Targeted Molecular Imaging Using Aptamers in Cancer
Abstract
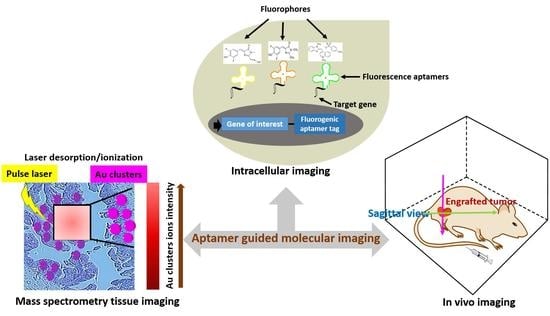
1. Introduction
2. Cell Imaging
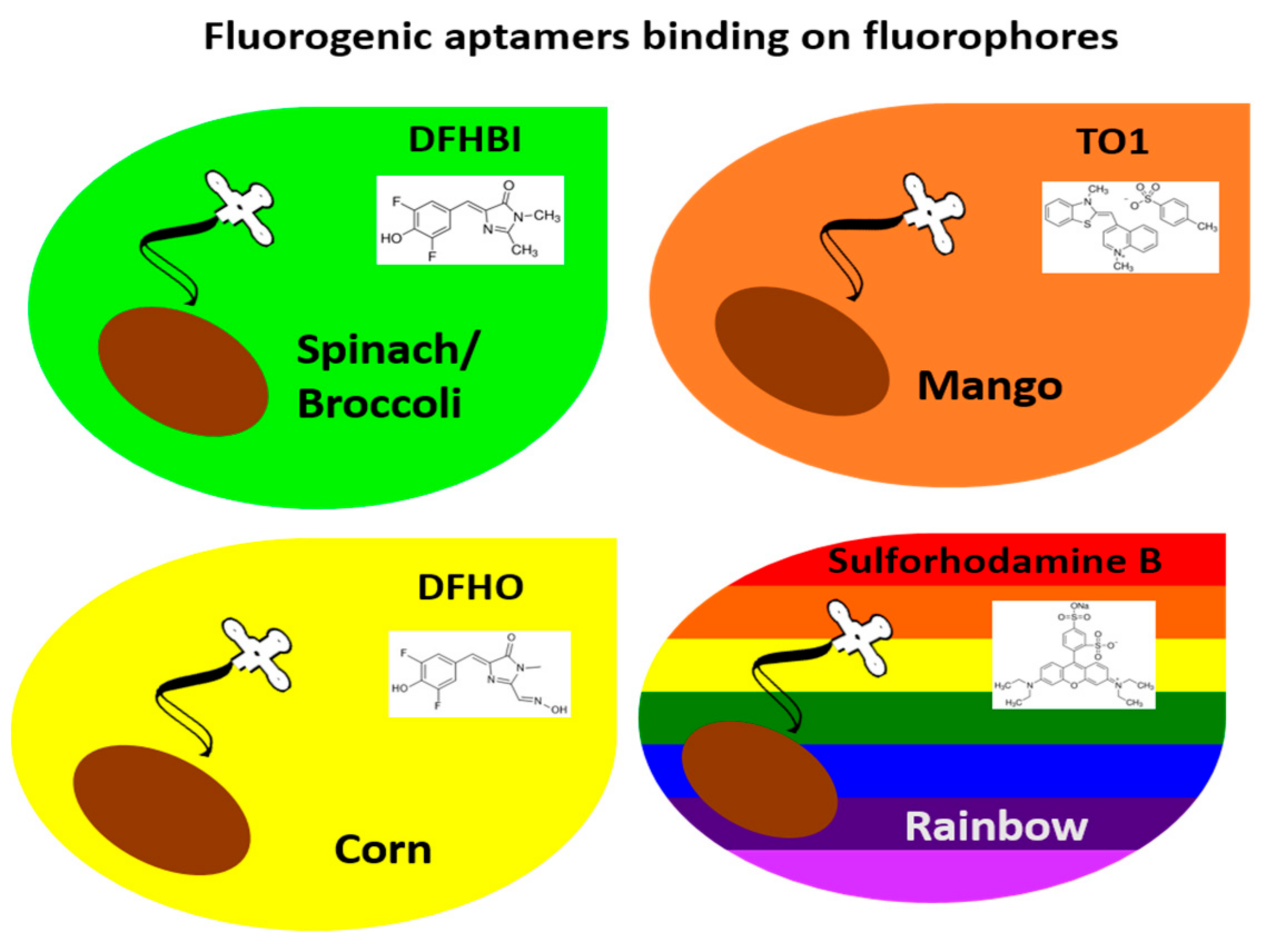
2.1. Green Fluorogenic Aptamers: Spinach and Broccoli
2.2. Orange Fluorogenic Aptamer: Mango
2.3. Yellow Fluorogenic Aptamer: Corn
2.4. Rainbow Fluorogenic Aptamer: SRB-2
3. Tissue Imaging
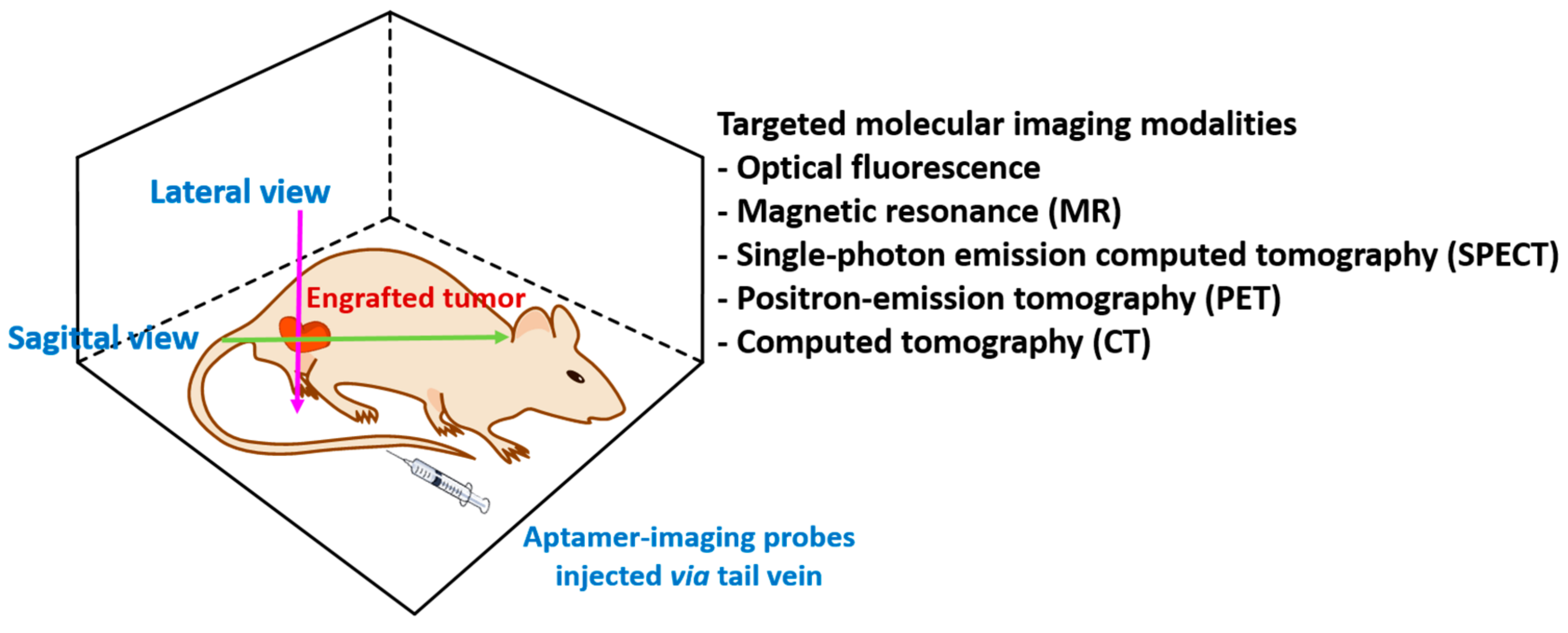
4. In Vivo Imaging
4.1. Fluorescence Imaging
4.2. Magnetic Resonance Imaging (MRI)
4.3. Single-Photon Emission Computed Tomography (SPECT)
4.4. Positron-Emission Tomography (PET)
4.5. Computed Tomography (CT)
4.6. Ultrasound (US)
4.7. Photoacoustic (PA) Imaging
4.8. Multimodal Imaging
5. Clinical Trials
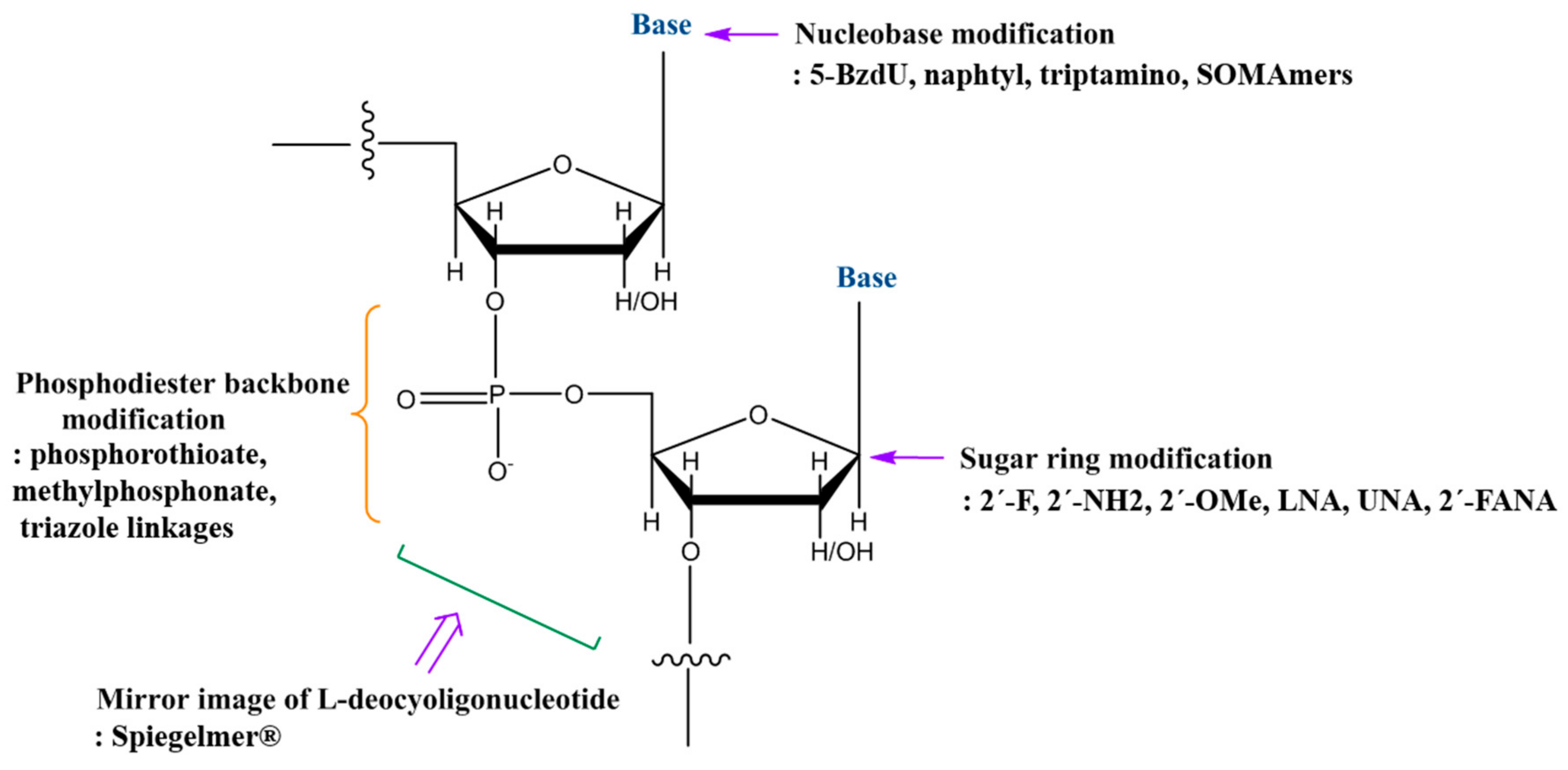
6. Chemical Modifications
7. Conclusions and Implications for Clinical Use
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weissleder, R.; Pittet, M.J. Imaging in the era of molecular oncology. Nature 2008, 452, 580–589. [Google Scholar] [CrossRef] [PubMed]
- James, M.L.; Gambhir, S.S. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol. Rev. 2012, 92, 897–965. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, R.; Goel, S.; Cai, W. Nanobody: The “magic bullet” for molecular imaging? Theranostics 2014, 4, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Warram, J.M.; de Boer, E.; Sorace, A.G.; Chung, T.K.; Kim, H.; Pleijhuis, R.G.; van Dam, G.M.; Rosenthal, E.L. Antibody-based imaging strategies for cancer. Cancer Metastasis Rev. 2014, 33, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Que-Gewirth, N.S.; Sullenger, B.A. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007, 14, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Gomes de Castro, M.A.; Hobartner, C.; Opazo, F. Aptamers provide superior stainings of cellular receptors studied under super-resolution microscopy. PLoS ONE 2017, 12, e0173050. [Google Scholar] [CrossRef] [PubMed]
- Melancon, M.P.; Zhou, M.; Zhang, R.; Xiong, C.; Allen, P.; Wen, X.; Huang, Q.; Wallace, M.; Myers, J.N.; Stafford, R.J.; et al. Selective uptake and imaging of aptamer- and antibody-conjugated hollow nanospheres targeted to epidermal growth factor receptors overexpressed in head and neck cancer. ACS Nano 2014, 8, 4530–4538. [Google Scholar] [CrossRef] [PubMed]
