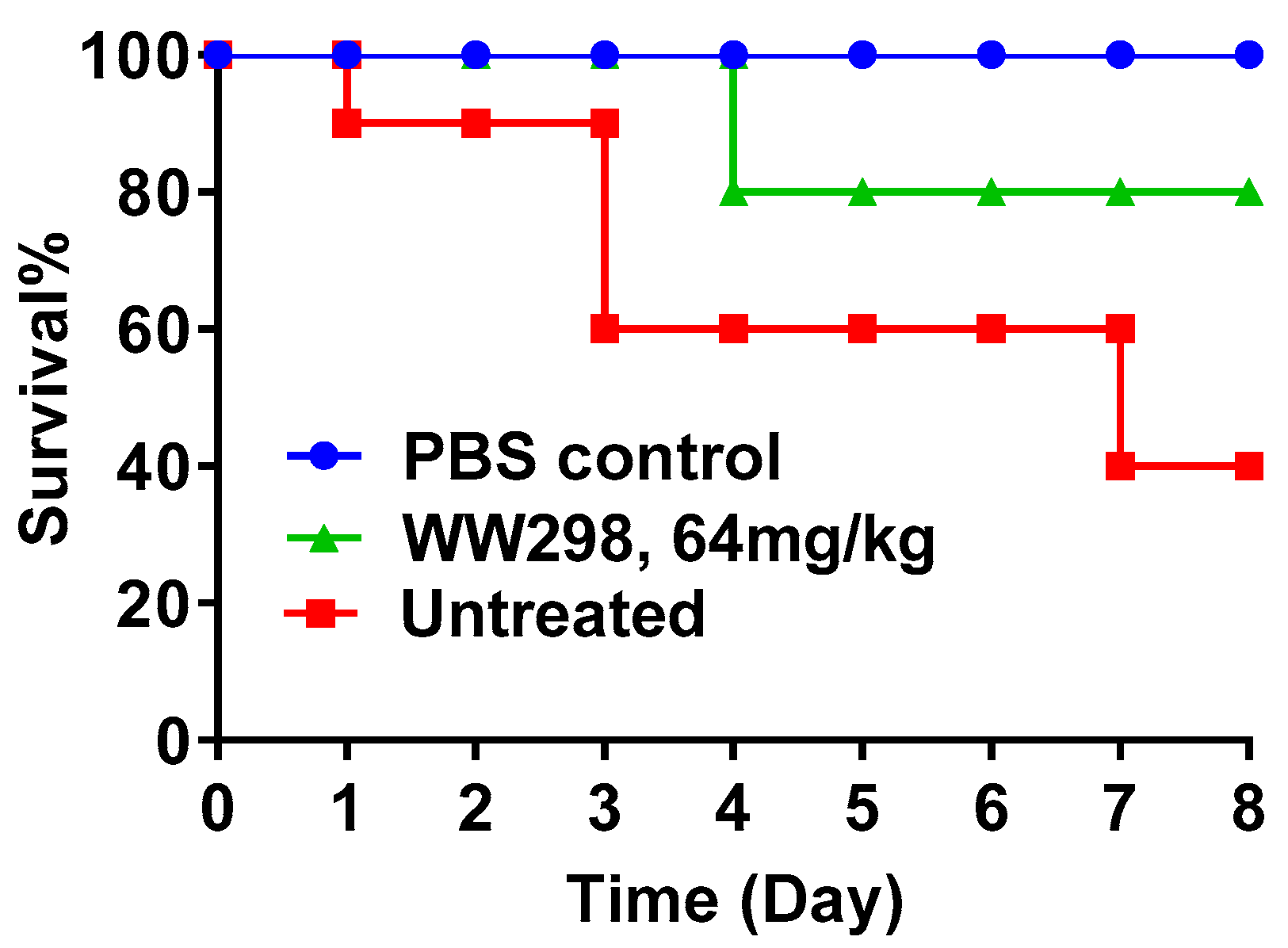
2.8. Protection of Galleria Mellonella Wax Moths from MRSA Infection
3. Discussion
4. Materials and Methods
4.1. Peptides and Property Calculations
4.2. Bacterial Strains and Growth Media
4.3. HPLC Retention Time Measurements
4.4. Antimicrobial Assays
4.5. Hemolytic Assays
4.6. Toxicity of Peptides on Mammalian Cells
4.7. Real-Time Fluorescence-Based Kinetics of Bacterial Killing
4.8. Simultaneous Penetration of the Bacterial Outer and Inner Bacterial Membranes
4.9. Membrane Depolarization of Bacteria
4.10. Peptide Effects on Biofilm Attachment
4.11. Antibiofilm Effects on 24 h Established Bacterial Biofilms
4.12. Confocal Microscopy Observation of Live and Dead Bacteria in Established Biofilms after Peptide Treatment
4.13. Protection of Invertebrates from Death
4.14. Statistics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Loannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Scheenstra, M.R.; van Harten, R.M.; Veldhuizen, E.J.A.; Haagsman, H.P.; Coorens, M. Cathelicidins Modulate TLR-Activation and Inflammation. Front. Immunol. 2020, 11, 1137. [Google Scholar] [CrossRef]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy Against Infection by Boosting Innate Immunity. Front Immunol. 2020, 11, 1209. [Google Scholar] [CrossRef]
- Quaglio, D.; Corradi, S.; Erazo, S.; Vergine, V.; Berardozzi, S.; Sciubba, F.; Cappiello, F.; Crestoni, M.E.; Ascenzioni, F.; Imperi, F.; et al. Structural Elucidation and Antimicrobial Characterization of Novel Diterpenoids from Fabiana densa var. ramulosa. ACS Med. Chem. Lett. 2020, 11, 760–765. [Google Scholar] [CrossRef]
- Hanson, M.A.; Lemaitre, B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, G. APD: The Antimicrobial Peptide Database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef]
- Wang, G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66. [Google Scholar]
- Wang, G. Bioinformatic Analysis of 1000 Amphibian Antimicrobial Peptides Uncovers Multiple Length-Dependent Correlations for Peptide Design and Prediction. Antibiotics 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- André, S.; Raja, Z.; Humblot, V.; Piesse, C.; Foulon, T.; Sereno, D.; Oury, B.; Ladram, A. Functional Characterization of Temporin-SHe, a New Broad-Spectrum Antibacterial and Leishmanicidal Temporin-SH Paralog from the Sahara Frog (Pelophylax saharicus). Int. J. Mol. Sci. 2020, 21, 6713. [Google Scholar] [CrossRef]
- Sani, M.A.; Le Brun, A.P.; Separovic, F. The antimicrobial peptide maculatin self assembles in parallel to form a pore in phospholipid bilayers. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183204. [Google Scholar] [CrossRef]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host defense antimicrobial peptides as antibiotics: Design and application strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; O′Connor, P.M.; Saalbach, G.; Walsh, C.J.; Hegarty, J.W.; Guinane, C.M.; Mayer, M.J.; Narbad, A.; Cotter, P.D. First evidence of production of the lantibiotic nisin P. Sci. Rep. 2020, 10, 3738. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-López, F.J.; Carretero-Molina, D.; Sánchez-Hidalgo, M.; Martín, J.; González, I.; Román-Hurtado, F.; de la Cruz, M.; García-Fernández, S.; Reyes, F.; Deisinger, J.P.; et al. Cacaoidin, First Member of the New Lanthidin RiPP Family. Angew. Chem. Int. Ed. Engl. 2020, 59, 12654–12658. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459, published correction appears in Nature 2015, 520, 388. [Google Scholar] [CrossRef]