- Enterina, J.R.; Wu, L.; Campbell, R.E. Emerging fluorescent protein technologies. Curr. Opin. Chem. Biol. 2015, 27, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Mishin, A.S.; Belousov, V.V.; Solntsev, K.M.; Lukyanov, K.A. Novel uses of fluorescent proteins. Curr. Opin. Chem. Biol. 2015, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Moon, J.D.; Svensen, N.; Jaffrey, S.R. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014, 136, 16299–16308. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Jeng, S.C.; Panchapakesan, S.S.S.; Cojocaru, R.; Chen, P.S.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA mango aptamer-fluorophore: A bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Filonov, G.S.; Kim, H.; Hirsch, M.; Li, X.; Moon, J.D.; Jaffrey, S.R. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol. 2017, 13, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Strack, R.L.; Disney, M.D.; Jaffrey, S.R. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods 2013, 10, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.D.; Chen, M.C.; Song, W.; Strack, R.L.; Thorn, A.; Jaffrey, S.R.; Ferré-D’Amaré, A.R. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014, 21, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Westhof, E.; Ryckelynck, M. iSpinach: A fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016, 44, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Nguyen-Duc, T.; Song, W.; Jaffrey, S.R. Fluorescence imaging of cellular metabolites with RNA. Science 2012, 335, 1194. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, C.A.; Wilson, S.C.; Sales-Lee, J.; Hammond, M.C. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135, 4906–4909. [Google Scholar] [CrossRef] [PubMed]
- Trachman, R.J., 3rd; Demeshkina, N.A.; Lau, M.W.; Panchapakesan, S.S.S.; Jeng, S.C.; Unrau, P.J.; Ferré-D’Amaré, A.R. Structural basis for high-affinity fluorophore binding and activation by RNA Mango. Nat. Chem. Biol. 2017, 13, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Jeng, S.; Cawte, A.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S.S.; Rueda, D.; Ryckelynck, M.; Unrau, P.J. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018, 9, 656. [Google Scholar] [CrossRef] [PubMed]
- Sunbul, M.; Jaschke, A. SRB-2: A promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Rossi, J.J. Emerging cancer-specific therapeutic aptamers. Curr. Opin. Oncol. 2017, 29, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Huang, K.W.; Reebye, V.; Mintz, P.; Tien, Y.W.; Lai, H.S.; Sætrom, P.; Reccia, I.; Swiderski, P.; Armstrong, B.; et al. Targeted Delivery of C/EBPα-saRNA by Pancreatic Ductal Adenocarcinoma-specific RNA Aptamers Inhibits Tumor Growth In Vivo. Mol. Ther. 2016, 24, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Long, Y.; Yang, C.; Wu, X.; Sun, Y.; Li, J.; Hu, X.; Lin, W.; Han, D.; Zhao, Y.; et al. Selection and characterization of DNA aptamer for metastatic prostate cancer recognition and tissue imaging. Oncotarget 2016, 7, 36436–36446. [Google Scholar] [CrossRef] [PubMed]
- Chughtai, K.; Heeren, R.M. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 2010, 110, 3237–3277. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xiong, C.; Liu, H.; Wan, Q.; Hou, J.; He, Q.; Badu-Tawiah, A.; Nie, Z. Mass spectrometry imaging reveals the sub-organ distribution of carbon nanomaterials. Nat. Nanotechnol. 2015, 10, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.T.; Harroun, S.G.; Wu, C.W.; Mao, J.Y.; Chang, H.T.; Huang, C.C. Satellite-like Gold Nanocomposites for Targeted Mass Spectrometry Imaging of Tumor Tissues. Nanotheranostics 2017, 1, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Tang, Z.; Kim, Y.; Nie, H.; Huang, Y.F.; He, X.; Deng, K.; Wang, K.; Tan, W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem. Asian J. 2010, 5, 2209–2213. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Cui, W.; He, X.; Guo, Q.; Wang, K.; Ye, X.; Tang, J. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE 2013, 8, e70476. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Yan, X.P. Doped quantum dots for chemo/biosensing and bioimaging. Chem. Soc. Rev. 2013, 42, 5489–5521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ji, X.; Zhang, Y.; Zhou, G.; Ke, X.; Wang, H.; Tinnefeld, P.; He, Z. One-pot synthesized aptamer-functionalized CdTe:Zn2+ quantum dots for tumor-targeted fluorescence imaging in vitro and in vivo. Anal. Chem. 2013, 85, 5843–5849. [Google Scholar] [CrossRef] [PubMed]