- Donia, M.S.; Cimermancic, P.; Schulze, C.J.; Wieland Brown, L.C.; Martin, J.; Mitreva, M.; Clardy, J.; Linington, R.G.; Fischbach, M.A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 2014, 158, 1402–1414. [Google Scholar] [CrossRef]
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef]
- Wang, G.; Watson, K.M.; Peterkofsky, A.; Buckheit, R.W., Jr. Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob. Agents Chemother. 2010, 54, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Wang, G. Ab initio design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Narayana, J.L.; Lushinikova, T.; Wang, X.; Wang, G. Low cationicity is important for systemic in vivo efficacy of database-derived peptides against drug-resistant Gram-positive pathogens. Proc. Natl. Acad. Sci. USA 2019, 116, 13517–13522. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.; Lushnikova, T.; Golla, R.M.; Wang, X.; Wang, G. Design and surface immobilization of short anti-biofilm peptides. Acta Biomater. 2017, 49, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Zarena, D.; Mishra, B.; Lushnikova, T.; Wang, F.; Wang, G. The π Configuration of the WWW Motif of a Short Trp-rich Peptide Is Critical for Targeting Bacterial Membranes, Disrupting Preformed Biofilms and Killing Methicillin-resistant Staphylococcus aureus. Biochemistry 2017, 56, 4039–4043. [Google Scholar] [CrossRef]
- Lakshmaiah Narayana, J.; Mishra, B.; Lushinikova, T.; Wu, Q.; Chhonker, Y.S.; Zhang, Y.; Zarena, D.; Salnikov, E.; Dang, X.; Wang, F.; et al. Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc. Natl. Acad. Sci. USA 2020, 117, 19446–19454. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Clinical Laboratories Standards Institute (CLSI): M07-A10. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition; Clinical Laboratories Standards Institute (CLSI): M07-A10: Annapolis Junction, MD, USA, 2015. [Google Scholar]
- Lehrer, R.I.; Barton, A.; Daher, K.A.; Harwig, S.S.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 1989, 84, 553–561. [Google Scholar] [CrossRef]
- Desbois, A.P.; Coote, P.J. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef]
- Wang, X.; Mishra, B.; Lushnikova, T.; Narayana, J.L.; Wang, G. Amino Acid Composition Determines Peptide Activity Spectrum and Hot-Spot-Based Design of Merecidin. Adv. Biosyst. 2018, 2, 1700259. [Google Scholar] [CrossRef]
- Wang, G.; Waston, K.; Buckheit, R., Jr. Anti-human immunodeficiency virus type 1 activities of antimicrobial peptides derived from human and bovine cathelicidins. Antimicrob. Agents Chemother. 2008, 52, 3438–3440. [Google Scholar] [CrossRef]