- Greenall, S.A.; Donoghue, J.F.; Van Sinderen, M.; Dubljevic, V.; Budiman, S.; Devlin, M.; Street, I.; Adams, T.E.; Johns, T.G. EGFRvIII-mediated transactivation of receptor tyrosine kinases in glioma: Mechanism and therapeutic implications. Oncogene 2015, 34, 5277–5287. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, N.; Zhang, X.; Zhou, T.; Tan, Y.; Pi, J.; Pi, L.; Cheng, S.; Zheng, H.; Cheng, Y. Aptamer-conjugated PEGylated quantum dots targeting epidermal growth factor receptor variant III for fluorescence imaging of glioma. Int. J. Nanomed. 2017, 12, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.W.; Jeong, H.Y.; Kang, S.J.; Choi, M.J.; You, Y.M.; Im, C.S.; Lee, T.S.; Song, I.H.; Lee, C.G.; Rhee, K.J.; et al. Cancer-targeted Nucleic Acid Delivery and Quantum Dot Imaging Using EGF Receptor Aptamer-conjugated Lipid Nanoparticles. Sci. Rep. 2017, 7, 9474. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Hernandez, L.I.; Thomas, G.S.; Long, M.E.; Rockey, W.M.; Howell, C.A.; Chen, Y.; Hernandez, F.J.; Liu, X.Y.; Wilson, M.E.; et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol. Ther. 2014, 22, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, J.; Zhang, M.; Yang, H.; Ma, Y.; Gu, Y. MUC1 aptamer-based near-infrared fluorescence probes for tumor imaging. Mol. Imaging Biol. 2015, 17, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.E.; Papan, C.; Ruffins, S.; Tyszka, J.M.; Fraser, S.E. MRI: Volumetric imaging for vital imaging and atlas construction. Nat. Rev. Mol. Cell Biol. 2003, 4, SS10–SS16. [Google Scholar] [CrossRef]
- Sipkins, D.A.; Cheresh, D.A.; Kazemi, M.R.; Nevin, L.M.; Bednarski, M.D.; Li, K.C. Detection of tumor angiogenesis in vivo by alphaVbeta3-targeted magnetic resonance imaging. Nat. Med. 1998, 4, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Kim, B.; Choi, Y.; Ro, Y.; Cho, E.J.; Lee, J.H.; Ryu, S.H.; Suh, J.S.; Haam, S.; Huh, Y.M. Aptamer-conjugated