- Wang, G. Database-guided discovery of potent peptides to combat HIV-1 or Superbugs. Pharmaceuticals 2013, 6, 728–758. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Gao, B.; Zhu, S. Characterization of a chimeric antimicrobial peptide uncovers evolutionary significance of exon-shuffling. Biochem. Biophys. Res. Commun. 2012, 428, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Bahnsen, J.S.; Franzyk, H.; Sandberg-Schaal, A.; Nielsen, H.M. Antimicrobial and cell-penetrating properties of penetratin analogs: Effect of sequence and secondary structure. Biochim. Biophys. Acta 2013, 1828, 223–232. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51, published correction appears in Nat. Rev. Drug Discov. 2012, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Loose, C.; Jensen, K.; Rigoutsos, I.; Stephanopoulos, G. A linguistic model for the rational design of antimicrobial peptides. Nature 2006, 443, 867–869. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Han, H.; Miller, D.W.; Wang, G. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 2006, 128, 5776–5785. [Google Scholar] [CrossRef]
- Monroc, S.; Badosa, E.; Feliu, L.; Planas, M.; Montesinos, E.; Bardají, E. De novo designed cyclic cationic peptides as inhibitors of plant pathogenic bacteria. Peptides 2006, 27, 2567–2574. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.S.; Lee, B.J. Correlation between the activities of alpha-helical antimicrobial peptides and hydrophobicities represented as RP HPLC retention times. Peptides 2005, 26, 2050–2056. [Google Scholar] [CrossRef]
- Wang, G.; Narayana, J.L.; Mishra, B.; Zhang, Y.; Wang, F.; Wang, C.; Zarena, D.; Lushnikova, T.; Wang, X. Design of Antimicrobial Peptides: Progress Made with Human Cathelicidin LL-37. Adv. Exp. Med. Biol. 2019, 1117, 215–240. [Google Scholar] [PubMed]
- Martins, W.M.B.S.; Toleman, M.A.; Gales, A.C. Clinical utilization of bacteriophages: A new perspective to combat the antimicrobial resistance in Brazil. Braz. J. Infect. Dis. 2020, 24, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [PubMed]
- Varshavsky, A. The N-end rule pathway of protein degradation. Genes Cells 1997, 2, 13–28. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Wang, G. The Antimicrobial Peptide Database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci. 2020, 29, 8–18. [Google Scholar] [CrossRef]
- Marks, L.R.; Clementi, E.A.; Hakansson, A.P. Sensitization of Staphylococcus aureus to methicillin and other antibiotics in vitro and in vivo in the presence of HAMLET. PLoS ONE 2013, 8, e63158. [Google Scholar] [CrossRef]

| Peptide | Sequence | Boman Index a | GRAVY a,b | Aliphatic Index b | Half-Life b | Instability Index b | HPLC (tr, min) c |
|---|---|---|---|---|---|---|---|
| WW291 | WWWLRKIW | 0.16 | −0.46 | 97.5 | 2.8 h | 37.64 | ND d |
| WW292 | WWLRKIWW | 0.16 | −0.46 | 97.5 | 2.8 h | 37.64 | 11.849 |
| WW293 | WLRKIWWW | 0.16 | −0.46 | 97.5 | 2.8 h | 37.64 | 11.921 |
| WW294 | LRKIWWWW | 0.16 | −0.46 | 97.5 | 5.5 h | 22.21 | 12.202 |
| WW295 | RKIWWWWL | 0.16 | −0.46 | 97.5 | 1.0 h | 13.56 | 12.453 |
| WW296 | KIWWWWLR | 0.16 | −0.46 | 97.5 | 1.3 h | 37.64 | 11.941 |
| WW297 | IWWWWLRK | 0.16 | −0.46 | 97.5 | 20 h | 48.25 | 11.652 |