magnetic nanoparticles enable efficient targeted detection of integrin alphavbeta3 via magnetic resonance imaging. J. Biomed. Mater. Res. A 2014, 102, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Yang, J.; Hwang, M.; Choi, J.; Kim, H.O.; Jang, E.; Lee, J.H.; Ryu, S.H.; Suh, J.S.; Huh, Y.M.; et al. Aptamer-modified magnetic nanoprobe for molecular MR imaging of VEGFR2 on angiogenic vasculature. Nanoscale Res. Lett. 2013, 8, 399. [Google Scholar] [CrossRef] [PubMed]
- Paolicchi, E.; Gemignani, F.; Krstic-Demonacos, M.; Dedhar, S.; Mutti, L.; Landi, S. Targeting hypoxic response for cancer therapy. Oncotarget 2016, 7, 13464–13478. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, L.; Liu, Y.; Zhou, Y.; Wang, K.; Xie, X.; Song, L.; Wang, D.; Han, C.; Chen, Q. Aptamer-PEG-modified Fe3O4@Mn as a novel T1- and T2-dual-model MRI contrast agent targeting hypoxia-induced cancer stem cells. Sci. Rep. 2016, 6, 39245. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.M.; Yuan, Z. PET/SPECT molecular imaging in clinical neuroscience: Recent advances in the investigation of CNS diseases. Quant. Imaging Med. Surg. 2015, 5, 433–447. [Google Scholar] [PubMed]
- Ray, P.; Viles, K.D.; Soule, E.E.; Woodruff, R.S. Application of aptamers for targeted therapeutics. Arch. Immunol. Ther. Exp. 2013, 61, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Stephens, A.W.; Gould, T.; Chang, Y.; Lynott, C.K.; Heil, J.; Borkowski, S.; Hilger, C.; Cook, G.; Warren, S.; et al. Tumor targeting by an aptamer. J. Nucl. Med. 2006, 47, 668–678. [Google Scholar] [PubMed]
- Bonner, J.A.; De Los Santos, J.; Waksal, H.W.; Needle, M.N.; Trummel, H.Q.; Raisch, K.P. Epidermal growth factor receptor as a therapeutic target in head and neck cancer. Semin. Radiat. Oncol. 2002, 12, 1–20. [Google Scholar] [CrossRef]
- Hofmann, U.B.; Westphal, J.R.; van Muijen, G.N.; Ruiter, D.J. Matrix metalloproteinases in human melanoma. J. Investig. Dermatol. 2000, 115, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Kryza, D.; Debordeaux, F.; Azéma, L.; Hassan, A.; Paurelle, O.; Schulz, J.; Savona-Baron, C.; Charignon, E.; Bonazza, P.; Taleb, J.; et al. Ex Vivo and In Vivo Imaging and Biodistribution of Aptamers Targeting the Human Matrix MetalloProtease-9 in Melanomas. PLoS ONE 2016, 11, e0149387. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liang, H.; Tan, Y.; Yuan, C.; Li, S.; Li, X.; Li, G.; Shi, Y.; Zhang, X. Cell-SELEX aptamer for highly specific radionuclide molecular imaging of glioblastoma in vivo. PLoS ONE 2014, 9, e90752. [Google Scholar] [CrossRef] [PubMed]
- Gambhir, S.S. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer 2002, 2, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, H.; Bates, P.J.; Malik, T.; Li, X.F.; Trent, J.O.; Ng, C.K. Aptamer imaging with Cu-64 labeled AS1411: Preliminary assessment in lung cancer. Nucl. Med. Biol. 2014, 41, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massagué, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Leins, A.; Riva, P.; Lindstedt, R.; Davidoff, M.S.; Mehraein, P.; Weis, S. Expression of tenascin-C in various human brain tumors and its relevance for survival in patients with astrocytoma. Cancer 2003, 98, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, O.; Yan, X.; Niu, G.; Weiss, I.D.; Ma, Y.; Szajek, L.P.; Shen, B.; Kiesewetter, D.O.; Chen, X. PET imaging of tenascin-C with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015, 56, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Lee, T.S.; Song, I.H.; Cho, Y.L.; Chae, J.R.; Yun, M.; Kang, H.; Lee, J.H.; Lim, J.H.; Cho, W.G.; et al. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials 2016, 100, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jacobson, O.; Avdic, D.; Rotstein, B.H.; Weiss, I.D.; Collier, L.; Chen, X.; Vasdev, N.; Liang, S.H. Ortho-Stabilized (18) F-Azido Click Agents and their Application in PET Imaging with Single-Stranded DNA Aptamers. Angew. Chem. Int. Ed. Engl. 2015, 54, 12777–12781. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeong, Y.Y.; Jon, S. A drug-loaded aptamer-gold nanoparticle bioconjugate for combined CT imaging and therapy of prostate cancer. ACS Nano 2010, 4, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Kuo, T.R.; Su, H.J.; Lai, W.Y.; Yang, P.C.; Chen, J.S.; Wang, D.Y.; Wu, Y.C.; Chen, C.C. Fluorescence-Guided Probes of Aptamer-Targeted Gold Nanoparticles with Computed Tomography Imaging Accesses for in Vivo Tumor Resection. Sci. Rep. 2015, 5, 15675. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shen, M.; Xu, C.S.; Sun, Y.; Duan, Y.R.; Du, L.F. Biodegradable double-targeted PTX-mPEG-PLGA nanoparticles for ultrasound contrast enhanced imaging and antitumor therapy in vitro. Oncotarget 2016, 7, 80008–80018. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wang, Y.; Wang, Y.; Zhang, M.; Luo, Y.; Tang, J.; Wang, Z.; Wang, D.; Hao, L.; Wang, Z. Paclitaxel-loaded and A10-3.2 aptamer-targeted poly(lactide-co-glycolic acid) nanobubbles for ultrasound imaging and therapy of prostate cancer. Int. J. Nanomed. 2017, 12, 5313–5330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Smaga, L.P.; Satyavolu, N.S.R.; Chan, J.; Lu, Y. DNA Aptamer-Based Activatable Probes for Photoacoustic Imaging in Living Mice. J. Am. Chem. Soc. 2017, 139, 17225–17228. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.J.; Lee, J.; Lee, Y.S.; Cho, S.; Ali, B.A.; Al-Khedhairy, A.A.; Heo, H.; Kim, S. Multimodal imaging probe for targeting cancer cells using uMUC-1 aptamer. Colloids Surf. B Biointerfaces 2015, 136, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Kanwar, R.K.; Kanwar, J.R. LNA aptamer based multi-modal, Fe3O4-saturated lactoferrin (Fe3O4-bLf) nanocarriers for triple positive (EpCAM, CD133, CD44) colon tumor targeting and NIR, MRI and CT imaging. Biomaterials 2015, 71, 84–99. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, S.; Rossi, J.J. Targeted Molecular Imaging Using Aptamers in Cancer. Pharmaceuticals 2018, 11, 71. https://doi.org/10.3390/ph11030071
Yoon S, Rossi JJ. Targeted Molecular Imaging Using Aptamers in Cancer. Pharmaceuticals. 2018; 11(3):71. https://doi.org/10.3390/ph11030071
Chicago/Turabian StyleYoon, Sorah, and John J. Rossi. 2018. "Targeted Molecular Imaging Using Aptamers in Cancer" Pharmaceuticals 11, no. 3: 71. https://doi.org/10.3390/ph11030071
APA StyleYoon, S., & Rossi, J. J. (2018). Targeted Molecular Imaging Using Aptamers in Cancer. Pharmaceuticals, 11(3), 71. https://doi.org/10.3390/ph11030071