| WW298 | WWWWLRKI | 0.16 | −0.46 | 97.5 | 2.8 h | 37.64 | 11.892 |
| Peptide | Minimal Inhibitory Concentration (MIC, μM) | MBC (μM) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SA | SE | BS | VRE | EC | PA | KP a | AB | USA300 | |
| WW291 | 3.1 | 3.1–6.25 | 6.25 | 25 | 6.2–12.5 | 50 | 12.5 | 6.25 | 6.2 |
| WW292 | 12.5 | 6.2–12.5 | 12.5 | >50 | 25 | 25 | >50 | 12.5 | >25 |
| WW293 | 12.5 | 6.2 | 12.5 | 50 | 12.5–25 | 50 | 25 | 12.5 | 25 |
| WW294 | 12.5 | 6.2 | 25 | 25 | 12.5 | 50 | 6.2–12.5 | 12.5 | 25 |
| WW295 | 12.5 | 6.2 | 12.5 | 6.25 | 6.2 | 25–50 | 3.1–6.2 | 6.25–12.5 | 12.5 |
| WW296 | 3.1 | 3.1 | 12.5 | 25 | 6.2–12.5 | 50 | 12.5 | 12.5 | 6.2 |
| WW297 | 6.2 | 3.1 | 12.5 | 12.5 | 12.5–25 | 50 | 12.5–25 | 6.25–12.5 | 12.5 |
| WW298 | 3.1 | 1.5–3.1 | 6.25–12.5 | 12.5 | 12.5 | 50 | 6.2–12.5 | 6.25–12.5 | 3.1 |
| Peptide | TSB | +150 mM NaCl | +10% Human Serum |
|---|---|---|---|
| E. coli ATCC 25922 | |||
| WW291 | 6.2–12.5 | 12.5 | >25 |
| WW292 | 12.5 | >25 | >25 |
| WW293 | 12.5 | >25 | >25 |
| WW294 | 12.5 | 25 | >25 |
| WW295 | 3.1 | 6.2 | 12.5 |
| WW296 | 3.1–6.2 | 25 | >25 |
| WW297 | 12.5 | 12.5 | >25 |
| WW298 | 12.5 | 12.5 | >25 |
| S. aureus USA300 LAC | |||
| WW291 | 3.1 | 3.1 | 25 |
| WW292 | 6.2 | 6.2 | >25 |
| WW293 | 6.2 | 6.2 | >25 |
| WW294 | 6.2 | 6.2 | 25 |
| WW295 | 3.1 | 3.1 | 12.5 |
| WW296 | 1.6 | 1.6 | 12.5 |
| WW297 | 3.1 | 3.1 | 25 |
| WW298 | 1.6 | 1.6 | 12.5–25 |
| Peptide | Human RBC (µM) | Chicken RBC (µM) | Pig RBC (µM) | HaCaT (µM) | THP-1 (µM) | Vero Cells (µM) |
|---|---|---|---|---|---|---|
| WW291 | 30 | 50 | ND | 60 | <25 a | 100 |
| WW292 | 175 | >100 | >100 | >100 | >100 | >100 |
| WW293 | 224 | >100 | >100 | >100 | >100 | >100 |
| WW294 | 143 | >100 | >100 | >100 | >100 | >100 |
| WW295 | 57 | 100 | >100 | 80 | 50 | >100 |
| WW296 | 103 | 50 | >100 | 100 | 35 | >100 |
| WW297 | 134 | 100 | 110 | 100 | 38 | >100 |
| WW298 | 52 | 70 | 37 | 80 | 30 | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, B.; Lakshmaiah Narayana, J.; Lushnikova, T.; Zhang, Y.; Golla, R.M.; Zarena, D.; Wang, G. Sequence Permutation Generates Peptides with Different Antimicrobial and Antibiofilm Activities. Pharmaceuticals 2020, 13, 271. https://doi.org/10.3390/ph13100271
Mishra B, Lakshmaiah Narayana J, Lushnikova T, Zhang Y, Golla RM, Zarena D, Wang G. Sequence Permutation Generates Peptides with Different Antimicrobial and Antibiofilm Activities. Pharmaceuticals. 2020; 13(10):271. https://doi.org/10.3390/ph13100271
Chicago/Turabian StyleMishra, Biswajit, Jayaram Lakshmaiah Narayana, Tamara Lushnikova, Yingxia Zhang, Radha M. Golla, D. Zarena, and Guangshun Wang. 2020. "Sequence Permutation Generates Peptides with Different Antimicrobial and Antibiofilm Activities" Pharmaceuticals 13, no. 10: 271. https://doi.org/10.3390/ph13100271
APA StyleMishra, B., Lakshmaiah Narayana, J., Lushnikova, T., Zhang, Y., Golla, R. M., Zarena, D., & Wang, G. (2020). Sequence Permutation Generates Peptides with Different Antimicrobial and Antibiofilm Activities. Pharmaceuticals, 13(10), 271. https://doi.org/10.3390/ph13100271






